翻译后修饰(Post-translational modification,PTM)可以改变蛋白质的三维结构,进而影响蛋白质的定位、活性和稳定性,对蛋白质正常生物学功能的发挥起重要调节作用[1-2]。甲基化(Methyla-tion)、磷酸化(Phosphorylation)、乙酰化(Acetyla-tion)、糖基化(Glycosylation)、亚硝基化(Nitrosyla-tion)、泛素化(Ubiquitination)和SUMO化(Sumoy-lation)等都是重要的翻译后修饰途径[3-4],它们共同协调着真核生物体内不同的生理生化反应,对真核生物的生长发育和逆境适应等过程具有重要的调控作用[5]。
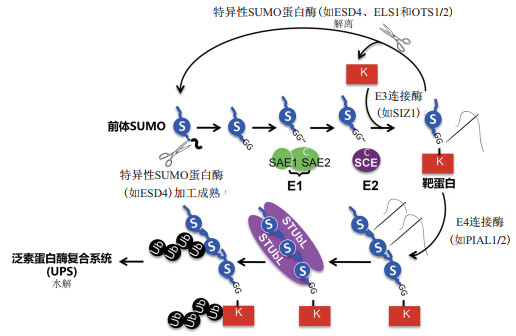
SUMO(Small ubiquitin-related modifier)是一种由100个左右的氨基酸组成的类泛素小分子多肽,在真核生物中高度保守且广泛存在。与泛素化过程非常类似,蛋白质的SUMO化修饰过程主要由3种酶通过分级作用来完成:首先,未成熟的前体SUMO蛋白Pre-SUMO在特异酶的作用下,其C端的2个甘氨酸(Gly)残基发生暴露,成为成熟的SUMO,接着在E1激活酶(E1 activating enzyme)作用下通过转酯反应被激活,然后转移到E2结合酶(E2 conjugating enzyme)上并通过硫酯键与E2的活性位点Cys连接而与E2结合,最后在E3连接酶(E3 ligase)作用下转移到靶蛋白上,完成SUMO化修饰[6-7]。完成修饰后的SUMO蛋白有2个去向,一是在SUMO蛋白酶的作用下与底物蛋白之间的肽键断裂直接形成游离的SUMO蛋白;二是在SUMO E4连接酶的作用下,继续征集SUMO蛋白,形成SUMO链,最后在泛素E3连接酶的作用下被降解[5](图 1)。与泛素化不同的是,SUMO化没有直接介导蛋白质的降解,而是通过调控目标蛋白的构象,影响该蛋白的活性和与其他蛋白的相互结合过程[6, 8]。由于SUMO化和泛素化大都靶向相同类型的氨基酸,因此,它们最初被认为是相互拮抗的过程。在酵母和后生动物中,SUMO化在细胞行使正常功能的很多方面都发挥着重要作用,包括染色质重塑[9]、DNA修复[10]、细胞核/细胞质间物质转运[11]、转录[11]和细胞周期调控[12]等。
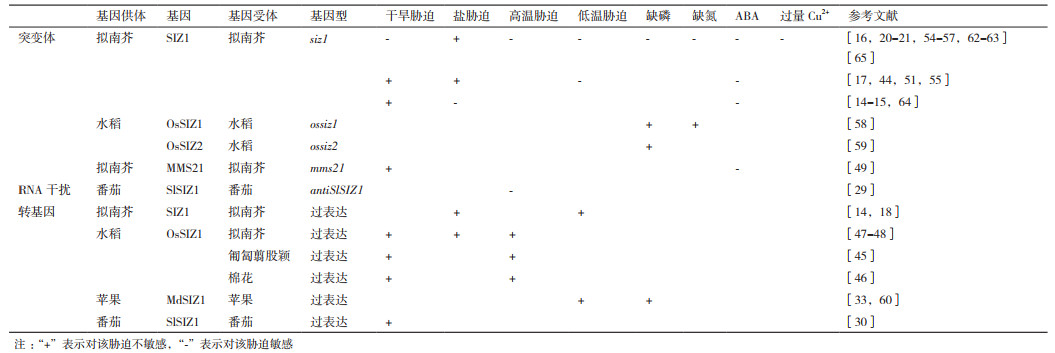
研究发现,SUMO化蛋白的积累可以增强拟南芥(Arabidopsis thaliana)对盐[14]、高温[6, 15-16]、寒冷[17-18]、干旱[19-20]、铜离子[21]和氧化胁迫[22-23]等多种环境胁迫的抗性。水稻(Oryza sativa)SUMO化蛋白的积累可以增强其对冷、高盐或ABA(abscisic acid)等非生物胁迫的抗性[24-26]。玉米(Zea mays)种子发育过程中SUMO化相关基因会大量表达,这可能对保持种子在胁迫条件下的存活率有重要影响。此外,热和氧化胁迫也会诱导SUMO化蛋白的大量积累[27-28]。SlSIZ1介导的HsfA1的SUMO化可以增强番茄(Solanum lycopersicum)的抗热性[29-30]。GmSIZ1a和GmSIZ1b的表达可以促进大豆(Glycine max)的生长并显著增加在各种非生物胁迫(如高盐、热或ABA)处理下的SUMO化水平[31-32]。低温条件下,MdSIZ1介导MdMYB1的SUMO化可以提高苹果(Malus domestica)花青素的含量[33]。以上表明SUMO化对植物适应多种非生物胁迫具有重要作用。在这些SUMO化修饰过程中,E3连接酶具有选择底物的作用,对SUMO化系统进行精准的调节[34-35]。在植物细胞中,少量的E3连接酶调控了大量靶蛋白的SUMO化修饰过程[36]。本文就SUMO E3连接酶在植物适应干旱、盐、高/低温、营养元素匮缺和重金属毒害等非生物胁迫过程中的作用研究进展进行综述。
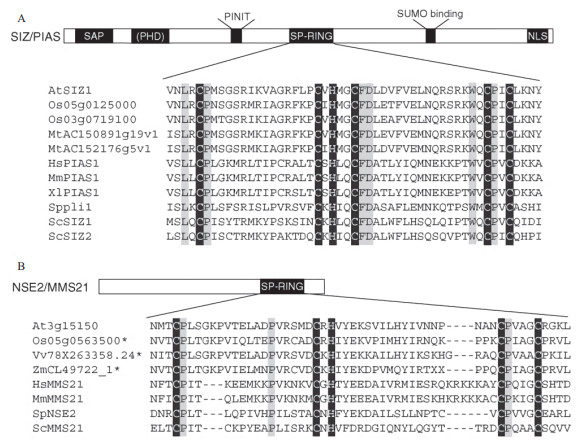
1 SUMO E3连接酶的结构与功能动物中已发现的多种SUMO E3连接酶都包含有SIMs结构域,非典型的SUMO E3连接酶如人类的RanBP2和ZNF451只有一个SIM结构域,而大多数保守的SUMO E3连接酶都属于Siz/PIAS家族,它们都含有一个Siz/PIAS RING(SP-RING)结构域和一个SAP结构域[37-38]。目前,已知植物体内主要有2种类型的SUMO E3连接酶,即SIZ1和MMS21,它们都属于SP-RING家族[36],其中对SIZ1的研究较为深入。拟南芥AtSIZ1也具有动物SUMO E3连接酶中发现的4个保守结构域[39-40],即SAP(Scaffold attachment factor(SAF)-A/B,acinus,PIAS for DNA binding)、PINIT(Nuclear retention of SP-RING protein/subcellular localization,Proline- isoleucine-asparagines-isoleucine-threonine)、SP-RING(Siz/PIAS RING)和SXS(serine-X-serine)结构域。除此之外,AtSIZ1还具有一个植物E3连接酶特有的PHD(plant homeodomain)结构域[41](图 2)。通过在siz1-2突变体中转入功能缺失突变的SIZ1sap、SIZ1phd、SIZ1pinit、SIZ1sp-ring和SIZ1sxs发现,SP-RING与SIZ1和SUMO结合的亚核结构的标记有关,PHD和PINIT可能与植物在响应糖信号和光信号过程中下胚轴的伸长有关,同时还发现,PHD对植物在热胁迫下的SUMO化响应是必需的[41]。植物中还有另外一种类型的SUMO E3连接酶MMS21(Methyl methanesulfonate-sensitive21),其只含有1个SP-RING结构域(图 2),主要参与细胞周期调控[42]、干细胞维持[34]和配子发育[43]等生物学过程。
2 SUMO E3连接酶在植物适应非生物胁迫中的作用 2.1 SUMO E3连接酶在植物适应干旱胁迫中的作用干旱是限制全球农业生产的最重要非生物胁迫之一。SIZ1可以通过ABA非依赖途径直接调控大量基因的表达并介导多种目标蛋白的SUMO化提高植物的抗旱性。Catala等[20]通过基于全基因组的表达分析发现,在拟南芥中总共有大约1 700个基因的表达受干旱胁迫诱导,其中,有262个基因的表达是受SIZ1调控且独立于DREB2A和ABA途径的,主要与花青素生物合成、茉莉酸信号转导和磷饥饿响应等有关。气孔开度的调节与植物对水分的利用效率和干旱响应有重要关系,Miura等[44]和Kim等[15]发现,SIZ1的缺失可以引起由SA介导的ROS积累,使气孔开度减小,从而增强siz1突变体的抗旱性。而在siz1突变体中过表达编码水杨酸羟化酶nahG后,SA的积累量减少,气孔开度增大,植株的抗旱性降低[15]。这与此前Catala等[20]提出的SIZ1通过介导大量蛋白SUMO化增强拟南芥抗旱性的结论正好相反,推测是与他们所用的实验材料和实验条件不同有关。
Li等[45]在匍匐翦股颖(Agrostis stolonifera)上的研究发现,过表达OsSIZ1后的转基因植株在干旱胁迫下SUMO化蛋白积累量增加,叶片保水能力更强且质膜完整性也更好[45],抗旱能力显著增强。Mishra等[46]在棉花(Gossypium sp.)中过表达OsSIZ1后转基因植株在干旱胁迫下的净光合效率和棉花产量显著增加。番茄SlSIZ1的表达受冷、NaCl、PEG、H2O2和ABA的诱导,在烟草中表达后,SlSIZ1转基因植株SUMO化蛋白含量显著增多,渗透胁迫下种子萌发力和植株抗旱性显著增强[30]。Mishra[47]和Esmaeili等[48]也发现在拟南芥中过表达OsSIZ1可以提高转基因植株在干旱条件下P5CS的表达量,增加脯氨酸的积累并提高转基因拟南芥的抗旱性。
另一种类型的SUMO E3连接酶MMS21也可以通过调控ABA合成途径相关基因的表达调控拟南芥对干旱胁迫的响应。Zhang等[49]发现ABA、PEG处理或干旱胁迫可以抑制拟南芥AtMMS21的表达,过表达MMS21后拟南芥抗旱性显著降低,而mms21突变体表现出更强的抗旱性。
2.2 SUMO E3连接酶在植物适应盐胁迫中的作用高浓度盐分会造成离子毒害、渗透胁迫和氧化损伤等次级胁迫[50]。siz1突变体可以通过SOS3非依赖途径减少拟南芥对Na+的吸收和积累,增强拟南芥的耐盐性。100 mmol/L NaCl能促进拟南芥siz1突变体的生长并降低Na+的吸收和积累[51]。sos3-1突变体对Na+和Li+超敏感,而突变SIZ1后的sos3-1 siz1-1双突变体对Na+的敏感性显著降低,而对Li+的敏感性无明显变化[51-52]。
与以上Miura等[51]发现拟南芥siz1突变体通过减少对Na+的吸收和积累提高抗盐性的结论不同的是,有学者研究发现SIZ1也可以正向调节植物对盐胁迫的响应。Conti等[14]发现,盐胁迫可以诱导拟南芥体内SUMO化蛋白的积累。Miura等[18]在拟南芥中过表达SIZ1发现,在125 mmol/L NaCl条件下,转基因株系根系生长、叶面积和种子产量均明显优于野生型,耐盐性显著增强。在盐胁迫条件下,拟南芥过表达OsSIZ1后地上部和根中的Na+积累显著减少,而K+积累显著增多,同时P5CS的表达量也显著增加,抗盐能力显著增强[47-48]。
2.3 SUMO E3连接酶在植物适应高温胁迫中的作用为了鉴定拟南芥中的SUMO化靶标蛋白,Rytz等[53]利用适用于质谱研究的工程化SUMO1对siz1和mms21突变体进行蛋白质组学分析,发现有超过1 000个靶标蛋白在正常条件或热胁迫刺激后被SUMO化。其中,大多是核定位蛋白,它们中大多数都可以和SIZ1结合,但是没有鉴定到可以和MMS21结合的靶标蛋白,这说明MMS21只能介导少量低丰度蛋白的SUMO化修饰。
SIZ1可以通过SA非依赖途径介导热激转录因子(Heat shock transcription factors,HSFs)的SUMO化,调控编码热激蛋白(Heat shock proteins,HSPs)相关基因的表达,从而影响植物对热胁迫的响应。Yoo等[16]发现,在热胁迫下,相较于野生型,siz1-2和siz1-3突变体SA积累量更多,SUMO化蛋白含量更少,热敏感性更强。而在过表达nahG后siz1-2突变体尽管SA积累量减少,但热敏感性更强[16]。
在匍匐翦股颖中过表达水稻OsSIZ1可以极大提高转基因株系的耐热性[45]。Zhang等[29]发现,番茄SlSIZ1干扰株系对热胁迫敏感,过表达SlSIZ1可以提高番茄HSPs和HSFs等基因的表达,同时降低热胁迫下ROS的积累,使转基因株系抗热性显著增强。热胁迫下SlSIZ1过表达株系HSP70、HSP90和HsfA2的表达量显著增加,酵母双杂交(Yeast two-hybrid,Y2H)和双分子萤光互补(Bimolecular fluorescence complementation,BiFC)试验发现,SlSIZ1可以和SlHsfA1互作并介导SlHsfA1的SUMO化[29]。随后在棉花[46]和拟南芥[47-48]中过表达水稻OsSIZ1的研究也都发现相似的结果。
2.4 SUMO E3连接酶在植物适应冷胁迫中的作用低温诱导下,SIZ1会介导ICE1的SUMO化,被SUMO化的ICE1活性和稳定性均显著增加,并促进CBF3/DREB1A的表达而抑制MYB15的表达,以增强植物的耐寒能力。Miura等[17, 54]发现,在拟南芥siz1突变体中,CBF3/DREB1A的表达被抑制;ICE1的K393位置被替换后ICE1K393R无法与SUMO蛋白结合,在野生型拟南芥中表达ICE1K393R后CBF3/DREB1A的表达被抑制,同时MYB15积累量增加,转基因株系对低温的敏感性增强[17]。低温下SIZ1还可以通过抑制SA积累,诱导CBF3/DREB1A表达,从而增强植物的抗寒性。siz1突变体对冷胁迫敏感,SA积累水平较高,CBF3/DREB1A、COR47和KIN1等关键低温诱导表达基因的表达量较低;而在siz1突变体中表达nahG后,SA积累量降低,转基因植株抗冷性显著增强[55]。Miura等[18]也发现在拟南芥中过表达SIZ1可提高转基因株系的耐寒性。此外,Zhou等[33]在苹果中的研究还发现,低温可以诱导MdSIZ1的表达,而MdSIZ1能直接介导MdMYB1的SUMO化以提高MdMYB1的活性和稳定性,促进花青素的合成。
2.5 SUMO E3连接酶与植物养分吸收和利用植物感应磷酸盐(Pi)匮缺并启动控制Pi获取所必需信号传导途径是植物磷元素吸收的重要步骤。Miura等[52]发现,siz1突变体表现出过度的原型磷饥饿反应(包括初生根停止生长,侧根和根毛增多,根冠比增大,花青素积累量增加,但细胞内Pi含量与野生型相近)。AtPT2、AtPS2和AtPS3是拟南芥磷饥饿响应基因,Pi足量条件下,它们在siz1中的表达量显著高于野生型,但在磷饥饿状态下,它们在siz1突变体和野生型中的表达量无显著差异[52]。PHR是作用于磷饥饿响应基因AtIPS1和AtRNS1的MYB类转录因子,可在SIZ1介导下发生SUMO化,在siz1突变体中,AtIPS1和AtRNS1在磷饥饿条件下的响应要显著慢于野生型[52]。
SIZ1还可以通过介导硝酸盐还原酶NA1和NA2的SUMO化而参与到拟南芥的氮素同化过程,发生SUMO化后的NA1和NA2硝酸盐还原酶活性显著提高。Park等[56]发现,siz1-2中硝酸盐还原酶NRs活性较低,补充铵盐(NH4NO3或(NH4)2SO4)可以恢复拟南芥siz1-2突变体早花、矮化、种子发育异常和高SA积累量等表型,但补充硝酸盐(KNO3或NaNO3)、磷酸盐(NaH2PO4)和钾盐(KCl或KNO3)均无此作用。同时Park等[57]还发现,尽管拟南芥中编码亚硝酸盐还原酶的基因NiR的表达量显著高于siz1突变体,但并没有发现SIZ1介导的NiR的SUMO化。
水稻OsSIZ1也与氮(N)和磷(P)的同化有关。OsSIZ1对多个编码Pi转运蛋白、Pi调控蛋白、硝酸盐或铵盐转运蛋白的基因和氮代谢途径相关的基因的表达也都有调控作用[58]。无论Pi是否足量,ossiz1中P、Pi和N的含量都要显著高于野生型,测定其他矿质营养元素(K、Ca、Mg、Mn、Si、Fe、Zn、Cu)发现,OsSIZ1调控的营养元素同化是对N和P特异性的[58]。Pei等[59]利用ossiz2对磷吸收的研究发现OsSIZ2和OsSIZ1也具有类似的磷吸收调控作用。Zhang等[60]发现苹果MdSIZ1与Pi的响应有关,磷饥饿会诱导MdSIZ1基因大量表达,SUMO化蛋白的迅速积累,同时参与苹果磷饥饿响应的MYB类转录因子MdPHR1也会被SUMO化;在磷饥饿条件下,antiMdSIZ1愈伤组织无法生长而过表达MdSIZ1的愈伤组织生长则几乎不受影响。
2.6 SUMO E3连接酶与ABA途径ABA是五大经典植物激素之一,对植物生长和发育的很多方面都有重要的调控作用,因其对植物响应各种生物和非生物胁迫的重要作用,还被称为“胁迫激素”[61]。SIZ1可以通过介导ABI5的SUMO化参与ABA信号响应。siz1突变体对ABA超敏感,外源ABA会抑制siz1种子的萌发和初生根的生长,并显著促进依赖ABI5的ABA信号响应基因如RD29A、RD29B、AtEm6、RAB18和ADH1等的表达[18]。ABI5被SUMO化后活性降低且不易被蛋白酶体降解,使得RD29A、RD29B、AtEm6、RAB18和ADH1等基因的表达受到抑制;相较于siz1-2,siz1-2 abi5-4双突变体对ABA的敏感性显著降低,ABI5蛋白的K391位置为SUMO蛋白和ABI5的特异性识别位点,在abi5-4中过表达ABI5K391R后,由于ABI5K391R无法被SUMO化,转基因株系对ABA超敏感[62-63]。
SIZ1还可通过介导MYB30发生SUMO化而调节与ABI5途径“相平行”的ABA信号响应途径。拟南芥myb30突变体对外源ABA超敏感,Zheng等[64]研究发现,MBY30的K283为发生SIZ1介导的SUMO化结合的特异位点,在myb30表达MYB30K283R只能部分回补其ABA超敏感表型;在野生型拟南芥中过表达MYB30后转基因株系对ABA的敏感性显著降低,而在siz1突变体中过表达MYB30并不会改变siz1的ABA超敏感表型。同时Zheng等[64]还发现,siz1-2 myb30-2双突变体比任意一个单突变体对ABA更为敏感。综上所述,依赖SIZ1介导的MYB30和ABI5的SUMO化修饰共同影响了拟南芥的ABA信号途径中相关基因的表达,进而影响了拟南芥对ABA的响应。
MMS21也与ABA的合成和响应有关。Zhang等[49]发现,ABA可以抑制MMS21的表达,mms21对ABA超敏感,COR15A、RD22和P5CS1等ABA途径相关的胁迫响应基因在mms21突变体的表达量均要显著高于野生型植株。
2.7 SUMO E3连接酶在植物适应重金属胁迫中的作用Cu是植物正常生命活动所必需的微量元素之一,在很多生物学过程中都有重要作用,但过量的Cu2+会诱导活性氧的产生而对植物生长造成毒害,SIZ1可以减少拟南芥对Cu2+的吸收和并促进其再分配,以降低其对植物的毒害作用。Chen等[21]发现,Cu2+处理会诱导拟南芥SUMO化水平的迅速增加。siz1-2和siz1-3突变体对外源Cu2+超敏感,且地上部Cu2+积累显著高于野生型;在Cu2+处理下,siz1突变体中金属转运蛋白YELLOW STRIPE-LIKE1(YSL1)和YSL3的编码基因的表达量要明显高于野生型,而siz1 ysl3和siz1 ysl1双突变体均可有效降低植株体内Cu2+的积累水平并显著提高对Cu2+胁迫的耐受性[21, 65](表 1)。
SUMO化是一种响应多重信号途径的复杂调控机制,在植物的正常生长发育、激素代谢和胁迫响应过程中发挥着重要的调控作用。众多研究表明,E3连接酶介导的SUMO化对植物抗逆性产生显著的影响,在此过程中SUMO E3连接酶SIZ1对植物适应干旱、高盐、高/低温、N/P缺乏和重金属毒害等非生物胁迫起了决定性作用,很多的SUMO化靶蛋白已经被鉴定出来。但迄今为止,以下问题还难以解答:SUMO E3连接酶是如何进行活性调节并特异性选择靶蛋白的;在SUMO化/去SUMO化的循环过程中SUMO蛋白酶是如何准确发挥作用的?启动SUMO化修的分子机制是什么,这些问题的解答将对阐明SUMO蛋白如何与不同底物蛋白相结合而响应单一或复合胁迫的机制至关重要。此外,对参与SUMO化修饰过程的众多重要蛋白的生化和结构特性和大量靶蛋白功能的鉴定等也将是未来SUMO化研究领域需要重点研究的课题。
| [1] |
Castro PH, Tavares RM, Bejarano ER, et al. SUMO, a heavyweight player in plant abiotic stress responses[J]. Cellular and Molecular Life Sciences, 2012, 69(19): 3269-3283. DOI:10.1007/s00018-012-1094-2 |
| [2] |
Tomanov K, Nukarinen E, Vicente J, et al. Sumoylation and phosphorylation:hidden and overt links[J]. Journal of Experimental Botany, 2018, 69(19): 4583-4590. DOI:10.1093/jxb/ery167 |
| [3] |
He Z, Huang T, Ao K, et al. Sumoylation, phosphorylation, and acetylation fine-tune the turnover of plant immunity components mediated by ubiquitination[J]. Front Plant Sci, 2017, 8: 1682. DOI:10.3389/fpls.2017.01682 |
| [4] |
Vertegaal AC. Uncovering ubiquitin and ubiquitin-like signaling networks[J]. Chemical Reviews, 2011, 111(12): 7923-7940. DOI:10.1021/cr200187e |
| [5] |
Elrouby N. Regulation of plant cellular and organismal development by SUMO[J]. Advances in Experimental Medicine and Biology, 2017, 963: 227-247. |
| [6] |
Kurepa J, Walker JM, Smalle J, et al. The small ubiquitin-like modifier(SUMO)protein modification system in Arabidopsis - Accumulation of SUMO1 and -2 conjugates is increased by stress[J]. Journal of Biological Chemistry, 2003, 278(9): 6862-6872. DOI:10.1074/jbc.M209694200 |
| [7] |
Benlloch R, Lois LM. Sumoylation in plants:mechanistic insights and its role in drought stress[J]. Journal of Experimental Botany, 2018, 69(19): 4539-4554. DOI:10.1093/jxb/ery233 |
| [8] |
Zhang ZY, Li JH, Liu HH, et al. Roles of ubiquitination-mediated protein degradation in plant responses to abiotic stresses[J]. Environmental and Experimental Botany, 2015, 114: 92-103. DOI:10.1016/j.envexpbot.2014.07.005 |
| [9] |
Ouyang J, Gill G. SUMO engages multiple corepressors to regulate chromatin structure and transcription[J]. Epigenetics, 2009, 4(7): 440-444. DOI:10.4161/epi.4.7.9807 |
| [10] |
Garvin AJ, Morris JR. SUMO, a small, but powerful, regulator of double-strand break repair[J]. Philos Trans R Soc Lond B Biol Sci, 2017, 28(372): 20160281. |
| [11] |
Melchior F, Schergaut M, Pichler A. SUMO:ligases, isopeptidases and nuclear pores[J]. Trends in Biochemical Sciences, 2003, 28(11): 612-618. DOI:10.1016/j.tibs.2003.09.002 |
| [12] |
Enserink JM. SUMO and the cellular stress response[J]. Cell Div, 2015, 10: 4. DOI:10.1186/s13008-015-0010-1 |
| [13] |
Elrouby N. Analysis of small ubiquitin-like modifier(SUMO)targets reflects the essential nature of protein sumoylation and provides insight to elucidate the role of SUMO in plant development[J]. Plant Physiology, 2015, 169(2): 1006-1017. |
| [14] |
Conti L, Price G, O'Donnell E, et al. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis[J]. Plant Cell, 2008, 20(10): 2894-2908. DOI:10.1105/tpc.108.058669 |
| [15] |
Kim JY, Song JT, Seo HS. Post-translational modifications of Arabidopsis E3 SUMO ligase AtSIZ1 are controlled by environmental conditions[J]. FEBS Open Bio, 2017, 7(10): 1622-1634. DOI:10.1002/2211-5463.12309 |
| [16] |
Yoo CY, Miura K, Jin JB, et al. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid[J]. Plant Physiology, 2006, 142(4): 1548-1558. |
| [17] |
Miura K, Jin JB, Lee J, et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis[J]. The Plant Cell, 2007, 19(4): 1403-1414. |
| [18] |
Miura K, Nozawa R. Overexpression of SIZ1 enhances tolerance to cold and salt stresses and attenuates response to abscisic acid in Arabidopsis thaliana[J]. Plant Biotechnology, 2014, 31(2): 167-172. DOI:10.5511/plantbiotechnology.14.0109a |
| [19] |
Castro PH, Couto D, Freitas S, et al. SUMO proteases ULP1c and ULP1d are required for development and osmotic stress responses in Arabidopsis thaliana[J]. Plant Mol Biol, 2016, 92(1/2): 143-159. |
| [20] |
Catala R, Ouyang J, Abreu IA, et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses[J]. The Plant Cell, 2007, 19(9): 2952-2966. DOI:10.1105/tpc.106.049981 |
| [21] |
Chen CC, Chen YY, Tang IC, et al. Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance[J]. Plant Physiology, 2011, 156(4): 2225-2234. DOI:10.1104/pp.111.178996 |
| [22] |
Miller MJ, Barrett-Wilt GA, Hua Z, et al. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis[J]. Proc Natl Acad Sci USA, 2010, 107(38): 16512-16517. DOI:10.1073/pnas.1004181107 |
| [23] |
Miller MJ, Scalf M, Rytz TC, et al. Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis[J]. Molecular & Cellular Proteomics, 2013, 12(2): 449-463. |
| [24] |
Chaikam V, Karlson DT. Response and transcriptional regulation of rice SUMOylation system during development and stress conditions[J]. BMB Rep, 2010, 43(2): 103-109. DOI:10.5483/BMBRep.2010.43.2.103 |
| [25] |
Srivastava AK, Zhang C, Caine RS, et al. Rice SUMO protease overly tolerant to salt 1 targets the transcription factor, OsbZIP23 to promote drought tolerance in rice[J]. Plant Journal, 2017, 92(6): 1031-1043. DOI:10.1111/tpj.13739 |
| [26] |
Srivastava AK, Zhang C, Yates G, et al. SUMO is a critical regulator of salt stress responses in rice[J]. Plant Physiology, 2016, 170(4): 2378-2391. DOI:10.1104/pp.15.01530 |
| [27] |
Augustine RC, York SL, Rytz TC, et al. Defining the SUMO system in maize:SUMOylation is up-regulated during endosperm development and rapidly induced by stress[J]. Plant Physiology, 2016, 171(3): 2191-2210. DOI:10.1104/pp.16.00353 |
| [28] |
Wang H, Wang M, Xia Z. Overexpression of a maize SUMO conjugating enzyme gene(ZmSCE1e)increases Sumoylation levels and enhances salt and drought tolerance in transgenic tobacco[J]. Plant Science, 2019, 281: 113-121. DOI:10.1016/j.plantsci.2019.01.020 |
| [29] |
Zhang S, Wang S, Lv J, et al. SUMO E3 ligase SlSIZ1 facilitates heat tolerance in tomato[J]. Plant Cell Physiol, 2018, 59(1): 58-71. |
| [30] |
Zhang S, Zhuang K, Wang S, et al. A novel tomato SUMO E3 ligase, SlSIZ1, confers drought tolerance in transgenic tobacco[J]. J Integr Plant Biol, 2017, 59(2): 102-117. DOI:10.1111/jipb.12514 |
| [31] |
Cai B, Kong X, Zhong C, et al. SUMO E3 ligases GmSIZ1a and GmSIZ1b regulate vegetative growth in soybean[J]. J Integr Plant Biol, 2017, 59(1): 2-14. DOI:10.1111/jipb.12504 |
| [32] |
Li Y, Wang G, Xu Z, et al. Organization and regulation of soybean SUMOylation system under abiotic stress conditions[J]. Front Plant Sci, 2017, 8: 1458. DOI:10.3389/fpls.2017.01458 |
| [33] |
Zhou LJ, Li YY, Zhang RF, et al. The SUMO E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low temperature conditions in apple[J]. Plant, Cell & Environment, 2017, 40: 2068-2080. |
| [34] |
Panglian X, Chengwei Y. Emerging role of SUMOylation in plant development[J]. Plant Signaling & Behavior, 2013, 8(7): e24727. |
| [35] |
Park HJ, Yun DJ. SUMO proteins grapple with biotic and abiotic stresses in Arabidopsis[J]. Journal of Plant Biology, 2013, 56(2): 77-84. |
| [36] |
Maria N, Konstantin T, Kay H, et al. Update on sumoylation:defining core components of the plant SUMO conjugation system by phylogenetic comparison[J]. New Phytologist, 2012, 195(1): 23-31. DOI:10.1111/j.1469-8137.2012.04135.x |
| [37] |
Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p[J]. Journal of Biological Chemistry, 1997, 272(43): 26799-26802. DOI:10.1074/jbc.272.43.26799 |
| [38] |
Yunus AA, Lima CD. Structure of the Siz/PIAS SUMO E3 ligase SIZ1 and determinants required for SUMO modification of PCNA[J]. Molecular Cell, 2009, 35(5): 669-682. DOI:10.1016/j.molcel.2009.07.013 |
| [39] |
Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants[J]. Current Opinion in Plant Biology, 2007, 10(5): 495-502. DOI:10.1016/j.pbi.2007.07.002 |
| [40] |
Reindle A, Belichenko I, Bylebyl GR, et al. Multiple domains in Siz SUMO ligases contribute to substrate selectivity[J]. Journal of Cell Science, 2006, 119(Pt 22): 4749-4757. |
| [41] |
Cheong MS, Park HC, Hong MJ, et al. Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes[J]. Plant Physiology, 2009, 151(4): 1930-1942. DOI:10.1104/pp.109.143719 |
| [42] |
Ishida T, Yoshimura M, Miura K, et al. MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development[J]. PLoS One, 2012, 7(10): e46897. DOI:10.1371/journal.pone.0046897 |
| [43] |
Liu M, Shi SF, Zhang SC, et al. SUMO E3 ligase AtMMS21 is required for normal meiosis and gametophyte development in Arabidopsis[J]. BMC Plant Biology, 2014, 14(1): 153. DOI:10.1186/1471-2229-14-153 |
| [44] |
Miura K, Okamoto H, Okuma E, et al. SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis[J]. Plant Journal, 2013, 73(1): 91-104. DOI:10.1111/tpj.12014 |
| [45] |
Li Z, Hu Q, Zhou M, et al. Heterologous expression of OsSIZ1, a rice SUMO E3 ligase, enhances broad abiotic stress tolerance in transgenic creeping bentgrass[J]. Plant Biotechnology Journal, 2013, 11(4): 432-445. DOI:10.1111/pbi.12030 |
| [46] |
Mishra N, Sun L, Zhu X, et al. Overexpression of the rice SUMO E3 ligase gene OsSIZ1 in cotton enhances drought and heat tolerance, and substantially improves fiber yields in the field under reduced irrigation and rainfed conditions[J]. Plant & Cell Physiology, 2017, 58(4): 735-746. |
| [47] |
Mishra N, Srivastava AP, Esmaeili N, et al. Overexpression of the rice gene OsSIZ1 in Arabidopsis improves drought-, heat-, and salt-tolerance simultaneously[J]. PLoS One, 2018, 13(8): e0201716. DOI:10.1371/journal.pone.0201716 |
| [48] |
Esmaeili N, Yang X, Cai Y, et al. Co-overexpression of AVP1 and OsSIZ1 in Arabidopsis substantially enhances plant tolerance to drought, salt, and heat stresses[J]. Sci Rep, 2019, 9(1): 7642. DOI:10.1038/s41598-019-44062-0 |
| [49] |
Zhang S, Qi Y, Liu M, et al. SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana(F)[J]. J Integr Plant Biol, 2013, 55(1): 83-95. DOI:10.1111/jipb.12024 |
| [50] |
Zhu JK. Abiotic stress signaling and responses in plants[J]. Cell, 2016, 167(2): 313-324. |
| [51] |
Miura K, Sato A, Ohta M, et al. Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2[J]. Planta, 2011, 234(6): 1191-1199. DOI:10.1007/s00425-011-1476-y |
| [52] |
Miura K, Rus A, Sharkhuu A, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses[J]. Proc Natl Acad Sci USA, 2005, 102(21): 7760-7765. DOI:10.1073/pnas.0500778102 |
| [53] |
Rytz TC, Miller MJ, McLoughlin F, et al. SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress[J]. The Plant Cell, 2018, 30(5): 1077-1099. DOI:10.1105/tpc.17.00993 |
| [54] |
Miura K, Hasegawa PM. Regulation of cold signaling by sumoylation of ICE1[J]. Plant Signal Behav, 2008, 3(1): 52-53. DOI:10.4161/psb.3.1.4865 |
| [55] |
Miura K, Ohta M. SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation[J]. Journal of Plant Physiology, 2010, 167(7): 555-560. DOI:10.1016/j.jplph.2009.11.003 |
| [56] |
Park BS, Song JT, Seo HS. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1[J]. Nat Commun, 2011, 2: 400. DOI:10.1038/ncomms1408 |
| [57] |
Park BS, Kim SI, Seo HS. AtSIZ1 regulates expression of nitrite reductase but not its activity[J]. Journal of the Korean Society for Applied Biological Chemistry, 2013, 56(2): 243-245. DOI:10.1007/s13765-012-3223-x |
| [58] |
Wang H, Sun R, Cao Y, et al. OsSIZ1, a SUMO E3 ligase gene, is involved in the regulation of the responses to phosphate and nitrogen in rice[J]. Plant & Cell Physiology, 2015, 56(12): 2381-2395. |
| [59] |
Pei W, Jain A, Sun Y, et al. OsSIZ2 exerts regulatory influences on the developmental responses and phosphate homeostasis in rice[J]. Sci Rep, 2017, 7(1): 12280. DOI:10.1038/s41598-017-10274-5 |
| [60] |
Zhang RF, Zhou LJ, Li YY, et al. Apple SUMO E3 ligase MdSIZ1 is involved in the response to phosphate deficiency[J]. Journal of Plant Physiology, 2019, 232: 216-225. DOI:10.1016/j.jplph.2018.11.012 |
| [61] |
Wu Y, Yu F, Xie Q. Abscisic acid:Metabolism, transport and signaling[M]. New York: Springer, 2014.
|
| [62] |
Miura K, Lee J, Jin JB, et al. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling[J]. Proc Natl Acad Sci USA, 2009, 106(13): 5418-5423. DOI:10.1073/pnas.0811088106 |
| [63] |
Miura K, Hasegawa PM. Sumoylation and abscisic acid signaling[J]. Plant Signal Behav, 2009, 4(12): 1176-1178. DOI:10.4161/psb.4.12.10044 |
| [64] |
Zheng Y, Schumaker KS, Guo Y. Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana[J]. Proc Natl Acad Sci USA, 2012, 109(31): 12822-12827. DOI:10.1073/pnas.1202630109 |
| [65] |
Chen CC, Chen YY, Yeh KC. Effect of Cu content on the activity of Cu/ZnSOD1 in the Arabidopsis SUMO E3 ligase siz1 mutant[J]. Plant Signal Behav, 2011, 6(10): 1428-1430. DOI:10.4161/psb.6.10.16933 |