2. 中国农业科学院作物科学研究所,北京 100081
2. Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081
花青素是植物呈色的一种关键物质,是天然水溶性色素,隶属于类黄酮类色素。植物色素具有广泛的生物学功能,不仅使植物呈现丰富色彩引诱昆虫和动物传粉,而且使植物抵御多种生物胁迫和非生物胁迫(干旱、冷、盐、强光、紫外线、生长调节剂等)[1-6]。花青素生物合成由一系列结构基因编码的酶催化完成,编码花青素合成的结构基因直接受MYB转录因子、碱性螺旋-环-螺旋(bHLH)转录因子、WD40转录因子及其互作形成的MBW复合体调控[7]。本文概述了MYB、bHLH和WD40类转录因子在植物花青素合成中的功能,探讨了转MYB、bHLH和WD40类转录因子对植物花青素合成的影响,并综述了花青素合成的生物过程和调控网络,旨为研究植物花青素形成的分子调控机理提供借鉴。
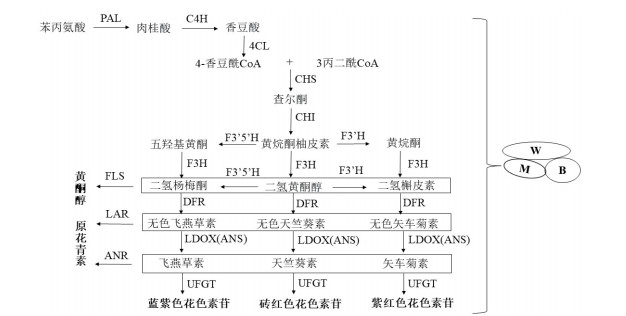
1 植物花青素合成途径植物花青素合成途径由结构基因催化完成(图 1)。结构基因编码一系列合成酶,是花青素形成的关键物质。如查尔酮合成酶(CHS)、查尔酮异构酶(CHI)、黄烷酮3-羟化酶(F3H)、类黄酮-3’-羟化酶(F3’H)、无色花青素双加氧酶(LDOX)、二氢黄酮醇4-还原酶(DFR)、花色素合成酶(ANS)和类黄酮3-O-葡糖基转移酶(UFGT)等。Li等[8]发现模式植物拟南芥(Arabidopsis thaiiana L.)UGT79B2和UGT79B3过表达显著增加了花青素的积累,同时增强了植物对低温以及干旱和盐胁迫的耐受性。Zohar等[9]发现石榴(Punica granatum L.)PgLDOX编码区中的碱基插入是导致“白色”石榴花青素含量较少的原因。Tan等[10]发现芒果(Mangifera indica L.)MiANRs在ban突变体中过度表达重建了种皮内原花青素(PAs)的生物合成途径。Wang等[11]证明SoCHS1在紫丁香(Syringa oblata Lindl.)类黄酮的生物合成中起着重要作用,过度表达可使花的颜色从淡粉色变成粉红色,可用于植物类黄酮成分的改良。
|
| 图 1 花青素生物合成途径 |
转录因子通过与结构基因启动子中的顺式元件结合,调控花青素生物合成途径。其中包括MYB转录因子、bHLH转录因子和WD40转录因子。Feng等[12]发现水芹(Oenanthe javanica)OjMYB1在拟南芥中异源表达可以提高花青素的含量,并上调与花青素生物合成相关的结构基因的表达水平。Wang等[13]发现PdMYB118在杨树(Populus deltoids)叶片原生质体中表达时,促进了花青素生物合成基因的表达。蒺藜苜蓿(Medicago truncatula)bHLH转录因子MtTT8是控制花青素和PAs生物合成的三元复合物的中心成分,MtTT8能恢复拟南芥tt8突变体中PAs和花青素的产生[14]。Gonzalez等[15]研究发现拟南芥AtTTG1与bHLH和MYB形成转录复合物直接靶向花青素晚期生物合成基因,促进幼苗花青素合成。
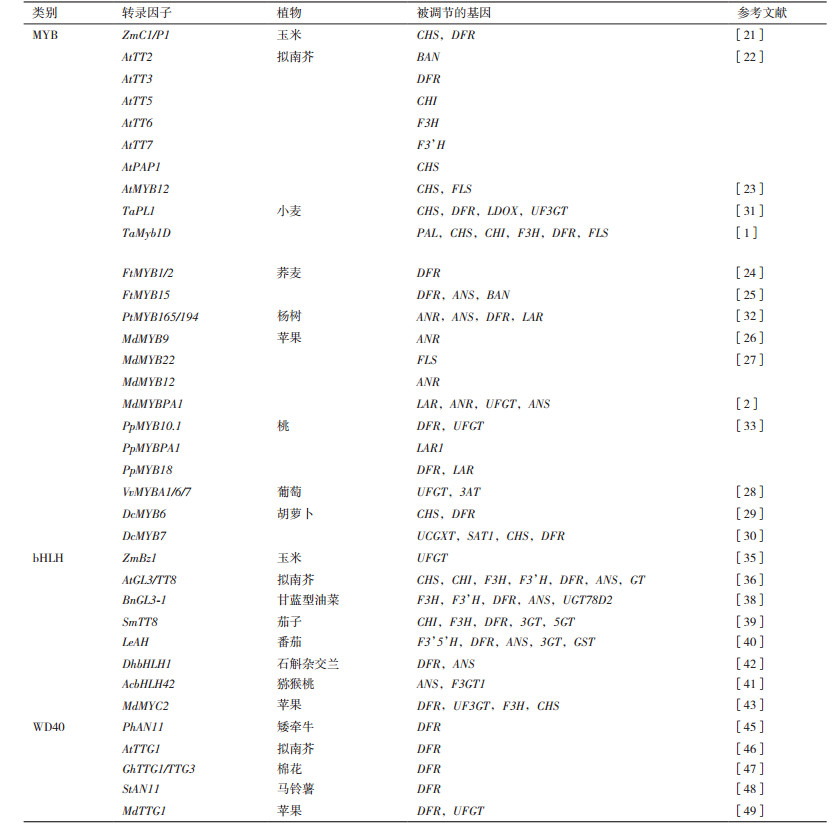
2 植物花青素合成途径的转录因子研究 2.1 MYB转录因子MYB转录因子存在于所有的真核生物中,N末端含一个高度保守的DNA结合结构域,由1-4个含51-53个氨基酸的不完全重复(R1、R2和R3)序列组成[16],是植物最大的转录因子家族之一。根据MYB结构域中不完全重复数目,MYB转录因子可以分为4个亚类:1R-MYB,R2R3-MYB,R1R2R3-MYB和4R-MYB[17]。其中,1R-MYB类蛋白是一类重要的端粒结合蛋白,主要维持染色体结构的完整性和调节基因转录;R2R3-MYB类蛋白是植物中数目最多的一类,形成包含100多个成员的MYB亚科,它们以N端含两个MYB结构域构成的DNA结合功能域为共同特征,参与植物主要和次生代谢合成[18];R1R2R3-MYB主要参与细胞周期的控制和细胞分化调节;4R-MYB类蛋白是植物中最少的一类。MYB转录因子家族各成员在植物中发挥着重要作用,包括次生代谢,激素信号转导与环境因子应答和对内外环境胁迫的反应等[16, 19]。
Paz-Ares等[20]从单子叶植物玉米(Zea mays L.)中克隆出与花青素合成有关的ZmC1基因,是植物中第一个被分离鉴定的MYB转录因子。随后大量MYB转录因子从各种植物中陆续得到分离和鉴定,这些MYB类转录因子能够参与调节植物组织着色,在花青素合成途径中发挥重要的调控作用(表 1)。
MYB转录因子对植物积累花青素具有重要意义。Hernandez等[21]发现R2R3-MYB类转录因子ZmC1/P1需要bHLH类转录因子ZmR才能转录共激活花青素合成途径的CHS和DFR的启动子以调控玉米花青素的合成。而在拟南芥花青素合成途径的研究中,部分MYB类转录因子(AtTT2、AtTT3、AtTT5、AtTT6、AtTT7、AtPAP1、AtMYB12等基因)已被克隆并阐明其作用模式,它们分别与不同的关键酶基因启动子结合来促进或抑制花青素的合成[22-23]。Bai等[24]从苦荞(Fagopyrum tataricum L.)中分离到R2R3-MYB型的转录因子基因FtMYB1、FtMYB2,过表达FtMYB1和FtMYB2显著增加了原花青素的积累。Luo等[25]在拟南芥中通过异位表达FtMYB15(R2R3-MYB)发现转基因拟南芥中叶片和种皮色素沉着增加,花青素和原花青素积累水平显著提高。
花青素浓度是许多水果颜色的重要决定因素,苹果(Malus pumila Mill.)MdMYB9是编码原花青素合成的正调控因子,与杨树(Populus spp.)PtMYB134功能相似,能够激活苹果和杨树花青素还原酶(ANR)启动子,参与调节花青素含量变化[26]。Wang等[2, 27]通过苹果愈伤组织过表达和拟南芥突变体异位表达MdMYB12,MdMYB22和MdMYBPA1,明确了它们在黄酮类化合物合成中的作用,这些研究为选育高含量花青素的优质苹果奠定了基础。Matus等[28]发现葡萄(Vitis vinifera spp.)中3个密切相关的R2R3-MYB基因VvMYBA1,VvMYBA6和VvMYBA7通过激活关键酶基因UFGT和3AT参与花青素合成途径。Xu等[29-30]从紫色胡萝卜(Daucus carota L.)品种中分离R2R3型MYB基因DcMYB6和DcMYB7,通过异位表达和敲除验证了它们是决定花青素合成的关键基因。
激活因子和抑制因子的协同作用可能对花青素生物合成在发育或应激后的精细调控具有重要意义。如小麦(Triticum aestivum L.)TaPL1作为正调控因子通过结合CHS、DFR、LDOX和UF3GT基因的启动子促进花青素合成;而TaMyb1D作为负调控因子参与类黄酮合成途径,另一方面增强了植物对干旱和氧化胁迫的耐受性[1, 31]。在杨树中,PtMYB165和PtMYB194在瞬时转录激活实验中均抑制PtMYB134对类黄酮途径酶基因启动子的激活,它们在杨树中的过表达使花青素和原花青素等化合物的积累量大大减少[32]。Zhou等[33]发现桃子(Prunus persica L.)花青素相关MYB转录因子PpMYB10.1和原花青素相关MYB转录因子PpMYBPA1均可激活抑制因子PpMYB18的转录,PpMYB18蛋白与MYB激活物相竞争,与bHLH转录因子bHLH3和bHLH33结合,激活反馈调节环以平衡花青素和原花青素的积累。
综上,MYB转录因子在植物色素积累和生理发育调控中均具有重要作用,MYB转录因子功能多样,继续挖掘其有效功能可为MYB家族转录因子的研究提供参考依据。
2.2 bHLH转录因子bHLH(basic Helix-Loop-Helix)转录因子家族,也参与了植物花青素的合成,bHLH转录基序约含60个氨基酸残基,包括两个功能区,由C末端的α螺旋-环-α螺旋(HLH)区域和N末端的(能与DNA顺式元件结合的)碱性氨基酸区域组成,HLH中环的长度依据不同bHLH蛋白而定,且区域之间可依赖疏水氨基酸相互作用形成同型或异型二聚体。通常bHLH蛋白运用保守的基序参与蛋白质间的相互作用,具有调控其核心的功能[34]。
植物bHLH转录因子能够参与调控多种生理代谢途径,如植物气孔开闭、激素应答、光形态建成、花器官发育、毛状体发生及体内离子平衡等,而调节花青素生物合成是植物bHLH转录因子最重要的功能之一,尤其是在植物次生化合物的合成和代谢途径发挥着重要的作用。1989年,Chandler等[35]在玉米中鉴定出调控类黄酮途径的第一个bHLH转录因子,随后bHLH转录因子序列在不同的生物中被鉴定出来(表 1)。Zhou等[36]研究发现AtGL3/TT8可与TTG1和PAP1形成复合物参与氮营养物质协同调节拟南芥花青素的合成。Xu等[37]在前人研究的基础上对AtTT8进行深入研究发现,AtTT8调节涉及至少6种不同的MBW复合物,形成正反馈调节回路以调控花青素合成。Gao等[38]发现甘蓝型油菜(Brassica napus L.)BnGL3-1基因在拟南芥gl3-3突变体中的异位表达导致真叶中花青素水平高于野生植物。Li等[39]在研究光诱导茄子(Solanum melongena L.)花青素合成的分子机理时发现转录因子SmTT8与光感受器共同参与花青素合成。Oiu等[40]验证了番茄(Lycopersicon esculentum Mill.)AH基因编码bHLH蛋白,过度表达AH显著提高了番茄下胚轴、叶片和果皮的花青素水平。Wang等[41]发现猕猴桃(Actinidia chinensis)AcbHLH42和AcMYB123能激活AcANS和AcF3GT1的启动子,调节猕猴桃内果皮组织特异性花青素的合成。Li等[42]从石斛杂交兰(Dendrobium hybrids orchid)中克隆出DhbHLH1基因,发现其能在唇部组织中形成明显的花青素色素沉着。An等[43]通过在苹果愈伤组织中过表达MdMYC2发现,花青素生物合成基因MdDFR、MdUF3GT、MdF3H和MdCHS的转录水平显著上调。
大量的研究数据表明bHLH转录因子在植物花青素合成途径中发挥着重要的调控作用,但花青素合成代谢过程是一个复杂的调控网络,目前对bHLH转录因子的研究还不能完全揭示它的全部功能,需要更充实的实验来为bHLH转录因子的功能揭示提供依据。
2.3 WD40转录因子WD40转录因子基序是由44-60个氨基酸组成的保守序列,N-末端(距N-末端11-24个残基)的GH起始和C-末端的WD结尾[44]。WD40蛋白不直接参与靶基因启动子的特异性识别,而在增强基因激活方面更具有积极作用,通常通过组蛋白的修饰来参与染色质的重塑,进而影响转录调控。矮牵牛(Petunia hybrida Vilm)PhAN11是植物中第一个分离出来的WD40蛋白,研究发现矮牵牛an11突变体中结构基因DFR的表达量下降[45]。之后,Walker等[47]从拟南芥中克隆出与AN11同源基因AtTTG1,其功能与AN11相似[46]。Humphries等[47]将棉花(Gossypium spp.)的两个WD-repeat基因GhTTG1和GhTTG3在白花紫罗兰(Matthiola incana L.)Mittg1突变体中异位表达,恢复了部分花青素的表型。Li等[48]从马铃薯(Solanum tuberosum L.)中克隆了一个WD40基因StAN11发现,该基因参与调控马铃薯花青素的合成。An等[49]发现苹果MdTTG1基因与bHLH转录因子和MYB转录因子相互作用形成复合物,然后启动下游花青素合成基因MdDFR和MdUFGT转录。Liu等[50]发现杨梅(Myrica rubra)MrWD40-1与MrMYB1和MrbHLH1相互作用参与杨梅花青素积累。Zhao等[51]发现蓝莓(Vaccinium spp.)WD40转录因子VcWDL2与VcMYBL1和VcbHLHL1相互作用参与蓝莓果实花青素积累和颜色发育(表 1)。
WD40类转录因子在植物组织花青素调控途径中具有重要作用,通过与bHLH和MYB转录因子作用形成复合物影响植物花青素和原花青素的合成。因此需要进一步挖掘WD40蛋白的功能以及深入的调控机理来揭示花青素合成网络。
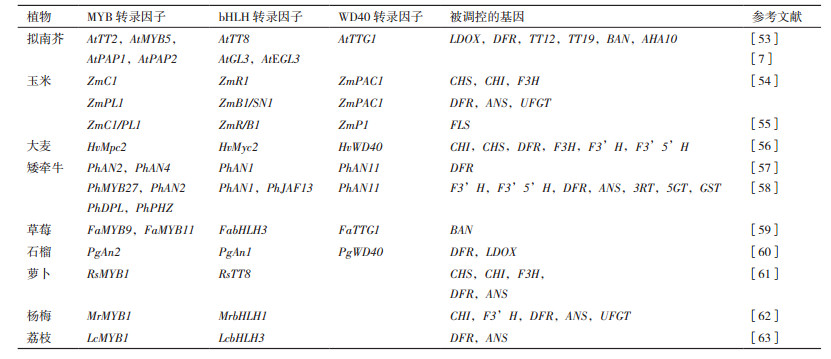
2.4 MBW复合体MBW复合体是MYB、bHLH和WD40三类转录因子互作形成的三元复合物,能够调控花青素合成,主要参与调节晚期类黄酮生物合成基因的表达(表 2)。如拟南芥在合成原花青素和花青素的过程中,MBW复合物直接参与晚期生物合成基因(LBGs)的表达[52]。MBW复合物在拟南芥种子原花青素合成的不同部位发挥着重要作用,如TT2-TT8-TTG1复合物通过调节关键酶基因DFR、LDOX、BAN、TT19、TT12和AHA10的表达在种子内皮细胞中具有活性;TT2-GL3/EGL3-TTG1复合物通过调控LDOX、BAN、AHA10和DFR的表达在种子的合点部位具有作用,这些结果突出了花青素调控机制的复杂性和精密性[7, 53]。此外,MBW复合物还能在激素茉莉酸酯(JA)的诱导下激活下游信号级联反应以调节花青素积累,从而增加植物应对外界胁迫的能力[6]。玉米中ZmPAC1基因编码WD40蛋白类似于花青素调控蛋白AN11和TTG1,能与玉米的ZmR1(bHLH)和ZmC1(MYB)互作共同控制花色素的合成[54]。Falcone Ferreyra等[55]通过染色质免疫沉淀(ChIP)实验证明了ZmFLS是P1、C1/PL1和R/B1调节复合物的直接靶标,证明其与玉米类黄酮早期合成途径相关。对大麦(Hordeum vulgare L.)籽粒糊粉层和果皮中不同色素积累研究发现,糊粉层中蓝色颗粒的色素积累与MBW复合物相关[56]。
Spelt等[57]在矮牵牛叶片中异位表达PhAN2(MYB)能够诱导PhAN1(bHLH)的表达,而在花药中PhAN1的表达又依赖于PhAN4(MYB),表明PhAN2和PhAN4均是PhAN1表达的激活因子;同时它们又与PhAN11(WD40)组合成复合物调控下游结构基因DFR的表达,从而激活花青素合成途径。而PhMYB27是矮牵牛中的一种抑制子,通过抑制MBW复合物的bHLH因子PhAN1的表达来抑制花青素生物合成基因的表达[58]。草莓(Fragaria ananassa Duch.)中FaMYB9/FaMYB11,FabHLH3和FaTTG1的功能与拟南芥AtTT2,AtTT8和AtTTG1相似,将草莓的基因转到拟南芥tt2-1,tt8-3和ttg1突变体中,发现能够激活拟南芥BAN启动子而参与合成花青素[59]。Ben-Simhon等[60]通过实验验证了石榴PgWD40基因能与PgAn1(bHLH)和PgAn2(MYB)共表达参与调节花青素生物合成的下游结构基因DFR和LDOX的表达。
在不依赖WD40蛋白的条件下,MYB和bHLH转录因子也能协同调控花青素合成基因的表达。如萝卜(Raphanus sativus L.)RsTT8和RsMYB1相互作用能显著增加内源性花青素生物合成基因的表达和花青素的积累[61]。杨梅MrbHLH1和MrMYB1的共表达以及荔枝(Litchi chinensis Sonn.)LcbHLH3和LcMYB1的共表达均能激活下游花青素生物合成基因,促进果实花青素合成[62-63]。
3 展望植物花青素的合成是一个复杂的过程,通过结构基因编码一系列的酶参与合成各种花青素途径相关的化合物,而各种调节基因则独自或相互作用结合成复合物与结构基因启动子中能被识别的顺式元件特异性结合,通过上调或下调结构基因的表达,最终调控花青素的合成。除了MYB、bHLH和WD40蛋白外,WADS、WRKY和WIP类蛋白也在植物花青素代谢途径中发挥着重要的调控作用,可能与MBW复合物相互作用或作用于其上下游参与花青素调控网络。
目前对植物花青素调控网络的研究已经从结构基因转向了调控基因,越来越多的花青素合成调控因子在转录水平上被鉴定出来。然而许多非模式植物或不同组织器官中花青素合成相关的转录因子还未广泛鉴定,迄今为止,MYB,bHLH和WD40转录因子及其组成的MBW复合体对基因表达的调控机制还远未揭示。未来需要借助基因组测序技术、转基因技术、CRISPR-Cas9技术、瞬时转录激活实验、酵母双杂交技术及蛋白质组学技术进一步对植物各类转录因子及其组成的MBW复合体进行鉴定,对转录因子间以及转录因子与结构基因间的相互作用及其表达调控模式进行深入研究。为进一步了解花青素生物合成途径的复杂调控过程和对培养具有优异农艺性状的蔬菜水果及观赏植物品种具有重要的价值意义。通过对花青素合成调控网络的深入研究,运用转基因或敲除技术控制转录因子的表达以定向改变植物颜色,增加植物的观赏价值,提高蔬菜水果的营养价值,为将来实现花青素合成的工业化生产奠定理论基础。同时也为抵御生物和非生物多样性胁迫,增强植物与环境之间的适应性,培育植物各种生理发育功能以及揭示植物品质资源丰富度提供必要的科学依据。
| [1] |
Wei Q, Zhang F, Sun F, et al. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants[J]. Plant Sci, 2017, 265: 112-123. DOI:10.1016/j.plantsci.2017.09.020 |
| [2] |
Wang N, Qu C, Jiang S, et al. The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low temperature conditions in red-fleshed apple[J]. Plant J, 2018, 96: 39-55. DOI:10.1111/tpj.14013 |
| [3] |
Kim J, Lee WJ, Vu TT, et al. High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance in Brassica napus L.[J]. Plant Cell Rep, 2017, 6: 1-10. |
| [4] |
Bai S, Tao R, Tang Y, et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear[J]. Plant Biotechnol J, 2019, 17(10): 1985-1997. DOI:10.1111/pbi.13114 |
| [5] |
Nguyen CT, Lim S, Lee JG, et al. VcBBx, VcMYB21, and VcR2R3MYB transcription factors are involved in UV-B-induced anthocyanin biosynthesis in the peel of harvested blueberry fruit[J]. J Agric Food Chem, 2017, 65(10): 2066-2073. DOI:10.1021/acs.jafc.6b05253 |
| [6] |
Qi T, Song S, Ren Q, et al. The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana[J]. Plant Cell, 2011, 23(5): 1795-1814. DOI:10.1105/tpc.111.083261 |
| [7] |
Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20(3): 176-185. DOI:10.1016/j.tplants.2014.12.001 |
| [8] |
Li P, Li YJ, Zhang FJ, et al. The Arabidopsis UDP-glycosyltransfe-rases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation[J]. The Plant Journal, 2016, 89: 85-103. |
| [9] |
Zohar BS, Sylvie J, Taly T, et al. A "white" anthocyanin-less pomegranate (Punica granatum L.) caused by an insertion in the coding region of the leucoanthocyanidin dioxygenase (LDOX; ANS) gene[J]. PLoS One, 2015, 10(11): e0142777-. DOI:10.1371/journal.pone.0142777 |
| [10] |
Tan L, Wang M, Kang Y, et al. Biochemical and functional characterization of anthocyanidin reductase (ANR) from Mangifera indica L.[J]. Molecules, 2018, 23(11): 2876. DOI:10.3390/molecules23112876 |
| [11] |
Wang Y, Dou Y, et al. Molecular characterization and functional analysis of chalcone synthase from Syringa oblata Lindl. in the fla-vonoid biosynthetic pathway[J]. Gene, 2017, 635(7): 16-23. |
| [12] |
Feng K, Xu ZS, Que F, et al. An R2R3-MYB transcription factor, OjMYB1, functions in anthocyanin biosynthesis in Oenanthe javanica[J]. Planta, 2017, 247: 301-315. |
| [13] |
Wang H, Wang X, Song W, et al. PdMYB118, isolated from a red leaf mutant of Populus deltoids, is a new transcription factor regulating anthocyanin biosynthesis in poplar[J]. Plant Cell Rep, 2019, 38: 927-936. DOI:10.1007/s00299-019-02413-1 |
| [14] |
Li P, Chen B, et al. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8[J]. New Phytol, 2016, 210(3): 905-921. DOI:10.1111/nph.13816 |
| [15] |
Gonzalez A, Zhao M, Leavitt JM, et al. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings[J]. Plant J, 2008, 53(5): 814-827. DOI:10.1111/j.1365-313X.2007.03373.x |
| [16] |
Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis[J]. Trends Plant Sci, 2010, 15(10): 573-581. DOI:10.1016/j.tplants.2010.06.005 |
| [17] |
Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants[J]. Molecular Plant, 2015, 8(5): 689-708. DOI:10.1016/j.molp.2015.03.012 |
| [18] |
Seo MS, Kim JS. Understanding of MYB transcription factors involved in glucosinolate biosynthesis in Brassicaceae[J]. Molecules, 2017, 22(9): 1549. DOI:10.3390/molecules22091549 |
| [19] |
Gao F, Zhou J, Deng RY, et al. Overexpression of a tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis[J]. J Plant Physiol, 2017, 214: 81-90. DOI:10.1016/j.jplph.2017.04.007 |
| [20] |
Paz-Ares J, Ghosal D, Wienand U, et al. The regulatory c1 locus of Zea mays encodes a protein with homology to MYB proto-oncogene products and with structural similarities to transcriptional activators[J]. EMBO J, 1987, 6(12): 3553-3558. DOI:10.1002/j.1460-2075.1987.tb02684.x |
| [21] |
Hernandez JM, Heine GF, Irani NG, et al. Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1[J]. J Biol Chem, 2004, 279(46): 48205-48213. DOI:10.1074/jbc.M407845200 |
| [22] |
Nesi N, Jond C, Debeaujon I, et al. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed[J]. Plant Cell, 2001, 13(9): 2099-2114. DOI:10.1105/TPC.010098 |
| [23] |
Mehrtens F, Kranz H, et al. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis[J]. Plant Physiol, 2005, 138(2): 1083-1096. DOI:10.1104/pp.104.058032 |
| [24] |
Bai YC, Li CL, et al. Characterization of two tartary buckwheat R2R3-MYB transcription factors and their regulation of proanthocyanidin biosynthesis[J]. Physiol Plant, 2014, 152(3): 431-440. DOI:10.1111/ppl.12199 |
| [25] |
Luo X, Zhao H, Yao P, et al. An R2R3-MYB transcription factor FtMYB15 involved in the synthesis of anthocyanin and proanthocyanidins from tartary buckwheat[J]. J Plant Growth Regul, 2017, 37(1): 1-9. |
| [26] |
Gesell A, Yoshida K, Tran LT, et al. Characterization of an apple TT2-type R2R3 MYB transcription factor functionally similar to the poplar proanthocyanidin regulator PtMYB134[J]. Planta, 2014, 240(3): 497-511. DOI:10.1007/s00425-014-2098-y |
| [27] |
Wang N, Xu H, Jiang S, et al. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana)[J]. Plant J, 2017, 90(2): 276-292. DOI:10.1111/tpj.13487 |
| [28] |
Matus JT, Cavallini E, Loyola R, et al. A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared to the berry color locus[J]. Plant J, 2017, 91: 220-236. DOI:10.1111/tpj.13558 |
| [29] |
Xu ZS, Feng K, Que F, et al. A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots[J]. Sci Rep, 2017, 7: 45324. DOI:10.1038/srep45324 |
| [30] |
Xu ZS, Yang QQ, Feng K, et al. Changing carrot color: insertions in DcMYB7 alter the regulation of anthocyanin biosynthesis and modification[J]. Plant Physiol, 2019, http://www.plantphysiol.org/content/early/2019/06/18/pp.19.00523.
|
| [31] |
Shin DH, Choi MG, Kang CS, et al. A wheat R2R3-MYB protein PURPLE PLANT1 (TaPL1) functions as a positive regulator of anthocyanin biosynthesis[J]. Biochem Biophys Res Commun, 2016, 469(3): 686-691. DOI:10.1016/j.bbrc.2015.12.001 |
| [32] |
Ma D, Reichelt M, Yoshida K, et al. Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar[J]. Plant J, 2018, 96: 949-965. DOI:10.1111/tpj.14081 |
| [33] |
Zhou H, Wang KL, Wang F, et al. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation[J]. New Phytol, 2019, 221: 1919-1934. DOI:10.1111/nph.15486 |
| [34] |
Feller A, Yuan L, Grotewold E. The BIF domain in plant bHLH proteins is an ACT-Like domain[J]. Plant Cell, 2017, 8(29): 1800-1802. |
| [35] |
Chandler VL, Radicella JP, Robbins TP, et al. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences[J]. Plant Cell, 1989, 1(12): 1175-1183. |
| [36] |
Zhou LL, Shi MZ, Xie DY. Regulation of anthocyanin biosynthesis by nitrogen in TTG1-GL3/TT8-PAP1-programmed red cells of Arabidopsis thaliana[J]. Planta, 2012, 236(3): 825-837. DOI:10.1007/s00425-012-1674-2 |
| [37] |
Xu W, Grain D, Gourrierec JL, et al. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis[J]. New Phytol, 2013, 198(1): 59-70. DOI:10.1111/nph.12142 |
| [38] |
Gao C, Guo Y, Wang J, et al. Brassica napus GLABRA3-1 promotes anthocyanin biosynthesis and trichome formation in true leaves when expressed in Arabidopsis thaliana[J]. Plant Biol, 2018, 20(1): 3-9. DOI:10.1111/plb.12633 |
| [39] |
Li J, Ren L, Gao Z, et al. Combined transcriptomic and proteomic analysis constructs a new model for light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.)[J]. Plant Cell Environ, 2017, 40: 3069-3087. DOI:10.1111/pce.13074 |
| [40] |
Qiu Z, Wang X, Gao J, et al. The tomato Hoffman's anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that is developmentally regulated and induced by low temperatures[J]. PLoS One, 2016, 11(3): e0151067. DOI:10.1371/journal.pone.0151067 |
| [41] |
Wang L, Tang W, Hu Y, et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang[J]. Plant J, 2019, 99: 359-378. |
| [42] |
Li C, Qiu J, Ding L, et al. Anthocyanin biosynthesis regulation of DhMYB2 and DhbHLH1 in Dendrobium hybrids petals[J]. Plant Physiol Biochem, 2017, 112: 335-345. DOI:10.1016/j.plaphy.2017.01.019 |
| [43] |
An JP, Li HH, Song LQ, et al. The molecular cloning and functional characterization of MdMYC2, a bHLH transcription factor in apple[J]. Plant Physiol Biochem, 2016, 108: 24-31. DOI:10.1016/j.plaphy.2016.06.032 |
| [44] |
Smith TF, et al. The WD repeat: a common architecture for diverse functions[J]. Trends Biochem Sci, 1999, 24(5): 181-185. DOI:10.1016/S0968-0004(99)01384-5 |
| [45] |
Vetten N, et al. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, pla-nts, and animals[J]. Genes Dev, 1997, 11(11): 1422-1434. DOI:10.1101/gad.11.11.1422 |
| [46] |
Walker AR, Davison PA, et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein[J]. Plant Cell, 1999, 11(7): 1337-1350. DOI:10.1105/tpc.11.7.1337 |
| [47] |
Humphries JA, Walker AR, Timmis JN, et al. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene[J]. Plant Mol Biol, 2005, 57(1): 67-81. DOI:10.1007/s11103-004-6768-1 |
| [48] |
Li W, Wang B, Wang M, et al. Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation[J]. J Integr Plant Biol, 2014, 56(4): 364-372. DOI:10.1111/jipb.12136 |
| [49] |
An XH, Tian Y, Chen KQ, et al. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation[J]. J Plant Physiol, 2012, 169(7): 710-717. DOI:10.1016/j.jplph.2012.01.015 |
| [50] |
Liu X, Feng C, Zhang M, et al. The MrWD40-1 gene of Chinese bayberry (Myrica rubra) interacts with MYB and bHLH to enhance anthocyanin accumulation[J]. Plant Molecular Biology Reporter, 2013, 31(6): 1474-1484. DOI:10.1007/s11105-013-0621-0 |
| [51] |
Zhao M, Li J, Zhu L, et al. Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development[J]. Genes, 2019, 10: E496. DOI:10.3390/genes10070496 |
| [52] |
Lloyd A, Brockman A, Aguirre L, et al. Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation[J]. Plant Cell Physiol, 2017, 58(9): 1431-1441. DOI:10.1093/pcp/pcx075 |
| [53] |
Xu W, et al. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed[J]. New Phytol, 2014, 202(1): 132-144. DOI:10.1111/nph.12620 |
| [54] |
Carey CC, Strahle JT, Selinger DA, et al. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana[J]. Plant Cell, 2004, 16: 450-464. DOI:10.1105/tpc.018796 |
| [55] |
Falcone, et al. Cloning and characterization of a UV-B-inducible maize flavonol synthase[J]. Plant J, 2010, 62(1): 77-91. DOI:10.1111/j.1365-313X.2010.04133.x |
| [56] |
Strygina KV, Börner A, et al. Identification and characterization of regulatory network components for anthocyanin synthesis in barley aleurone[J]. BMC Plant Biol, 2017, 17(S1): 184. DOI:10.1186/s12870-017-1122-3 |
| [57] |
Spelt C, et al. Anthocyanin1 of petunia encodes a basic Helix-Loop-Helix protein that directly activates transcription of structural anthocyanin genes[J]. Plant Cell, 2000, 12(9): 1619-1632. DOI:10.1105/tpc.12.9.1619 |
| [58] |
Albert NW, Davies KM, et al. A conserved network of transcrip-tional activators and repressors regulates anthocyanin pigmentation in eudicots[J]. Plant Cell, 2014, 26(9): 962-980. |
| [59] |
Schaart JG, Dubos C, Fuente IRDL, et al. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (fragaria × ananassa) fruits[J]. New Phytol, 2013, 197(2): 454-467. DOI:10.1111/nph.12017 |
| [60] |
Ben-Simhon Z, Judeinstein S, Nadler-Hassar T, et al. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development[J]. Planta, 2011, 234(5): 865-881. DOI:10.1007/s00425-011-1438-4 |
| [61] |
Lim SH, Kim DH, Kim JK, et al. A radish basic Helix-Loop-Helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis[J]. Front Plant Sci, 2017, 8: 1917. DOI:10.3389/fpls.2017.01917 |
| [62] |
Liu XF, Yin XR, Allan AC, et al. The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry (Myrica rubra) during anthocyanin biosynthesis[J]. Plant Cell Tiss Org, 2013, 115(3): 285-298. DOI:10.1007/s11240-013-0361-8 |
| [63] |
Lai B, Du LN, Liu R, et al. Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation[J]. Front Plant Sci, 2016, 7: 166. |