2. 吉林省人参基因资源开发与利用工程研究中心,长春 130118;
3. 吉林农业大学中药材学院,长春 130118
2. Research Center for Ginseng Genetic Resources Development and Utilization, Changchun 130118;
3. College of Chinese Medicinal Materials, Jilin Agricultural University, Changchun 130118
DNA binding with one finger(Dof)属于锌指蛋白家族中的一类亚家族,是植物中特有的一类转录因子,在酵母和线虫中尚未发现有Dof蛋白的存在[1]。由于Dof转录因子是植物特有、而且是一个较大的基因亚家族,其结构和功能具有多样性,是研究转录因子结构与进化理想的材料。本文重点综述Dof转录因子亚家族成员的结构、进化及在植物中的调控作用,以期对Dof转录因子深入研究奠定基础。
1 Dof转录因子在植物中的数量1993年,在玉米中首次发现了Dof转录因子,命名为ZmDof1(MNB1)[2]。截至目前,从简单的绿色单细胞藻类到复杂的高等植物都有Dof转录因子的报道(表 1)。
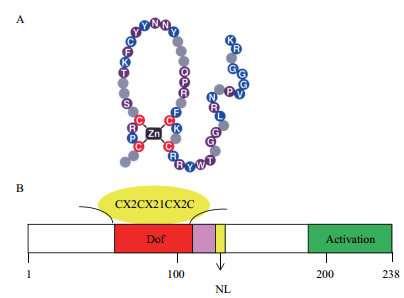
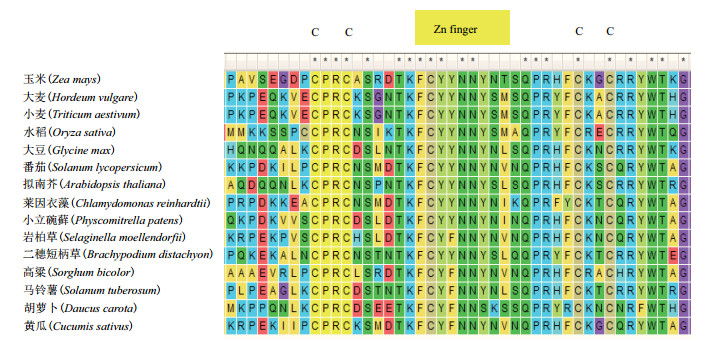
Dof转录因子一般由200-400个氨基酸组成,除了寡聚化位点和核定位信号外还具有两个功能结构域[24]:(1)位于N-末端的DNA结合区。在该区内具有一个富含半胱氨酸(Cysteine,Cys)残基的单锌指保守结构域[25](图 1),此结构域由52个氨基酸组成,其中包含4个保守的Cys残基和一个Zn2+,它们通过共价结合形成CX2CX21CX2C基序(Motif)(图 2)[1]。该基序中4个半胱氨酸任何一个发生突变,都会影响锌指结构使其不能与DNA结合,导致Dof转录因子活性丧失[26]。(2)位于C-末端的转录调控区。该区由一个色氨酸的单锌指保守结构域组成,其稳定性对DNA结合也非常重要。Dof转录因子的核心识别位点是AAAG序列(或它的互补序列,CTTT)[27-29],但南瓜的识别位点是AAGT序列[30]。Dof核心识别位点两侧序列不仅可以与DNA相互作用,也可与蛋白质相互作用,是双重功能区域。如拟南芥的Dof蛋白OBP1(OBF binding protein)可以通过该功能区域促进OBF4(Ocs element binding factor)与病原菌特异元件OCS(Octopine synthase)的结合[31]。在玉米中,Dof蛋白PBF(Prolamin-box binding factor)也可以通过该功能区域介导Dof与bZIP蛋白O2(Opaque 2)相互作用[32]。这些相互作用可能是Dof转录因子具有多种功能的因素[33]。
|
| 图 1 Dof转录因子典型特征与锌指结构相关的4个半胱氨酸残基 |
进行Dof转录因子系统分类研究不仅可以揭示Dof转录因子的起源与进化,还可以揭示它们相似的结构和进化关系,提供基因功能信息。因此,进化分析对基因的的结构和功能研究具有重要意义。
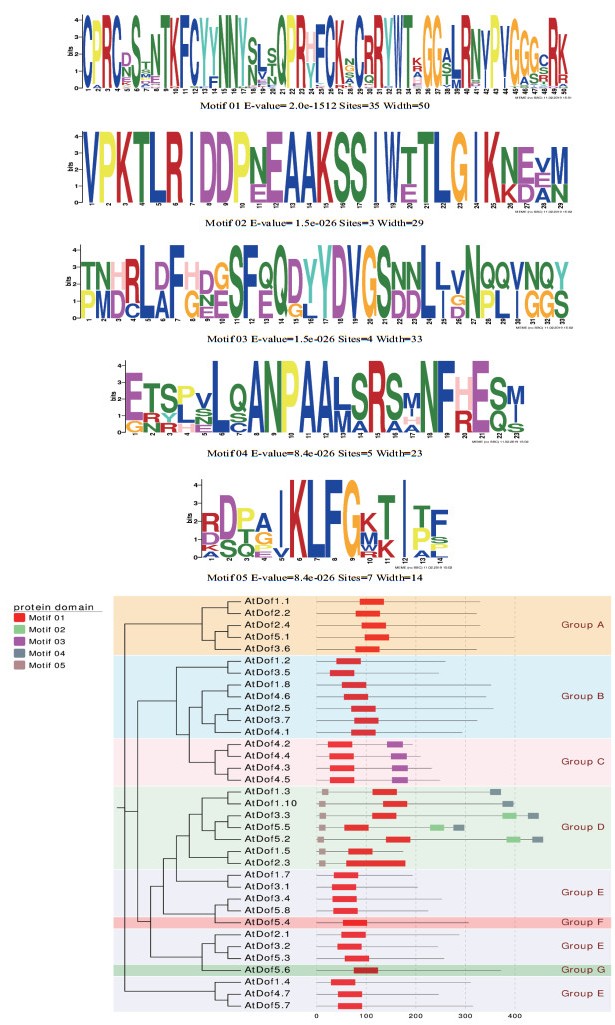
Yanagisawa等[34]对拟南芥中的36个Dof转录因子进行了进化分析,根据它们的蛋白序列将这些Dof转录因子分成了7个组。这7个组除了高度保守的Dof结构域外还存在其他的保守基序。从进化树上看蛋白结构相似并且高度同源的Dof转录因子形成一簇(Cluster),如DAG1(Dof affecting germination,AtDof3.7)和DAG2(Dof affecting germination,AtDof2.5),但是它们的功能相反,DAG1抑制种子萌发,DAG2促进种子萌发[35-38]。这些研究说明结构相似的基因,其功能有可能不同。因此用进化树邻近同一簇的基因做未知基因功能预测时,有可能得不到理想的结果。
Moreno-Risueno等[4]对拟南芥、水稻、大麦、绿藻、苔藓和裸子植物等6个物种的116个Dof转录因子进行了系统分类。结果这些Dof转录因子分成了7个组,分别是A、B、C、D、E、F和G,其结果与Yanagisawa的研究结果相似。Moreno-Risueno等又进一步分析了Dof转录因子的保守氨基酸序列,结果显示各组内具有相似的保守氨基酸序列,并且部分氨基酸序列在各组内高度保守,说明组内的基因在功能上有可能相似。内含子分析结果表明:A、F和G组含有内含子,而B、C和E组中没有内含子,D组中有2个基因(HvDof3和OsDof17)有内含子。绿藻CrDof1有4个内含子,其中有2个内含子是绿藻特有的。这些研究说明Dof转录因子的内含子随着植物的进化而逐渐减少或丢失。
由前人的结果笔者总结了36条拟南芥Dof蛋白的motif与进化树(图 3),找到motif共计5个,根据Yanagisawa等构建进化树的结果将36条蛋白序列分为7个组,与Yanagisawa等不同的结果是第6组和第7组串在第5组之间。进化树与Motif的组合图进一步说明了各组内具有相似的保守氨基酸序列,与Moreno-Risueno等结果一致。
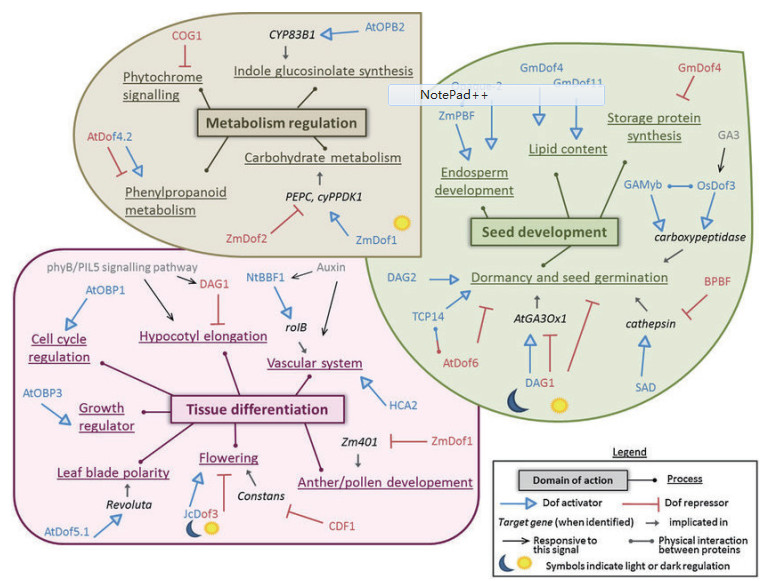
4 Dof转录因子在植物中的调控作用Dof转录因子在许多生理生化反应中起着关键的调控作用并且其功能多样化。尤其在种子发育、组织分化和代谢调控方面起着非常重要的调控作用(图 4)[39]。
4.1 种子发育赤霉素作为一种重要的植物激素,它能够调控种子的萌发,经研究证明一定量的赤霉素可以促进种子的萌发,而Dof转录因子通过调控赤霉素的生物合成实现对种子萌发的影响。有研究证明,控制拟南芥种子萌发的基因为DAG1(Dof affecting germination,AtDof3.7),该基因在脉管系统中特异表达,Papi等[35]在T-DNA插入突变体DAG1,结果发现种子一直保持休眠状态,不能在黑暗中萌发。Gabriele等[36]发现DAG1基因位于主阻遏子PIL5的下游,通过直接抑制GA3ox1基因的生物合成从而减少赤霉素的含量,抑制了种子的萌发,证明了在种子光诱导的萌发过程DAG1基因中是一个阻遏子。Gualberti等[37]又在拟南芥中发现一个与DAG1高度同源并具有相似结构特征的基因,将其命名为DAG2基因。DAG2基因的表达受到环境因素的影响,并且PIL5和DAG1可以抑制该基因的表达[38]。在大麦中,种子萌发的过程依赖激活组织蛋白酶基因,位于糊粉层中的Dof转录因子SAD通过激活该基因的表达促进种子的萌发,而另外一个转录因子BPBF却能抑制种子的萌发,其功能是通过抑制组织蛋白酶基因的表达而实现的[40]。此外,BPBF对于Amy32b基因(α-淀粉酶的功能基因)的调节过程与对组织蛋白酶基因的调节过程相似,Amy32b基因在种子发育阶段被抑制,但是在种子萌发阶段被激活[41]。拟南芥中的Dof转录因子AtDof6(AtDof3.2)基因能与TCP14蛋白相互作用,促进种子的萌发[42]。水稻中的Dof转录因子OsDof3同样可以促进种子的萌发,该转录因子在赤霉素的作用下可以与CPD3(type3羧肽酶)基因启动子的AAAG序列结合,激活该基因的表达,促进种子的萌发[43]。
有些Dof蛋白不仅能够调节贮藏蛋白的合成,还参与其他蛋白的表达。在水稻中,OsDof24和OsDof25能够调节谷蛋白GluB-1基因的表达,该蛋白属于种子贮藏蛋白[44]。有一些学者将某些Dof转录因子命名为PBFs,如玉米PBF、水稻RPBF、小麦WPBF、大麦BPBF,他们能够特异识别醇溶蛋白基因(胚乳表达基因)启动子上的TGTAAAG序列,从而激活其表达。如大麦的Dof基因BPBF能反式激活大麦胚乳醇溶蛋白基因hordein的表达。Mena等[45]在小麦中克隆到Dof家族基因小麦醇溶-谷蛋白盒结合因子(Wheat prolamin-box binding factor,WPBF),该基因在胚乳中特异表达,能激活醇溶蛋白的表达; Dong等[46]研究表明WPBF与TaQM基因相互作用,参与小麦胚乳特异α-醇溶蛋白基因的表达调控; 玉米的Dof蛋白PBF可以和胚乳特异表达玉米醇溶蛋白基因的启动子结合并激活其表达,从而调控玉米胚乳蛋白含量,影响种子的粒重和品质。研究表明PBFs还能够与Opaque-2(bZIPs转录因子)相互作用,并且PBFs在种子的发育阶段能够激活醇溶蛋白基因的表达[47]。PBFs和Opaque-2两个基因除了能调节醇溶蛋白基因以外,还可以调节其他的胚乳表达基因,如赖氨酸-酮戊二酸还原酶基因LKR,该基因的表达需要PBF和Opaque-2两个转录因子共同作用,而醇溶蛋白基因的表达只需要其中一个转录因子[48]。以上研究说明Dof转录因子对于植物种子的萌发和发育有着重要作用。
4.2 组织分化在拟南芥中AtDof4.2能调控植物的分支发育,这种调控机制是AtDof4.2能与AtEXPA9启动区结合从而调控AtEXPA9基因的表达来参与植物分支发育的[49]。在拟南芥中REV基因能够建立叶片远—近轴面极性,AtDof5.1基因可与REV基因的启动子区域结合,激活REV基因的表达,影响极性分化使叶片向上卷曲[50]。AtDof2.4和AtDof5.8能调控维管束的发育,其启动子分别在维管束形成初期的不同阶段发挥功能,AtDof2.4在形成层细胞中表达,而AtDof5.8在叶脉和花蕾的维管组织中表达[51]。ZmDof1通过负调控Zm401基因控制玉米的花粉发育[52]。
Dof转录因子可参与植物对激素应答基因的表达调控和对生长调节物质的响应过程,Dof转录因子NtBBF1(rolB domain B factor 1)能够特异性的结合到癌基因rolB启动子的ACTTTA基序,从而调控该基因在顶端分生组织和微管组织中的特异性表达和生长素诱导表达,促进根的生长[53-54]。在生长素诱导的条件下,Dof蛋白可以结合到南瓜抗坏血酸盐氧化酶基因(Ascorbate oxidase promoter-binding protein,AOBP)的启动子上,抑制该基因在生长组织中的表达[30]。生长素也可以调控拟南芥中OBP3(AtDof3.6)的表达,在过表达该基因后,发现其能够促进拟南芥的生长。在烟草中过表达Dof转录因子GmDof17-1基因,烟草的生长缓慢[55]。
Dof转录因子还可以保卫植物细胞的发育,在马铃薯中,StDof1可以激活与保卫细胞K+流通道蛋白调控的关键基因KST1基因,使其在保卫细胞中特异表达,从而调控保卫细胞对CO2的吸收和水分的扩散[56]。拟南芥中SCAP1基因在成熟的保卫细胞中表达,调控与气孔发育相关基因的表达,如K+通道蛋白、MYB转录因子、果胶甲酯酶等编码基因[57]。拟南芥中Dof转录因子OBP1(OBP-binding factor-1)参与细胞周期的调控。OBP1与Dof转录因子的另一个基因AtDof2.3共同作用可以调控周期蛋白基因CYCD3,使细胞周期缩短,但同时也减少了细胞总分裂数。在拟南芥中过表达OBP1会导致植株矮小[58]。拟南芥中的AtCDFs(Cycling Dof factor),它能够参与光周期反应调控过程,其中AtCDF1能够结合到成花因子FLOWERING LOCUS T(FT)的调控因子CONSTANS(CO)的启动子区域,抑制CO的表达,进而抑制FT的表达,最终抑制拟南芥的开花。AtCDF1是FKF1(Flavin-binding,Kelchrepeat,F-box-1)的靶基因,FKF1能够通过泛素依赖的途径抑制光反应,因此CDF1与开花响应过程有密切的关系[59]。在马铃薯中发现的Dof基因与CDF具有高度的同源性,研究表明它能够调控马铃薯块茎的发育。有报道称,在拟南芥中过表达番茄Dof转录因子SlCDF3,它能通过调节成花基因如CO和FT推迟开花[60]。拟南芥中AtDof4.1作为一个转录阻遏子也能够延迟开花,抑制生殖器官发育,并且能够使叶片、花和角果等器官变小[61]。AtDof4.7在植物的离区(Abscission zone)中高度表达,抑制了对花器官脱落非常重要的多聚半乳糖醛酸酶基因PGAZAT的表达。研究表明,AtDof4.7能够与AtZFP2(锌指类转录因子)相互作用,该转录因子也抑制了花器官的脱落,这两个转录因子具有协同作用[62]。水稻Dof转录因子OsDof12能通过调控Hd3a和OsMADS14的表达来调节水稻的开花机制[63]。以上研究结果表明,Dof转录因子在花粉发育,叶片极性,维管系统发育,保卫细胞发育,花器官的脱落及开花响应等生理过程中具有重要作用。
4.3 Dof转录因子在初级代谢中的调控作用在碳代谢过程中,Dof转录因子调控其相关基因的表达。在玉米中ZmDof1能够结合OsCS4PPDK启动区的AAAG基序使细胞质中的C4磷酸烯醇式丙酮酸羧化酶和丙酮酸激酶基因的表达提高,但是ZmDof2抑制C4磷酸烯醇式丙酮酸羧化酶基因的表达[64]。研究发现:将ZmDof1的转录水平降低到原来的80%,玉米中C4途径特异性基因的表达也不会受到影响[65]。在猕猴桃中,AdBAM3L基因是Dof转录因子AdDof3的靶基因,AdBAM3L基因控制该水果在成熟过程中淀粉降解成单糖的过程,可提高甜度及口感[66]。在甘薯中,根中的SRF1基因超量表达时可显著降低Ibβfruct2基因的转录水平从而使蔗糖转化酶的积累量降低,并导致了单糖的浓度减少,增加了块茎中淀粉含量,从而调控了碳代谢[67]。Dof蛋白不仅可以调控碳代谢还可以提高植物的氮利用率增加氮含量。在拟南芥中过表达ZmDof1基因后,阳性植株氮含量增加,能够在低氮条件下良好生长,同样,ZmDof1转基因水稻在低氮条件下也生长良好,证明了ZmDof1基因在氮代谢中的作用[39, 68]。在拟南芥中超量表达OsDof25基因,促进了高亲和、低亲和铵转运蛋白基因AtAMT1.1和AtAMT2.1的表达抑制了高亲和硝酸盐转运蛋白基因AtNRT2.1的表达,结果总的氨基氮含量提高,丙酮酸激酶(PK1和PK2),磷酸烯醇丙酮酸羧基酶(PEPC1、PEPC2)及NADP依赖的和NAD依赖的异柠檬酸脱氢酶基因的表达量增加。同时,谷氨酸脱氢酶活性增强,导致总氨基酸含量增加[69]。以上研究结果表明,无论是碳代谢还是氮代谢,Dof转录因子都起着至关重要的作用。
4.4 Dof转录因子在次生代谢中的调控作用Dof转录因子还参与植物次生代谢产物的合成。在拟南芥中,Dof转录因子与苯基丙酸合成途径和类黄酮合成途径两个次生代谢过程有关。据Skirycz的研究报道,超量表达AtDof4.2可以增加植株在低温下对光的敏感性,从侧面反映了AtDof4.2能够影响芥子酸酯和黄酮醇的变化。在正常条件下,转基因植株中黄酮醇没有发生显著变化,但是将其转移到4℃或者强光下,植物中积累的黄酮醇会越来越少,说明在低温和强光下,AtDof4.2可以负调控黄酮类物质的合成。之后该作者又在苯基丙酸合成途径中发现AtDof4.2可以正调控肉桂酸的合成[49]。
Dof转录因子还可以调控油脂的合成,并控制脂肪酸的含量。拟南芥中超表达的OBP2可通过增加P450基因(CYP83B1)的表达调控吲哚芥子油的生物合成[49]。Tian等[70]从大豆中鉴定出了28个Dof转录因子,其中GmDof4和GmDof11基因在油脂合成过程中起着重要作用。GmDof4和GmDof11基因产物分别特异性结合到乙酰辅酶A羧化酶基因(Acetyl CoA carboxylase)和长链脂酰辅酶A合成酶(Long-chain-CoA synthase)基因的启动子上,激活这两个基因的表达。乙酰辅酶A羧化酶和长链脂酰辅酶A合成酶是油脂合成过程的两个关键酶。在大豆中超表达GmDof11,可以提高大豆种子的含油量,降低蛋白质的含量,但对各脂肪酸组成没有显著影响[71]。在拟南芥中,超量表达GmDof4和GmDof11后,转基因株系种子的总油脂含量和油酸含量与野生型相比都显著提高,而种子贮藏蛋白基因CRA1的表达下调[72]。在椭圆小球藻(Chlorella ellipsoidea)中超量表达GmDof4基因后,细胞中油脂含量提高了44.4%-52.9%[73]。在油菜中干扰BnDof5.6基因的表达,后代种子胚变小,油分含量降低。转录组分析表明,干扰BnDof5.6基因的表达影响脂肪酸的合成和脂肪酸去饱和酶相关基因的表达[74]。
5 问题与展望Dof转录因子广泛参与植物种子发育和萌发、组织分化和代谢过程,这说明了Dof转录因子在植物的生长发育起着关键的作用。到目前为止,有关Dof转录因子的研究主要集中在模式植物和主要农作物中,如拟南芥、大豆和玉米等,但在药用植物中的研究少之甚少。研究领域主要集中在植物的发育及组织分化上,在次生代谢物合成中的作用仅报道了脂肪酸和类黄酮,其他的次生代谢物如甾醇类化合物并没有相关报道。该转录因子的靶基因尚不明确,与其他转录因子或蛋白的相互作用机制尚不清楚,对植物代谢调控机制还需要进一步研究。近几年随着基因组测序的陆续完成,Dof转录因子也依次在其他植物中被挖掘。因此,在今后的研究中可以在药用植物中挖掘转录因子并预测其功能,并利用生物技术手段将克隆到的基因转化到药用植物中,用以研究Dof转录因子调控的次生代谢合成,这将有助于提高具有药理活性的次生代谢产物的合成并阐明合成的分子机理及互作网络机制。
| [1] |
Gupta S, Malviya N, Kushwaha H, et al. Insights into structural and functional diversity of Dof(DNA binding with one finger)transcription factor[J]. Planta, 2015, 241(3): 549-562. DOI:10.1007/s00425-014-2239-3 |
| [2] |
Yanagisawa S, Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif[J]. J Biol Chem, 1993, 268(21): 16028-16036. |
| [3] |
Shigyo M, Tabei N, Yoneyama T, et al. Evolutionary processes during the formation of the plant-specific Dof transcription factor family[J]. Plant Cell Physiol, 2007, 48(1): 179-185. DOI:10.1093/pcp/pcl044 |
| [4] |
Moreno-Risueno MÁ, Martínez M, Vicente-Carbajosa J, et al. The family of DOF transcription factors:from green unicellular algae to vascular plants[J]. Mol Genet Genomics, 2007, 277(4): 379-390. DOI:10.1007/s00438-006-0186-9 |
| [5] |
Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families[J]. BMC Evolutionary Biology, 2003, 3(1): 17. |
| [6] |
Hernando-Amado S, González-Calle V, Carbonero P, et al. The family of DOF transcription factors in Brachypodium distachyon:phylogenetic comparison with rice and barley DOFs and expression profiling[J]. BMC Blant Biology, 2012, 12(1): 202. DOI:10.1186/1471-2229-12-202 |
| [7] |
Jiang Y, Zeng B, Zhao H, et al. Genome-wide transcription factor gene prediction and their expressional tissue-specificities in maize F[J]. Journal of Integrative Plant Biology, 2012, 54(9): 616-630. DOI:10.1111/j.1744-7909.2012.01149.x |
| [8] |
Shaw LM, Mcintyre CL, Gresshoff PM, et al. Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation[J]. Functional & Integrative Genomics, 2009, 9(4): 485-498. |
| [9] |
Moreno-Risueno MÁ, Díaz I, Carrillo L, et al. The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone[J]. The Plant Journal, 2007, 51(3): 352-365. DOI:10.1111/j.1365-313X.2007.03146.x |
| [10] |
Guo Y, Qiu LJ. Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics[J]. PLoS One, 2013, 8(9): e76809. DOI:10.1371/journal.pone.0076809 |
| [11] |
Kushwaha H, Gupta S, Singh VK, et al. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis[J]. Molecular Biology Reports, 2011, 38(8): 5037-5053. DOI:10.1007/s11033-010-0650-9 |
| [12] |
Venkatesh J, Park SW. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato[J]. Plant Physiology and Biochemistry, 2015, 94: 73-85. DOI:10.1016/j.plaphy.2015.05.010 |
| [13] |
Huang W, Huang Y, Li M, et al. Dof transcription factors in carrot:genome-wide analysis and their response to abiotic stress[J]. Biotechnology Letters, 2016, 38(1): 145-155. DOI:10.1007/s10529-015-1966-2 |
| [14] |
Cai X, Zhang Y, Zhang C, et al. Genome-wide analysis of plant-specific Dof transcription factor family in tomato[J]. Journal of Integrative Plant Biology, 2013, 55(6): 552-566. DOI:10.1111/jipb.v55.6 |
| [15] |
Wen C, Cheng Q, Zhao L, et al. Identification and characterisation of Dof transcription factors in the cucumber genome[J]. Scientific Reports, 2016, 6: 23072. DOI:10.1038/srep23072 |
| [16] |
Wu Z, Cheng J, Cui J, et al. Genome-wide identification and expression profile of dof transcription factor gene family in pepper(Capsicum annuum L.)[J]. Frontiers in Plant Science, 2016, 7: 574. |
| [17] |
Ma J, Li MY, Wang F, et al. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage[J]. BMC Genomics, 2015, 16(1): 33. DOI:10.1186/s12864-015-1242-9 |
| [18] |
Gupta S, Kushwaha H, Singh VK, et al. Genome wide in silico characterization of Dof transcription factor gene family of sugarcane and its comparative phylogenetic analysis with Arabidopsis, rice and sorghum[J]. Sugar Tech, 2014, 16(4): 372-384. DOI:10.1007/s12355-013-0288-8 |
| [19] |
Malviya N, Gupta S, Singh VK, et al. Genome wide in silico characterization of Dof gene families of pigeonpea(Cajanus cajan(L)Millsp.)[J]. Molecular Biology Reports, 2015, 42(2): 535-552. DOI:10.1007/s11033-014-3797-y |
| [20] |
Yang X, Tuskan GA. Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication[J]. Plant Physiol, 2006, 142(3): 820-830. DOI:10.1104/pp.106.083642 |
| [21] |
Li H, Huang W, Liu ZW, et al. Transcriptome-based analysis of Dof family transcription factors and their responses to abiotic stress in tea plant(Camellia sinensis)[J]. International Journal of Genomics, 2016. ID: 5614142. https://www.ncbi.nlm.nih.gov/pubmed/27872842
|
| [22] |
Song A, Gao T, Li P, et al. Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium[J]. Frontiers in Plant Science, 2016, 7: 199. |
| [23] |
Zhang Z, Yuan L, Liu X, et al. Evolution analysis of Dof transcription factor family and their expression in response to multiple abiotic stresses in Malus domestica[J]. Gene, 2018, 639: 137-148. DOI:10.1016/j.gene.2017.09.039 |
| [24] |
Diaz I, Vicente-Carbajosa J, Abraham Z, et al. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development[J]. The Plant Journal, 2002, 29(4): 453-464. DOI:10.1046/j.0960-7412.2001.01230.x |
| [25] |
Yanagisawa S. Dof domain proteins:plant-specific transcription factors associated with diverse phenomena unique to plants[J]. Plant Cell Physiol, 2004, 45(4): 386-391. DOI:10.1093/pcp/pch055 |
| [26] |
Umemura Y, Ishiduka T, Yamamoto R, et al. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain[J]. The Plant Journal, 2004, 37(5): 741-749. DOI:10.1111/tpj.2004.37.issue-5 |
| [27] |
Cominelli E, Galbiati M, Albertini A, et al. DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter[J]. BMC Plant Biology, 2011, 11(1): 162. DOI:10.1186/1471-2229-11-162 |
| [28] |
Yang J, Yang MF, Zhang WP, et al. A putative flowering-time-related Dof transcription factor gene, JcDof3, is controlled by the circadian clock in Jatropha curcas[J]. Plant Science, 2011, 181(6): 667-674. DOI:10.1016/j.plantsci.2011.05.003 |
| [29] |
Chen X, Wang D, Liu C, et al. Maize transcription factor Zmdof1 involves in the regulation of Zm401 gene[J]. Plant Growth Regulation, 2012, 66(3): 271-284. DOI:10.1007/s10725-011-9651-5 |
| [30] |
Kisu Y, Ono T, Shimofurutani N, et al. Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene[J]. Plant Cell Physiol, 1998, 39(10): 1054-1064. DOI:10.1093/oxfordjournals.pcp.a029302 |
| [31] |
Zhang B, Chen W, Foley RC, et al. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences[J]. The Plant Cell, 1995, 7(12): 2241-2252. DOI:10.1105/tpc.7.12.2241 |
| [32] |
Vicente-Carbajosa J, Moose SP, Parsons RL, et al. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2[J]. Proceedings of the National Academy of Sciences, 1997, 94(14): 7685-7690. DOI:10.1073/pnas.94.14.7685 |
| [33] |
Yanagisawa S. The transcriptional activation domain of the plant-specific Dof1 factor functions in plant, animal, and yeast cells[J]. Plant and Cell Physiol, 2001, 42(8): 813-822. DOI:10.1093/pcp/pce105 |
| [34] |
Yanagisawa S. The Dof family of plant transcription factors[J]. Trends in Plant Science, 2002, 7(12): 555-560. DOI:10.1016/S1360-1385(02)02362-2 |
| [35] |
Papi M, Sabatini S, Bouchez D, et al. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination[J]. Genes & Development, 2000, 14(1): 28-33. |
| [36] |
Gabriele S, Rizza A, Martone J, et al. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1[J]. The Plant Journal, 2010, 61(2): 312-323. |
| [37] |
Gualberti G, Papi M, Bellucci L, et al. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds[J]. The Plant Cell, 2002, 14(6): 1253-1263. DOI:10.1105/tpc.010491 |
| [38] |
Rizza A, Boccaccini A, Lopez-Vidriero I, et al. Inactivation of the ELIP1 and ELIP2 genes affects Arabidopsis seed germination[J]. New Phytologist, 2011, 190(4): 896-905. DOI:10.1111/nph.2011.190.issue-4 |
| [39] |
Noguero M, Atif RM, Ochatt S, et al. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants[J]. Plant Science, 2013, 209: 32-45. DOI:10.1016/j.plantsci.2013.03.016 |
| [40] |
Isabel-LaMoneda I, Diaz I, Martinez M, et al. SAD:a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds[J]. The Plant Journal, 2003, 33(2): 329-340. DOI:10.1046/j.1365-313X.2003.01628.x |
| [41] |
Zou X, Neuman D, Shen QJ. Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells[J]. Plant Physiol, 2008, 148(1): 176-186. DOI:10.1104/pp.108.123653 |
| [42] |
Rueda-Romero P, Barrero-Sicilia C, Gómez-Cadenas A, et al. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14[J]. J Exp Bot, 2011, 63(5): 1937-1949. |
| [43] |
Washio K. Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains[J]. Biochimica et Biophysica Acta(BBA)-Gene Structure and Expression, 2001, 1520(1): 54-62. DOI:10.1016/S0167-4781(01)00251-2 |
| [44] |
Zhang Y. Functional analysis of Dof transcription factors controlling heading date and PPDK gene expression in rice[M]. Institute of Biology(IBL), Faculty of Science, Leiden University, 2015.
|
| [45] |
Mena M, Vicentecarbajosa J, Schmidt RJ, et al. An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm[J]. Plant J, 2010, 16(1): 53-62. |
| [46] |
Dong G, Ni Z, Yao Y, et al. Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development[J]. Plant Mol Biol, 2007, 63(1): 73-84. |
| [47] |
Marzábal P, Gas E, Fontanet P, et al. The maize Dof protein PBF activates transcription of gamma-zein during maize seed development[J]. Plant Mol Biol, 2008, 67(5): 441-454. DOI:10.1007/s11103-008-9325-5 |
| [48] |
Kawakatsu T, Takaiwa F. Differences in transcriptional regulatory mechanisms functioning for free lysine content and seed storage protein accumulation in rice grain[J]. Plant & Cell Physiology, 2010, 51(12): 1964-1974. |
| [49] |
Skirycz A, Jozefczuk S, Stobiecki M, et al. Transcription factor AtDOF4; 2 affects phenylpropanoid metabolism in Arabidopsis thaliana[J]. New Phytologist, 2010, 175(3): 425-438. |
| [50] |
Kim HS, Kim SJ, Abbasi N, et al. The DOF transcription factor Dof5. 1 influences leaf axial patterning by promoting revoluta, transcription in Arabidopsis[J]. Plant J, 2010, 64(3): 524-535. DOI:10.1111/tpj.2010.64.issue-3 |
| [51] |
Konishi M, Yanagisawa S. Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana[J]. Plant Physiology and Biochemistry, 2007, 45(8): 623-629. DOI:10.1016/j.plaphy.2007.05.001 |
| [52] |
Chen X, Wang D, Liu C, et al. Maize transcription factor Zmdof1 involves in the regulation of Zm401 gene[J]. Plant Growth Regulation, 2012, 66(3): 271-284. DOI:10.1007/s10725-011-9651-5 |
| [53] |
De Paolis A, Sabatini S, De Pascalis L, et al. A rolB regulatory factor belongs to a new class of single zinc finger plant proteins[J]. The Plant Journal, 1996, 10(2): 215-223. DOI:10.1046/j.1365-313X.1996.10020215.x |
| [54] |
Baumann K, De Paolis A, Costantino P, et al. The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants[J]. The Plant Cell, 1999, 11(3): 323-333. DOI:10.1105/tpc.11.3.323 |
| [55] |
Yu J, Shi G, Yu D. Constitutive overexpression of GmDof17-1, a putative DOF transcription factor from soybean causing growth inhibition in tobacco[J]. Scientia Agricola, 2014, 71(1): 44-51. DOI:10.1590/S0103-90162014000100006 |
| [56] |
Fatima T, Snyder CL, Schroeder WR, et al. Fatty acid composition of developing sea buckthorn(Hippophae rhamnoides L.)berry and the transcriptome of the mature seed[J]. PLoS One, 2012, 7(4): e34099. DOI:10.1371/journal.pone.0034099 |
| [57] |
Negi J, Moriwaki K, Konishi M, et al. A Dof transcription factor, SCAP1, is essential for the development of functional stomata in Arabidopsis[J]. Current Biology, 2013, 23(6): 479-484. DOI:10.1016/j.cub.2013.02.001 |
| [58] |
Skirycz A, Radziejwoski A, Busch W, et al. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana[J]. The Plant Journal, 2008, 56(5): 779-792. DOI:10.1111/tpj.2008.56.issue-5 |
| [59] |
Song YH, Smith RW, To BJ, et al. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering[J]. Science, 2012, 336(6084): 1045-1049. DOI:10.1126/science.1219644 |
| [60] |
Corrales AR, Nebauer SG, Carrillo L, et al. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses[J]. Journal of Experimental Botany, 2014, 65(4): 995-1012. DOI:10.1093/jxb/ert451 |
| [61] |
Ahmad M, Rim Y, Chen H, et al. Functional characterization of Arabidopsis Dof transcription factor AtDof4. 1[J]. Russian Journal of Plant Physiology, 2013, 60(1): 116-123. DOI:10.1134/S1021443712060027 |
| [62] |
Wei PC, Feng T, Gao XQ, et al. Overexpression of AtDOF4. 7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis[J]. Plant Physiol, 2010, 153(3): 1031-1045. DOI:10.1104/pp.110.153247 |
| [63] |
Li D, Yang C, Li X, et al. Functional characterization of rice OsDof12[J]. Planta, 2009, 229(6): 1159-1169. DOI:10.1007/s00425-009-0893-7 |
| [64] |
Yanagisawa S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize[J]. The Plant Journal, 2000, 21(3): 281-288. DOI:10.1046/j.1365-313x.2000.00685.x |
| [65] |
Cavalar M, Phlippen Y, Kreuzaler F, et al. A drastic reduction in DOF1 transcript levels does not affect C4-specific gene expression in maize[J]. J Plant Physiol, 2007, 164(12): 1665-1674. DOI:10.1016/j.jplph.2006.09.008 |
| [66] |
Wang W, Tong Y, Li M, et al. Transcriptome analysis identifies a zinc finger protein regulating starch degradation in kiwifruit[J]. Plant Physiol, 2018, 178(2): 850-863. DOI:10.1104/pp.18.00427 |
| [67] |
Tanaka M, Takahata Y, Nakayama H, et al. Altered carbohydrate metabolism in the storage roots of sweetpotato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor[J]. Planta, 2009, 230(4): 737-746. DOI:10.1007/s00425-009-0979-2 |
| [68] |
Kurai T, Wakayama M, Abiko T, et al. Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions[J]. Plant Biotechnology Journal, 2011, 9(8): 826-837. DOI:10.1111/j.1467-7652.2011.00592.x |
| [69] |
Santos LA, de Souza SR, Fernandes MS. OsDof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply[J]. Plant Biotechnol Rep, 2012, 6(4): 327-337. DOI:10.1007/s11816-012-0227-2 |
| [70] |
Tian AG, Wang J, Cui P, et al. Characterization of soybean genomic features by analysis of its expressed sequence tags[J]. Theoretical and Applied Genetics, 2004, 108(5): 903-913. DOI:10.1007/s00122-003-1499-2 |
| [71] |
王志坤, Arun Sebastian, 常健敏, 等. 转GmDof11基因高油转基因大豆的鉴定及主要农艺性状调查[J]. 作物杂志, 2014(2): 39-42. |
| [72] |
Wang HW, Zhang B, Hao YJ, et al. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants[J]. Plant J, 2010, 52(4): 716-729. |
| [73] |
Zhang J, Hao Q, Bai L, et al. Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea[J]. Biotechnol Biofuels, 2014, 7(1): 128. |
| [74] |
Deng W, Yan F, Zhang X, et al. Transcriptional profiling of canola developing embryo and identification of the important roles of BnDof5. 6 in embryo development and fatty acids synthesis[J]. Plant Cell Physiol, 2015, 56(8): 1624-1640. DOI:10.1093/pcp/pcv074 |