马铃薯是我国第四大粮食作物和重要的工业原料作物,我国是马铃薯生产的第一大国,已形成四大主产区,2017年全国马铃薯种植面积598万hm2,总产量1.2亿t,占全国粮食产量的20%,占世界马铃薯总产量的1/4左右。马铃薯是C3作物,其CO2补偿点很高,光合效率较低。与甘薯、山药、芋头等块茎类作物相比,马铃薯淀粉含量较低,一般在12%-20%,这可能与其淀粉合成和降解过程有关。淀粉有直链淀粉(Amylose)和支链淀粉(Amylopectin),其中直链淀粉由1 000-6 000全葡萄糖残基通过1,4-α糖苷键相连而成,分子量从几千至百万,微溶于热水;其余为支链淀粉,由连接在1,6-α糖苷键分支点上的25-30个葡萄糖残基以1,4-α糖苷键连接的1-10万个葡萄糖链组成,分子量超过百万,只能在热2008水中膨胀形成稳定的胶体。马铃薯淀粉黏性大、质地细腻、白色有光泽、糊化温度低、成膜性好,特别是直链淀粉与支链淀粉之比远大于玉米、小麦等禾谷类作物的淀粉比例,但吸水性差。一般中晚熟品种淀粉含量更高。马铃薯淀粉合成与降解的基因克隆对马铃薯品种遗传改良和贮藏具有重要意义。本文总结了马铃薯淀粉合成与降解的研究进展,并提出今后研究建议,以期为深入开展马铃薯淀粉改良工程研究提供参考。
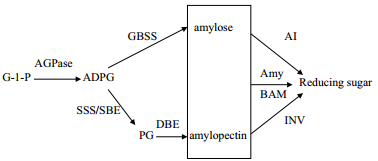
1 马铃薯淀粉合成淀粉有转运淀粉和贮藏淀粉两种存在形式。转运淀粉是马铃薯叶片通过光合作用在叶绿体中合成的临时储备碳源,可以长距离运输到植物的其他部位,而贮藏淀粉多贮藏于淀粉体中,作为块茎发芽时的营养物质。与淀粉合成的相关酶包括腺苷二磷酸葡萄糖焦磷酸化酶(ADP-glucose pyrophosphorylase,AGPase)、淀粉合成酶(Starch synthase,SS)、淀粉分支酶(Starch branching enzy-me,SBE)和淀粉去分支酶(Starch debranching enz-yme,DBE)[1](图 1)。目前,马铃薯中已鉴定的与淀粉合成相关基因,见表 1。
|
| 图 1 马铃薯淀粉合成与降解途径 ADPG:ADP-glucose; AGPase:ADP-glucose pyrophosphorylase; Amy:α-amylase; AI :amylase inhibitor; BAM : β-amylase; DBE :debranching enzyme; G-1-P : α-glucose 1-P; GBSS; INV :invertase; PG :phytoglycogen; SBE :starch branching enzyme; SSS :soluble starch synthase |
AGPase(EC 2.7.7.27)是由2个大亚基和2个小亚基组成的异源四聚体,其作用主要是催化ATP与葡萄糖-1-磷酸(G-1-P)生成ADP-葡萄糖(ADPG)并释放出焦磷酸[2]。作为淀粉合成的限速酶,AGPase活性与小亚基有关,大亚基主要起调节作用,受昼夜周期和Pi调控[3]。马铃薯块茎淀粉含量(Starch content,SC)和AGPase活力(AGPase activity,AA)间呈显著正相关(SC=2.47AA+5.45,R2=0.651)[4]。低温贮藏条件下,AGPase活性与还原糖含量负相关[5]。
1.2 淀粉合成酶马铃薯淀粉合成酶(Starch synthase,SS)(EC 2.4.1.21)包括颗粒结合淀粉合成酶(Granule-bound starch synthase,GBSS)和可溶性淀粉合成酶(Soluble starch synthase,SSS),主要通过在ADPG上加葡萄糖催化α-1,4-葡聚糖链的延伸,其活性影响淀粉的直链淀粉/支链淀粉比例、淀粉链长及结构等品质[6]。马铃薯块茎直链淀粉约占天然淀粉的20%-25%,主要与GBSS有关,GBSS主要存在于淀粉颗粒中,包括GBSSI和GBSSII。从马铃薯栽培种“东农303”中克隆到5 428 bp的GBSS(包括5'和3'侧翼区),并发现5'侧翼区存在较多的茎环结构[7]。通过PCR技术从马铃薯品种“大西洋”中克隆了马铃薯GBSS基因5'侧翼序列并证实其具有启动基因表达功能[8]。从马铃薯栽培种“甘农薯2号”中克隆到1 824 bp的GBSSI基因,虽然与其他植物同源性较低,但具有淀粉合成功能。宋东光等[9]在研究马铃薯GBSS基因5'侧翼区调控作用时,发现1.6 kb、2.9 kb GBSS-GUS的表达最高,蔗糖可诱导GBSS-GUS的表达,而光抑制其表达。SSS存在于造粉体的可溶性基质中,可分为SSI、SSII和SSIII三种,但马铃薯块茎中只有SSII和SSIII两种,其中SSIII占80%的酶活性[10]。用RT-PCR方法从“陇薯3号”中克隆了3 967 bp的SSIII基因并进行生物信息学分析[11]。
1.3 淀粉分支酶淀粉分支酶(Starch branching enzyme,SBE)(EC 2.4.1.18)可分为SBEI、SBEIIa和SBEIIb三种,主要在ADPG线性链间引入α-1,6-糖苷键形成分支结构。SBEI和SBEIIb主要存在于胚乳中,SBEIIa存在于叶片中。
1.4 淀粉去分支酶淀粉去分支酶(Starch debranching enzyme,DBE)有异淀粉酶(Isoamylase,ISA)和限制性糊精酶(Pullulanase,RE)两种,主要对分支链进行修饰并合成具有一定结构特性的淀粉结晶体[12]。SBE和DBE之间的活性平衡决定支链淀粉的最终分支程度,当SBE活性增强时,该平衡向PG方向移动;当DBE活性增强时,该平衡向支链淀粉方向移动[13]。
2 马铃薯淀粉降解淀粉可以在糖酶(淀粉酶、转化酶、乳糖酶、纤维素酶和果胶酶等)作用下水解为红糊精、无色糊精、麦芽糖和葡萄糖。马铃薯常低温贮藏,容易发生糖化,导致还原糖积累。为了减少低温糖化带来的损失,除了要增强碳水化合物从己糖到淀粉的流动,还要阻止蔗糖和淀粉水解成还原糖。淀粉降解可以通过淀粉磷酸化途径和淀粉水解途径(图 1),其过程包括可溶性葡聚糖的释放、可溶性和线性葡聚糖的代谢和麦芽糖代谢3个阶段。葡聚糖水双激酶(Glucan water dikinase,GWD)和磷酸葡聚糖水双激酶(Phosphoglucan water dikinase,PWD)可以磷酸化葡聚糖,降解淀粉[24]。淀粉磷酸酶(EC 2.4.1.1)也参与淀粉磷酸化途径。水解途径主要涉及α-amylase(Amy,EC 3.2.1.1)和β-amylase(BAM,EC 3.2.1.2)[25]。
2.1 淀粉酶淀粉酶有两种:α-淀粉酶和β-淀粉酶。α-淀粉酶有3个亚家族,分别位于胚乳、细胞质和叶绿体中,可以特异切断α-1,4-糖苷键,生成寡糖。拟南芥有3个α-淀粉酶,只有拥有叶绿体转运肽的AtAMY3具有α-淀粉酶活性[26]。β-淀粉酶可以从多聚糖非还原性末端切断α-1,4-糖苷键,生成麦芽糖。拟南芥有9个β-淀粉酶,不同淀粉酶亚型的亚细胞定位和功能也不一样[27]。利用基因芯片从马铃薯块茎中筛选到低温响应的StBMY7和PCT-BMY1[28]。马铃薯α-淀粉酶活性主要由StAmy23引起,β-淀粉酶活性主要由StBAM1和StBAM7引起,冷藏块茎中的淀粉酶抑制剂可以调节淀粉酶的活性[29]。
2.2 淀粉磷酸激酶GWD是催化ATP上的磷酸盐向支链淀粉葡萄糖基C3或C6位转移。PWD可对C3位置的葡萄糖基单元进行磷酸化[30]。马铃薯淀粉磷酸化约35%发生在葡萄糖基C3位置上,65%是在C6位置上。GWD和PWD对淀粉粒的半结晶结构有不同的影响。
2.3 转化酶转化酶(b-fructofuranosidaes,EC 3.2.1.26)可以将蔗糖不可逆水解成葡萄糖和果糖,根据最适pH可分为酸性转化酶(Acid invertase,AI)和中/碱性转化酶(Neutral/alkaline invertase,NI),AI存在于细胞壁和液泡,NI位于细胞质、线粒体和质体。马铃薯有6个酸性转化酶基因,4个定位于细胞壁,2个定位于液泡,其中StvacINV1表达受低温诱导,是造成马铃薯块茎低温糖化的主要原因[31]。
3 马铃薯淀粉特性改进为了有效改良马铃薯淀粉品质,可以深入研究马铃薯淀粉合成酶基因功能并采用基因工程技术来改进淀粉特性(表 2)。在增加淀粉含量方面,转glgC16(大肠杆菌AGPase基因)表达提高了马铃薯AGPase酶活性,增加了块茎淀粉含量[32]。过表达ATP/ADP转运子提高马铃薯块茎淀粉,而RNA干扰ATP/ADP转运子则降低淀粉含量[33]。超量表达sAGP(AGPase小亚基基因)提高了马铃薯AGPase酶活性,增加块茎淀粉,降低的还原糖含量,能有效改善马铃薯的加工品质[34]。转AGPase基因改变了马铃薯淀粉在不同溶剂和不同盐溶液中的特性黏度[35]。与对照相比,转AGPase马铃薯株系的AGPase活力、叶片光合速率和块茎淀粉含量显著提高,说明AGPase可反馈调控上游光合速率[4]。降低GWD的表达可以推迟冷藏转基因马铃薯块茎的还原糖积累[36]。通过RNA干扰马铃薯Amy23和BAM1表达可以降低还原糖含量[37]。将反义GBSSI和双拷贝glgC融合基因导入野生型马铃薯可以显著增加淀粉含量[38]。反义AGPase基因抑制AGPase酶活性,马铃薯块茎中只有糖而无淀粉[39]。把AGPase反义基因转入马铃薯中,降低了AGPase酶活性和淀粉产量[40]。反义RNA技术干涉马铃薯BMY1表达降低转基因株系叶片淀粉含量[41]。
在淀粉改性方面,降低马铃薯ISA1/ISA2表达可以增加块芭中可溶性葡聚糖含量[42]。同时抑制ISA1/ISA2/ISA3表达也会升高块茎葡聚糖含量[43]。用反义RNA技术抑制GBSSI基因表达可以使马铃薯丧失直链淀粉合成能力[44]。抑制GBSSI基因表达,可以大幅降低直链淀粉含量,从而增加支链淀粉[45]。AGPase基因不仅影响淀粉含量也改变淀粉的组成[46]。反义技术可以增加SSSIII基因表达[47]。反义抑制SSSIII基因表达,降低支链淀粉含量,改变淀粉结构和淀粉粒形态[48]。SBE也影响支链淀粉的合成、分子结构和淀粉粒形态[49]。
在改变淀粉组分方面,反义抑制SSIII表达可以改变马铃薯淀粉颗粒形态和磷酸含量[19]。通过抑制3种马铃薯淀粉合成酶基因可获得短支链淀粉[22]。反义抑制StGBSSI表达可以影响四倍体马铃薯淀粉组成[50]。构建GBSSI基因的ihpRNAi载体并转入马铃薯可以显著提高支链淀粉含量[51]。杜宏辉等[52]以马铃薯栽培种“克新1号”和“克新4号”为材料,通过农杆菌介导法获得了转SSIII基因的马铃薯植株,为马铃薯淀粉品质改良奠定基础。RNAi技术抑制马铃薯Sbe1和Sbe2a基因可获得直链淀粉含量高的马铃薯植株并提高冻融稳定性[53]。超量表达GWD对马铃薯淀粉粒形态结构、直链淀粉含量、糊化作用等有显著影响[54]。
4 展望马铃薯块茎淀粉包括直链淀粉和支链淀粉,两者都含有葡萄糖但大小形态不同[55]。淀粉合成与降解过程决定最终的淀粉含量,马铃薯淀粉合成与降解基因克隆对马铃薯品种遗传改良具有重要意义,目前部分克隆的马铃薯淀粉合成与降解基因已在增加马铃薯淀粉含量、淀粉改性和改变淀粉组分方面得到广泛应用。为了提高马铃薯块茎淀粉含量和培育特色马铃薯品种,今后要着力于做好以下几项工作:一是要加快铃薯淀粉合成与降解相关基因的克隆与功能鉴定,分析淀粉代谢关键基因启动子序列调控元件,阐明关键基因功能和酶活性的调节机制,弄清楚控制马铃薯淀粉结构的关键因素,为深入开展马铃薯淀粉改良工程研究提供参考。二是要加强马铃薯转基因技术、RNAi技术和基因组编辑技术研究,特别是启动子的选择,选育出抗病、抗逆、高产、优质的马铃薯新品种,使之更好地服务生产。马铃薯块茎特异性启动子主要有patatin基因启动子和GBSS基因启动子[56],选择适合的启动子可以更有效地调控马铃薯块茎中淀粉合成酶基因的表达。利用CRISPR技术获得Inv基因敲除等位基因的马铃薯品系,其冷藏块茎中还原糖含量和丙烯酰胺含量显著低于野生型[57]。三是根据基因功能和定位,发展一批可用于改良马铃薯的基因标记或候选基因。使用关联作图方法发现了GWD、SBEI、SSIII和SBEII的等位基因变异,可以作为与马铃薯淀粉磷酸化程度相关的遗传标记[58]。四是深入研究农艺措施对马铃薯块茎淀粉含量的影响,阐明其作用机理。增施氮素和钾肥可以提高马铃薯块茎的AGPase和淀粉合成酶的活性,相反,遮阴处理显著降低淀粉合成关键酶的活性。
| [1] |
Fernie AR, Willmitzer L, Trethewey RN. Sucrose to starch:a transition in molecular plant physiology[J]. Trends Plant Sci, 2002, 7(1): 35-41. DOI:10.1016/S1360-1385(01)02183-5 |
| [2] |
Sweetlove LJ, Müller-Röeber B, Willmitzer L, Hill SA. The contribution of adenosine 5'-diphosphoglucose pyrophosphorylase to the control of starch synthesis in potato tubers[J]. Planta, 1999, 209: 330-337. DOI:10.1007/s004250050640 |
| [3] |
Stitt M. Progress in understanding and engineering primary plant metabolism[J]. Curr Opin Biotechnol, 2013, 24(2): 229-238. DOI:10.1016/j.copbio.2012.11.002 |
| [4] |
白桦, 崔雪琼, 白少星, 姚新灵. 马铃薯AGPase活力反馈调控光合速率定量分析[J]. 生物技术通报, 2014(11): 125-129. |
| [5] |
成善汉, 苏振洪, 谢从华, 柳俊. 淀粉-糖代谢酶活性变化对马铃薯块茎还原糖积累及加工品质的影响[J]. 中国农业科学, 2004, 37(12): 1904-1910. DOI:10.3321/j.issn:0578-1752.2004.12.019 |
| [6] |
陈国梁, 张金文, 王蒂. 马铃薯gbss、ssÒ和ssÓ基因片段的融合及其RNAi载体的构建[J]. 中国生物工程杂志, 2008, 28(8): 51-56. |
| [7] |
戴卫列, 邓炜, 崔伟英, 等. 马铃薯GBSS基因的克隆与DNA顺序分析[J]. 植物学报, 1996, 38(10): 777-784. |
| [8] |
李淑洁, 张金文, 王煜, 等. 一个新的马铃薯GBSS基因5'侧翼序列克隆及调控活性研究[J]. 中国马铃薯, 2005, 19(3): 129-133. DOI:10.3969/j.issn.1672-3635.2005.03.001 |
| [9] |
宋东光, 孙国枫, 单海燕, 等. 马铃薯GBSS基因5'侧翼区调控作用的研究[J]. 植物学报, 1998, 40(9): 796-802. DOI:10.3321/j.issn:1672-9072.1998.09.004 |
| [10] |
Edwards A, Fulton DC, Hylton CM, et al. A combined reduction in activity of starch synthases Ⅱ and Ⅲ of potato has novel effects on the starch of tubers[J]. Plant J, 1999, 17(3): 251-261. DOI:10.1046/j.1365-313X.1999.00371.x |
| [11] |
杨涛, 张宁, 栗亮, 等. 马铃薯可溶性淀粉合成酶SSⅢ基因克隆及生物信息学分析[J]. 分子植物育种, 2009, 7(3): 545-549. |
| [12] |
Kristensen M, Lok F, Planchot V, et al. Isolation and characterisation of the gene encoding the starch debranching enzyme limit dextrinase from germinating barley[J]. Biochim Biophys Acta, 1999, 1431: 538-546. DOI:10.1016/S0167-4838(99)00077-1 |
| [13] |
Kim KN, Fisher DK, Gao M, Guiltinan MJ. Genome organization and promoter activity of the maize starch branching enzyme I gene[J]. Gene, 1998, 216: 233-243. DOI:10.1016/S0378-1119(98)00339-4 |
| [14] |
Bae JM, Liu JR. Molecular cloning and characterization of two novel iosoforms of the small subunit of ADPglucose pyrophosphorylase from sweet potato[J]. Mol Gen Genet, 1997(2): 179-185. |
| [15] |
Harn CH, Bae JM, Lee SS, et al. Presence of multiple cDNAs encoding an isoform of ADP-glucose pyrophosphorylase large subunit from sweet potato and characterization of expression levels[J]. Plant Cell Physiol, 2000, 41(11): 1235-1242. DOI:10.1093/pcp/pcd049 |
| [16] |
Dry I, Smith A, Edward A, et al. Characterization of cDNAs encoding two isoforms of granule-bound starch synthase which show differential expression in developing storage organs of pea and potato[J]. Plant J, 1992, 2(2): 193-202. |
| [17] |
Kossmann J, Abel GJW, Springer F, Lloyd JR, Willmitzer L. Cloning and functional analysis of a cDNA encoding a starch synthase from potato(Solanum tuberosum L.)that is predominantly expressed in leaf tissue[J]. Planta, 1999, 208: 503-511. DOI:10.1007/s004250050587 |
| [18] |
Larsson CT, Khoshnoodi J, Ek B, et al. Molecular cloning and characterization of starch-branching enzyme Ⅱ from potato[J]. Plant Mol Biol, 1998, 37(3): 505-511. DOI:10.1023/A:1005908305456 |
| [19] |
Abel GJW, Springer F, Willmitzer L, Kossmann J. Cloning and fuctional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato(Solanum tuberosum L.)[J]. Plant J, 1996, 10(6): 981-991. DOI:10.1046/j.1365-313X.1996.10060981.x |
| [20] |
Marshall J, Sidebottom C, Debet M, et al. Identification of the major starch synthase in the soluble fraction of potato tubers[J]. Plant Cell, 1996, 8(7): 1121-1135. DOI:10.1105/tpc.8.7.1121 |
| [21] |
Yang T, Zhang N, Li L, et al. Cloning and bioinformatics analysis of soluble starch synthase SSⅢ gene in potato[J]. Mol Plant Breed, 2009, 7: 545-549. |
| [22] |
Jobling SA, Westcott RJ, Tayal A, Jeffcoat R, Schwall GP. Production of a freeze-thaw-stable potato starch by antisense inhibition of three starch synthase genes[J]. Nature Biotechnol, 2002, 20(3): 295-299. DOI:10.1038/nbt0302-295 |
| [23] |
Lee SS, Bae JM, Oh MS, et al. Isolation and characterization of polymorphic cDNAs parially encoding ADP-glucose pyrophosphorylase(AGPase)large subunit from sweet potato[J]. Mol Cells, 2000, 10(1): 108-112. DOI:10.1007/s10059-000-0108-3 |
| [24] |
Silver DM, Kotting O, Moorhead GBG. Phosphoglucan phosphatase function sheds light on starch degradation[J]. Trends Plant Sci, 2014, 19(7): 471-478. DOI:10.1016/j.tplants.2014.01.008 |
| [25] |
Preiss J. Regulation of the biosynthesis and degradation of starch[J]. Annu Rev Plant Physiol, 1982, 33: 431-454. DOI:10.1146/annurev.pp.33.060182.002243 |
| [26] |
Streb S, Zeeman SC. Starch metabolism in Arabidopis[J]. The Arabidopsis Book, 2012, e0160. |
| [27] |
Monroe JD, Storm AR, Badley EM, et al. β-amylase 1 and β-amylase 3 are plastidic starch hydrolases in Arabidopsis that seem to be adapted for different thermal, pH, and stress conditions[J]. Plant Physiol, 2014, 166: 1748-1763. DOI:10.1104/pp.114.246421 |
| [28] |
Bagnaresi P, Moschella A, Beretta O, et al. Heterologous microarray experiments allow the identification of the early events associated with potato tuber cold sweetening[J]. BMC Genomics, 2008, 9: 176-198. DOI:10.1186/1471-2164-9-176 |
| [29] |
Zhang H, Hou J, Liu J, et al. Amylase anlaysis in potato starch degradation during cold storage and sprouting[J]. Potato Res, 2014, 57: 47-58. DOI:10.1007/s11540-014-9252-6 |
| [30] |
Ritte G, Heydenreich M, Mahlow S, et al. Phosphorylation of C6-and C3-position of glycosyl residues in starch is catalysed by distinct dikinases[J]. FEBS Lett, 2006, 580: 4872-4876. DOI:10.1016/j.febslet.2006.07.085 |
| [31] |
Liu X, Zhang C, Ou YB, et al. Systematic analysis of potato acid invertase genes reveals that a cold-responsive member, StvacINV1, regulates cold-induced sweetening of tubers[J]. Mol Genet Genomics, 2011, 286: 109-118. DOI:10.1007/s00438-011-0632-1 |
| [32] |
Sweetlove LJ, Burrell MM, Rees T. Characterization of transgenic potato(Solanum tuberosum)tubers with increased ADP-Glucose pyrophosphorylase[J]. Biochemistry J, 1996, 320: 478-492. |
| [33] |
Tjaden J, Mohlmann T, Kampfenkel K. Altered plastidic ATP/ADP-transporter activity influence potato(Solanum tuberosum L.)tuber morphology, yield and composition of tuber starch[J]. Plant J, 1998, 16: 531-540. DOI:10.1046/j.1365-313x.1998.00317.x |
| [34] |
宋波涛, 谢从华, 柳俊. 马铃薯sAGP基因表达对块茎淀粉和还原糖含量的影响[J]. 中国农业科学, 2005, 38(7): 1439-1446. DOI:10.3321/j.issn:0578-1752.2005.07.025 |
| [35] |
刘廷国, 李斌, 谢笔钧. 转AGPase基因马铃薯淀粉溶液行为及热特性比较研究[J]. 作物学报, 2006, 32(2): 310-312. DOI:10.3321/j.issn:0496-3490.2006.02.028 |
| [36] |
Lorbeth R, Ritte G, Willmitzer L, et al. Inhibition of a starch-gran-ule-bound protein leads to modified starch and repression of cold sweetening[J]. Nat Biotechnol, 1998, 16: 473-477. DOI:10.1038/nbt0598-473 |
| [37] |
Zhang H, Liu J, Hou J, et al. The potato amylase inhibitor gene SbAI regulates cold-induced sweetening in potato tubers by modulating amylase activity[J]. Plant Biotechnol J, 2014, 12: 984-993. DOI:10.1111/pbi.2014.12.issue-7 |
| [38] |
崔雪琼. 重组glgC基因调控块茎淀粉生物合成研究[M]. 银川: 宁夏大学, 2012.
|
| [39] |
Vardy KA, Emes MJ, Burrell MM. Starch synthesis in potato tubers transformed with the wheat genes for ADPglucose pyrophosphorylase[J]. Functional Plant Biol, 2002, 29(8): 975-985. DOI:10.1071/PP01161 |
| [40] |
Muller-Rober B, Sonnewald U, Willmitzer L. Inhibition of AGPase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber-storage protein genes[J]. EMBO J, 1992, 11: 1229-1238. DOI:10.1002/embj.1992.11.issue-4 |
| [41] |
Scheidig A, Frohlich A, Schulze S, et al. Downregulation of a chloroplast-targeted β-amylase leads to a starch-excess phenotype in leaves[J]. Plant J, 2002, 30: 581-591. DOI:10.1046/j.1365-313X.2002.01317.x |
| [42] |
Bustos R, Fahy B, Hylton CM, et al. Starch granule initiation is controlled by a heteromultimeric isoamylase in potato tubers[J]. Proc Natl Acad Sci USA, 2004, 101: 2215-2220. DOI:10.1073/pnas.0305920101 |
| [43] |
Ferreira SJ.Transcriptome based analysis of starch metabolism in Solanum tuberosum[D]. Friedrich-Alexander-Universitat Erlangen-Nurnberg, 2011.
|
| [44] |
Holen T, Amarzguioui M, Wiiger MT, et al. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor[J]. Nucleic Acids Res, 2002, 30(8): 1757-1766. DOI:10.1093/nar/30.8.1757 |
| [45] |
Heilersig HJB, Loonen A, Bergervoet M, et al. Post-transcriptional gene silencing of GBSSI in potato:effects of size and sequence of the inverted repeats[J]. Plant Mol Biol, 2006, 60: 647-662. DOI:10.1007/s11103-005-5280-6 |
| [46] |
Baba T. Identification, cDNA cloning, and gene expression of soluable starch synthase in rice(Oryza sativa L.)immature seeds[J]. Plant Physiol, 1993, 103: 565-573. DOI:10.1104/pp.103.2.565 |
| [47] |
杜宏辉.可溶性淀粉合成酶SSⅢ基因对马铃薯的遗传转化[D].兰州: 甘肃农业大学, 2011: 7-10.
|
| [48] |
Abel GJW, Springer F, Willmitzer L, et al. Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato(Solanum tuberosum L.)[J]. Plant J, 1996, 10(6): 981-991. DOI:10.1046/j.1365-313X.1996.10060981.x |
| [49] |
Blauth SL, Yao Y, Klucinec JD, et al. Identification of mutator insertional mutants of starch-branching enzymes in corn[J]. Plant Physiol, 2001, 125(3): 1396-1405. DOI:10.1104/pp.125.3.1396 |
| [50] |
Wolters AMA, Janssen EM, Rozeboom-Schippers MGM, et al. Composition of endogenous alleles can influence the level of antisense inhibition of granule-bound starch synthase gene expression in tetraploid potato plants[J]. Mol Breeding, 1998, 4(4): 343-358. DOI:10.1023/A:1009697725778 |
| [51] |
蔺琰东.马铃薯GBSSI基因的ihpRNAi载体转化试管薯片及转基因块茎相关生理分析[D].兰州: 甘肃农业大学, 2011.
|
| [52] |
杜宏辉, 文义凯, 张宁, 等. 可溶性淀粉合成酶SSⅢ基因对马铃薯的遗传转化[J]. 基因组学与应用生物学, 2011, 30(3): 303-307. DOI:10.3969/gab.030.000303 |
| [53] |
Andersson M, Melander M, Pojmark P, et al. Targeted gene suppression by RNA interference:An efficient method for production of highamylose potato lines[J]. J Biotechnol, 2006, 123(2): 137-148. DOI:10.1016/j.jbiotec.2005.11.001 |
| [54] |
Xu X, Dees D, Dechesne A, et al. Starch phosphorylation plays an important role in starch biosynthesis[J]. Carbohyd Polym, 2017, 157: 1628-1637. DOI:10.1016/j.carbpol.2016.11.043 |
| [55] |
Zhang H, Hou J, Liu J, et al. The roles of starch metabolic pathways in the cold-induced sweetening process in potatoes[J]. Starch-Starke, 2016. DOI:10.1002/star.201600194 |
| [56] |
Liu XY, Rocha-Sosa M, Hummel S, et al. A detailed study of the regulation and evolution of the two classes of patatin genes in Solanum tuberosum L[J]. Plant Mol Biol, 1991, 17(6): 1139-1154. DOI:10.1007/BF00028731 |
| [57] |
Clasen BM, Stoddard TJ, Luo S, et al. Improving cold storage and processing traits in potato through targeted gene knockout[J]. Plant Biotechnol J, 2016, 14: 169-176. DOI:10.1111/pbi.12370 |
| [58] |
Carpenter MA, Joyce NI, Genet RA, et al. Starch phosphorylation in potato tubers is influenced by allelic variation in the genes encoding glucan water dikinase, starch branching enzymes Ⅰ and Ⅱ, and starch synthase Ⅲ[J]. Front Plant Sci, 2015, 6: 143. |





