2. 广西壮族自治区农业科学院园艺研究所,南宁 530007;
3. 亚热带农业生物资源保护与利用国家重点实验室,南宁 530004
2. Institute of Horticulture, Guangxi Academy of Agricultural Science, Nanning 530007;
3. State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, Nanning 530004
转录因子是一种具有特殊结构并且能行使调控基因表达功能的蛋白质分子。植物的转录因子有2种,一种是非特异性转录因子,它们可以非选择性地调控基因的转录表达,如大麦中的HvCBF2(C-repeat/DRE binding factor 2)[1];另一种为特异性转录因子,它们能够选择性调控某种或某些基因的转录表达,如WRKY、bHLH、bZIP、MYB、NAC、HMG、HSF、zinc-finger蛋白、AP2/ERF(乙烯响应因子)等,它们在调控植物的特异性方面发挥着重要而独特的作用[2-3]。其中WKRY基因家族是高等植物中最大的转录因子家族之一,在整个植物谱系中均有发现。Ishiguro等[4]从甘薯中克隆出世界上第一个WRKY基因SPF1(Swet potato factor 1),随后在多种植物中成功分离鉴定到WRKY转录因子。WRKY基因家族在进化过程中,从绿藻中的一个或几个基因,到最早陆生苔藓中的30多个基因,再到高等植物中的100多个基因,显示了其基因扩张的过程(表 1)。
Rushton等[5]从欧芹中鉴定出WRKY1、WRKY2和WRKY3,将其命名为WRKY(发音为“worky”),并首次证明了WRKY蛋白在调节植物对病原体的反应中起着重要的作用。同时在调控蔗糖(SPF1)或萌发过程(ABF1和ABF2)基因表达方面也发挥着潜在的作用[4-5]。自2000年有学者发表了关于WRKY转录因子文章以来,在过去的十几年里,科学家们对WRKY蛋白的研究取得了重大进展。以往的研究综述主要集中在WRKY转录因子在防御反应中的作用,本文将从WRKY转录因子在植物中调控作用、WRKY蛋白功能机制、涉及WRKY转录因子信号传递中自调节和交叉调节的调控网络以及基于WRKY基因组测序的基因进化等方面总结植物WRKY转录因子最新研究进展,以便更全面地了解它们在植物中的作用。
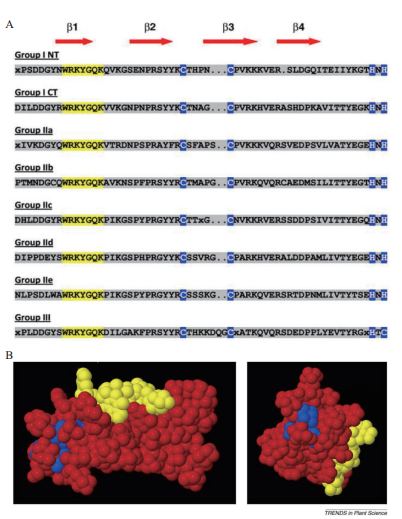
1 WRKY转录因子的结构特征与分类 1.1 WRKY转录因子的结构特征WRKY转录因子具有非常显著的结构特点,其蛋白结构基本含有1-2个WRKY结构域(图 1),为DNA结合域(DBD),约由60个高度保守的氨基酸残基组成,包括位于N端的七肽和位于C端的锌指结构。位于N末端的七肽WRKYGQK绝对保守,是核心序列,位于C端的序列由C2H2(C-X4-5-C-X22-23-H-X-H)或C2HC(C-X7-C-X23-H-X-C)型锌指结构组成[23-25]。WRKY转录因子可通过WRKY结构域与靶基因启动子区的顺式作用元件TTGAC(C/T)核苷酸序列(W-box)特异性结合,以此激活或抑制转录,进而调控下游基因的表达[15, 26]。Yamasaki等[27]报道了拟南芥WRKY4结构域由一个四链β片层组成,在β片层的C端由保守的半胱氨酸/组氨酸(Cys/His)残基形成一个锌结合袋,WRKYGQK残基对应于序列的N端链,在序列的中间被Gly残基扭折,使得涉及Trp(色氨酸)残基的广泛疏水作用,进而促使β链的结构具有稳定性。β链的WRKYGQK基序可以接触一个大约6bp区域,这在很大程度上与W-box(TTGACY)的长度是一致的。表明WRKYGQK的基序可以与靶基因启动子区的W-box结构特异性结合。
WRKY转录因子的结构域在不同植物中存在多种变异。水稻WRKY家族成员有19个WRKY结构域的变体,其中WRKYGEK和WRKYGKK是7个域和5个域共有的常见突变体[28]。在中间锦鸡儿转录组数据鉴定的53个CiWRKY基因中,CiWRKY蛋白既含有高度保守的WRKYGQK基序,同时又包含2个变异的WRKYGKK和WKKYEEK基序[29]。有研究表明WRKYGQK序列突变可显著降低WRKY转录因子与DNA的结合活性[25]。烟草NtWRKY转录因子的结构域中,C端结构域活性较强,而N端WRKY结构域与W-box的结合活性较弱,其C2H2型锌指状基序中保守的半胱氨酸和组氨酸残基被取代,也能使WRKY转录因子与DNA结合遭到破坏[25]。
除WRKY结构域之外,WRKY转录因子还包含其他结构域,包括TIR-NBS-LRR(Toll/interleukin-1 receptor-nucleotide binding site-leucine-rich repeat)、激酶结构域、脯氨酸富集区、谷氨酰胺富集区、丝氨酸-苏氨酸富集区、亮氨酸拉链、核定位结构域等[30]。拟南芥AtWRKY7同时含有一个WRKY结构域和一个钙调蛋白CaM结合结构域[31]。WRKY转录因子多样的结构域表明,拥有特殊结构的WRKY转录因子可以在基因表达调控中发挥重要的特殊功能作用。
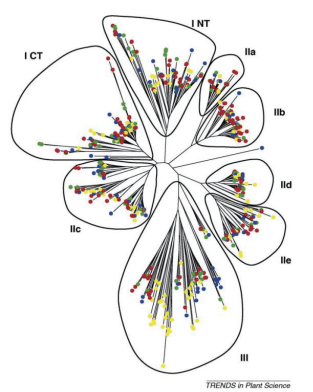
1.2 WRKY转录因子的分类从植物中获得完整的WRKY基因家族序列之前,Eulgem等[23]把WRKY家族成员分为Group Ⅰ、GroupⅡ和GroupⅢ 3个亚家族。Group Ⅰ含有2个WRKY保守结构域,其C端的锌指结构为C2H2型;Group Ⅱ只含有1个WRKY保守结构域,锌指结构与I类相同,为C2H2型;Group Ⅲ锌指结构为C2HC型,包含1个WRKY保守结构域(图 1-A)。但也有少部分WRKY蛋白的结构类型与这3类特征都不匹配,如AtWRKY10,其结构可能是N末端的WRKY保守结构域发生了丢失,只含有1个WRKY保守结构域,与Group Ⅰ的蛋白结构特征相似但又不一致[24];如苹果的WRKY蛋白C末端因缺少完整的类似锌指蛋白结构而被分类为Group Ⅳ[13]。Zhang等[28]认为Eulgem等对拟南芥WRKY家族的分类并不完全基于系统发育分析,为了反映WRKY域的演化过程,通过系统发育分析,WRKY转录因子分为Ⅰ、Ⅱa+Ⅱb、Ⅱc、Ⅱd+Ⅱe和Ⅲ,而Ⅱ类并不单独分为一类。随后,Tamura等[34]通过纯系统发育数据分析也证实了这一点(图 2)。WRKY转录因子的进化分析对理解植物生物多样性的整体机制,以及WRKY基因在植物调控网络中发挥的特殊功能具有重要意义。
WRKY转录因子家族成员众多,在植物不同发育时期和多种环境因素诱导下,激活或抑制目的基因表达特定的目的蛋白,发挥着各种非常重要的生物学功能,主要涉及对植物的生长发育和衰老调控、非生物和生物胁迫应答等过程。
2.1 生长发育调控植物基因组基因的有序表达是植物生长发育的基础。大量研究表明,WRKY转录因子在不同的组织中发挥非常重要的功能作用,调控植物的生长发育。许多研究已证实WRKY转录因子在植株生长、根系发育、果实成熟、衰老等代谢过程等多种生理过程中的重要作用。
2.1.1 植株生长许多转录因子具有调节植物发育和矮化的作用(包括WRKY、AP2/EREBP、bHLH、C2H2和ARF)。其中,已知最多的是WRKY家族[35-36]。WRKY转录因子具有诱导植株矮化的作用[37]。OsWRKY11过表达降低了转基因水稻植株的高度,导致植株矮小[38-39]。矮化表型主要是由于细胞分裂和细胞伸长的变化导致的,这些过程受细胞分裂素(CTK)、生长素(IAA)、赤霉素(GA)和油菜素类固醇(BR)的调控[40]。IAA、CTK和GA被广泛认为在植物矮化中具有重要作用[41-42]。另外,BR在植物矮化中也具有重要作用[40]。在拟南芥中,存在BR生物合成基因缺陷的cpd、cbb3、dwf4突变体和BR6ox1、BR6ox2双突变体均表现为矮化表型[43]。Zheng等[44]研究表明,WRKY可以直接调控BR的生物合成。苹果转录因子MdWRKY9是通过直接抑制油菜素类固醇限制合成酶MdDWF4的转录,减少BR的产生,正向调控植株的矮小[44]。以上证明WRKY转录因子在调控植物发育和矮化中发挥着重要的作用。
2.1.2 根系发育据报道WRKY基因在调节植物激素合成中发挥作用[45-46]。一些WRKY基因通过影响植物激素信号或基因表达来影响根系结构[47]。与野生型相比,OsWRKY31的过表达导致侧根更少更短,这可能是通过干扰生长素的响应或转运来实现的[47]。
类似的研究表明,OsWRKY28功能缺失突变体可能是通过降低了JA生物合成基因的表达从而影响根生长[48]。另外,乙烯(ETH)在根系的发育中可诱导根毛和不定根的形成。在小麦试验中,乙烯合成的1-氨基环丙烯-1-羧酸合酶基因在过表达系TaWRKY51-OE中下调,而在基因沉默TaWRKY51-RNAi系中上调,进一步研究发现,TaWRKY51是ETH合成的负调控因子,TaWRKY51通过与启动子区存在的W-box顺式元件结合抑制ETH合成基因ACS的表达,协调小麦乙烯合成和侧根形成[49]。说明WRKY转录因子可调节植物的相应的激素合成,然后通过这些植物激素来调控根系的发育。
2.1.3 果实成熟WRKY转录因子在果实成熟的生理途径中发挥重要的调控作用。通过表达模式分析显示,近60% CaWRKY在辣椒成熟过程中表达[50]。ClWRKY在西瓜果实组织中均较高表达[51]。在鳄梨[52]和草莓[53]中也成功筛选到与成熟过程相关的WRKY转录因子,表明其在果实成熟过程中可能发挥一定的调控作用。其中,FaWRKY转录因子参与了脱落酸(ABA)的信号通路,通过调控ABA的合成来促进草莓果实的成熟[53]。
2.1.4 衰老WRKY转录因子同样能参与叶片衰老的调控。WRKY转录因子是拟南芥衰老转录组中第二大转录因子家族[54]。AtWRKY6在衰老过程中明显上调,通过对AtWRKY6靶基因的分析,确定衰老诱导了受体激酶和受体样激酶(SIRK/FRK1)基因的表达。SIRK/FRK1编码一种受体样蛋白激酶,该蛋白激酶在叶片衰老过程中被强特异性诱导表达[55-56]。在其他物种中,调节衰老的WRKY基因(包括GhWRKY42、CiWRKY40-4和BrWRKY6)相继得到验证。如GhWRKY42的过表达导致导致衰老相关基因表达升高,促进了叶片早衰[57]。CiWRKY40-4过表达至拟南芥延缓了叶片衰老,是衰老的负调控因子[29]。WRKY转录因子也可以通过调控植物激素的合成来调控叶片的衰老。GA可以抑制叶片衰老,而BrWRKY6通过W-box顺式元件与衰老相关基因BrSAG12、BrNYCl、BrSGR1的启动子结合,抑制GA生物合成基因BrKAO2和BrGA20ox2的表达,加速了叶片的衰老[58]。
2.2 参与植物的非生物和生物胁迫调控植物的生存环境复杂多变,经常遭受非生物因素和生物因素的逆境胁迫,影响植物的生长发育,严重会直接造成植物死亡。植物通过基因的表达调控可以抵抗逆境胁迫的侵害,其中WRKY转录因子在植物对逆境胁迫响应过程中的调节具有重要作用。
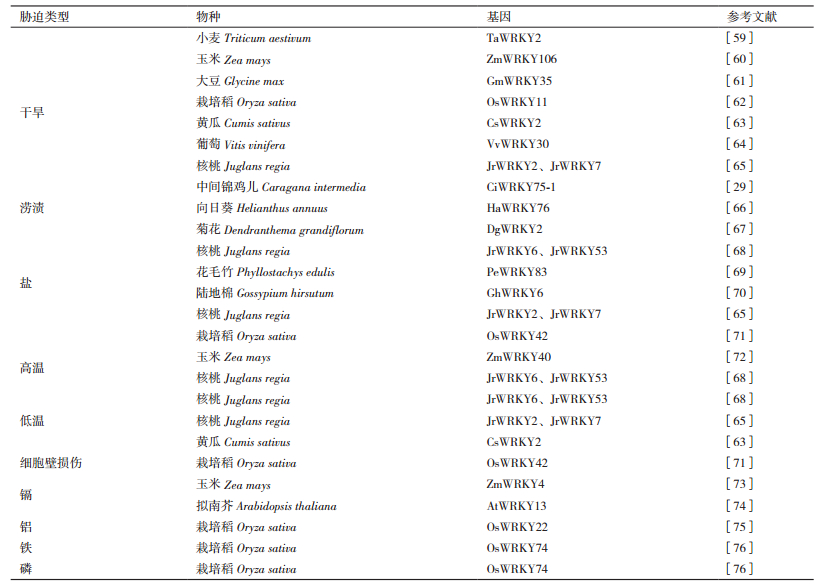
2.2.1 参与植物的非生物调控WRKY是一类锌指型转录因子,主要存在于植物中,它在调节植物的许多非生物应激反应(如干旱、涝渍、高盐、高温、低温、机械损伤、金属元素等)中起关键作用。近年来,发现越来越多的WRKY基因参与非生物逆境应答,不仅有模式植物烟草和拟南芥,还有水稻、小麦、大豆、玉米、棉花、黄瓜、向日葵、菊花、竹子、葡萄、核桃、野生树种等多种植物,且大多数已经通过基因过表达或敲除的方式进一步验证了其在植物非生物胁迫中的调控作用。WRKY转录因子作为干旱、低温等胁迫应答的主要成分,可与下游基因启动子中的顺式作用元件特异性结合,调节一系列依赖该顺式作用元件的抗逆功能基因以特定的强度在特定的时间与空间表达,进而增强植物对干旱、低温及高盐等逆境的抗性。目前,越来越多响应非生物胁迫的植物WRKY基因组序列被鉴定报道(表 2)。
WRKY转录因子的表达受多种环境和内部因子强烈而迅速地诱导,尤其是生物胁迫相关的因子。植物对生物胁迫的反应依赖存在于细胞膜和细胞内部种类繁多的受体蛋白。一般而言,处于细胞膜上的受体通过识别病原菌上保守的特征序列,从而启动抗性反应。这类保守的特征序列简称病原体相关分子模式(Pathogen associated molecular pattern,PAMP),识别这一序列的受体被称为模式识别受体(Pattern recognition rec-eptor,PRR),其介导的抗性反应称之为PAMP触发的免疫反应(PAMP triggered immunity,PTI)[77-79]。另外,细胞内部的受体蛋白多为NBS(Nucleotide binding site)-LRR(Leucine-rich repeat)类抗病基因所编码,其直接或间接识别病原菌释放到细胞内部的效应因子(Effector),启动抗病反应,这一反应被称为效应因子触发性免疫(Effector triggered immunity,ETI)。这两种反均需要WRKY转录因子介导[77, 79]。植物在受到各种微生物或病原体侵害时,通过启动这些复杂免疫系统来保护自己免受攻击。近年来,越来越多的WRKY转录因子在这些免疫系统中发挥着重要的调控作用(表 3)。
WRKY蛋白可以激活或抑制转录,通常富含潜在的转录激活和抑制域。一些WRKY因子具有两种功能。例如,在酵母中,AtWRKY53根据启动子上下游序列激活或抑制报告基因的转录。AtWRKY6在转录激活SIRK基因(编码与衰老有关的受体样蛋白激酶)的同时,负向自调节自身的启动子[56]。通过瞬时表达研究发现,OsWRKY72和OsWRKY77在糊粉蛋白细胞中是ABA信号的激活因子,同时又是GA信号的抑制因子[45]。CaWRKY40b既是自身启动子的激活因子,也是免疫相关基因HSC70的转录的抑制因子[97]。以上表明,WRKY转录因子在多种信号通路中既可以是激活因子,又可以成为抑制因子。
3.1.1 转录激活研究表明,许多基因受到与其启动子相关的WRKY因子的激活。BhWRKY1与BhGolS1(半乳糖醇合成酶)启动子的W-box结合,转录激活BhGolS1的表达,可以提高拟南芥的耐旱性[98]。AtWRKY50可以和TGA2或TGA5作用与PR1启动子的W-box结合,协同激活PR1的表达,增强拟南芥的抗性[99]。
MAP激酶(MAPK)参与调控WRKY转录因子的结合活性。例如,在拟南芥中,MEKK1蛋白激酶是双功能蛋白,在拟南芥衰老诱导的信号通路中,它既可以与WRKY53的启动子在W-box上游的一个位点(WP1)结合,又可以诱导AtWRKY53转录因子磷酸化,促进AtWRKY53转录因子和与其自身编码基因启动子的结合,调控拟南芥的衰老[100]。MAPK通过磷酸化激活WRKY46转录因子与PAMP响应基因NHL10启动子的结合活性,增加NHL10基因的表达,调控拟南芥的对细菌鞭毛蛋白病原体的防御反应[101]。
WRKY功能的另一种机制是通过小RNA(smRNA)(微小RNA,miRNA和小干扰RNA,siRNA)起作用,它们已成为调控基因表达的基本模式[102]。由于多个miRNA的预测靶点编码WRKY转录因子[102],因此,WRKY不仅可以调节smRNA的数量,而且WRKY转录因子本身也是smRNA的靶向因子。
3.1.2 转录的抑制和去抑制除了含有丰富的转录激活区,WRKY转录因子还存在重要的转录抑制区。大量的证据表明,许多基因被与其启动子结合的WRKY因子所抑制。从WRKY蛋白在抑制中的作用程度可以了解WRKY的功能。
其中,一种作用方式是靶基因被其他转录因子与启动子的调控位点结合,阻止自身转录因子与靶基因启动子的结合。例如,PcWRKY1可以结合PcPR10基因启动子的W-box,丝裂原活化蛋白激酶在细胞核中可以修饰已结合的PcWRKY1转录因子,这种修饰作用导致PcWRKY1转录因子的变构释放,并可能被其他WRKY转录因子从同源的W-box元件中取代,从而解除对PcPR10和PcWRKY1的抑制[15]。另一种方式是通过作用于其他转录因子来抑制其他转录因子的作用。例如,bZIP28的转录可以上调内质网ER蛋白基因的表达,对植物的抗病具有积极的作用,但bZIP28启动子中存在顺式作用元件W-box,WRKY7/WRKY11/WRKY17可以通过与bZIP28启动子中的W-box元件结合,在转录上抑制了bZIP28的上调,使得拟南芥对丁香假单胞细菌的抗性减弱[103]。
还有一种转录抑制方式是通过改变DNA或组蛋白的高级结构阻止转录的发生。如表观遗传修饰的DNA甲基化和去甲基化、组蛋白乙酰化和去乙酰化,是诱导应激转录关闭或开启的关键机制。DNA甲基化能引起DNA与蛋白质相互作用方式的改变,从而抑制基因表达。例如,水稻叶片组织中2种基因WRKY50和WRKY72启动子上的DNA甲基化,降低了WRKY50和WRKY72的表达水平[104]。相反,DNA的去甲基化可以实现WRKY转录因子的去抑制过程,使WRKY转录因子的抑制作用转为激活作用。DBR2的启动子区在4个CG-、4个CHH-和2个CHG-位点发生了去甲基化,使得青蒿素生物合成的关键调控基因DBR2表达上调,表明WRKY启动子上的去甲基化促进了ABI5青蒿素的表达[105]。同样,组蛋白的去乙酰化可以抑制转录因子与DNA结合位点特异性结合,而组蛋白的乙酰化则发挥相反的作用。组蛋白去乙酰化酶19(HDA19)通过去除组蛋白尾部的乙酰基抑制AtWRKY38和AtWRKY62的转录,负调控基底防御[106]。而另外一项乙酰化的研究表明,拟南芥WRKY40启动子上的组蛋白H3K9(组蛋白H3第9位赖氨酸)乙酰化,可以增加WRKY40基因的表达,提高拟南芥对镰刀菌的抗性[107]。以上这些研究表明,通过表观遗传修饰的DNA甲基化和去甲基化、组蛋白乙酰化和去乙酰化作用,可以诱导WRKY基因转录的关闭或开启。
4 WRKY转录因子的调控网络在DNA水平上,WRKY转录因子通过识别并结合其目标基因中的W-box,识别自身或其他目标基因的启动子来激活或抑制转录,实现调控作用。通过蛋白质-DNA相互作用,WRKY蛋白的上游调控因子和下游靶基因之间的相互作用和交叉作用构成了复杂的WRKY调控网络。在蛋白质水平上,通过蛋白质-蛋白质的相互作用,包括WRKY转录因子之间与多种调控蛋白的相互作用,共同调控植物的生长发育和响应环境中的各种应激反应。
4.1 WRKY转录因子的自调控和交叉调控WRKY信号网络的一个特点是通过WRKY转录因子与其自身启动子相互作用的自调控和其他WRKY转录因子作用的交叉调控来实现调节,这是通过识别并结合目标基因中的W-box启动子来实现的。例如,CaWRKY40b通过直接靶向自身启动子中的W-box,在转录水平上表现出正反馈调控[97],从而实现了WRKY转录因子的自调控。欧芹的PcWRKY1在启动子中具有3个协同作用的W-box的保守排列[108],在PAMP诱导后,PcWRKY1转录产物增加[109],通过染色质免疫沉淀分析显示,这3个W-box是由WRKY转录因子结合的,但PcWRKY1与自身的启动子结合时PcWRKY1的转录下调,表明PcWRKY1启动子位点的W-box被其他WRKY转录因子结合激活转录[110]。基于生物信息学和植物启动子的功能研究发现,许多WRKY基因启动子在统计上富集了W-box[111]。例如,在欧芹PcWRKY1中,存在多个W-box,其多个W-box对转录有协同作用[108];大麦HvWRKY38的转录需要2个相邻的W-box来有效绑定[112];水稻2个OsWRKY45-DBD分子交换β4-β5链形成二聚体,包含2个与W-box相互作用的DNA结合域[113]。WRKY基因的多个W-box表明自调节和交叉调节是WRKY转录因子调控网络的特征。
4.2 WRKY转录因子在蛋白水平的调控网络尽管WRKY转录因子具有功能多样性,而且几乎所有分析的WRKY蛋白都能识别W-box序列,但是除了可以识别核心W-box启动子元件外,还存在其他机制可以实现WRKY转录因子的特异性调控。例如,WRKY转录因子与多种蛋白相互作用,在信号和转录中发挥的调控作用。目前正在研究WRKY的信号网络和转录调控机制,通过蛋白质-蛋白质的相互作用,除了前文提到的钙调蛋白、MAP激酶、去乙酰化酶,还有抗性R蛋白和多种转录因子等,揭示WRKY蛋白的调控功能网络。
4.2.1 WRKY-MAPK的相互作用丝裂原活化蛋白激酶(Mitogen-activated protein kinase,MAPK)存在于所有真核生物中,是一个高度保守的模块。在植物中,MAPK通路参与调控发育、生长、程序性细胞死亡,以及对多种环境刺激的反应[114]。MAPK信号级联通过多个磷酸化作用将上游受体与下游目标连接起来,通过磷酸化放大和转导膜受体感知到的病原体衍生信号,并将这些信号转导改变相关的基因表达[115]。
MAPK信号传导途径响应植物的MTI(由识别微生物的保守分子MAMP引发的免疫)或PTI防御信号通路反应,植物在免疫反应中通过细胞内的MAPK级联信号可以感知到MAMP和PAMP,并刺激WRKY转录因子的诱导[24]。在拟南芥中,转录因子AtWrky33在没有病原体感染的情况下与MAP激酶4(MPK4)形成MAMP或PAMP复合物。MAMP或PAMP复合物被病原体感染激活MEKK1-MKK1/2-MPK4模块,激活的MPK4磷酸化MKS1,导致MPK4-MKS1-Wrky33复合物的核离解,MKS1和AtWrky33被释放,激活PAD3(合成抗菌复合物所需的酶)的表达,提高了拟南芥的抗病性[116]。随后一项研究中,通过磷蛋白迁移转移试验,表明AtWrky33可以被MPK3/MPK6磷酸化,并促进AtWrky33调控植物抗毒素的生物合成[117]。相反,WRKY34中MPK3/MPK6磷酸化位点的缺失影响了WRKY34在体内的功能[118]。OsWRKY53作为MPK3/MPK6的负反馈调节器发挥作用,从而起到诱导防御的早期抑制作用[119]。由MPK3/MPK6、Wrky33、类AGD2防御反应蛋白1(ALD1)和哌啶酸(PiP)组成的正调控环存在于系统获得性抗性(SAR)诱导过程中,推测在PiP生物合成水平上存在不同的SAR激活途径[120]。
4.2.2 WRKY-抗性R蛋白的相互作用植物防御信号通路的ETI是基于抗性R蛋白对效应蛋白的识别而产生的特异性抗性。当病原菌入侵植物后,植物分泌抗性R蛋白识别病原菌效应蛋白,WRKY转录因子和抗性R蛋白互作形成蛋白复合体,解除对基本防御途径的抑制作用[121]。在大麦对白粉病的免疫中,涉及ETI免疫途径,细胞质中的抗病R蛋白MLA能够识别白粉病效应子AVR10,并在细胞核中与HvWRKY1和HvWRKY2结合,解除HvWRKY1/2对抗病的抑制作用,从而达到抗病目的[122]。研究发现,细胞内存在一种典型的嵌合蛋白,如R蛋白NBS-LRR和WRKY转录因子嵌合而成的组合蛋白,其在免疫调控中发挥着重要的作用。例如,WRKY转录因子与RRS1(一种NBS-LRR蛋白)相互作用形成的嵌合蛋白AtWRKY52/RRS1,通过与细菌效应体PopP2的相互作用对细菌致病菌产生免疫[123-124]。拟南芥的RRS1-R、RPS4蛋白与WRKY转录因子结合形成受体复合物,该复合物可以识别细菌效应因子AvrRps4或PopP2,然后激活防御系统[121]。以上研究证明,WRKY转录因子通过与抗病R蛋白结合,识别病原微生物产生的效应蛋白,引发植物产生特异性的防卫反应,在ETI免疫系统中发挥着重要的调控作用。
4.2.3 WRKY与多种转录因子的相互作用蛋白质,尤其是调节蛋白,很少单独起作用,通常情况下,它们在生理上或短暂或永久地相互作用,以承担生命系统中的生物功能。例如,WRKY转录因子可以与多种转录因子相互作用,共同对植物的生长发育进行调控。AtWRKY50可以和TGA2或TGA5作用与PR1启动子结合,与TGA转录因子协同激活PR1的表达[99]。VviWRKY03通过与VviMYB14转录因子的组合效应发挥作用,共同激活VviSTS29启动子,调控白藜芦醇生物合成[125]。除了可以与其他家族的转录因子结合,WRKY转录因子还可以与自身家族中其他转录因子相互结合,发挥相应的调控作用。WRKY转录因子在结合DNA前可以通过蛋白质-蛋白质相互作用形成二聚体或多聚体。例如,在病原菌感染过程中,OsWRKY45可与OsWRKY62形成异二聚体,该二聚体可激活DPF基因(二萜植物抗毒素生物合成基因)的转录[114]。同时,WRKY转录因子还可以与自身形成二聚体发挥作用,例如,激活蛋白复合物WRKY60-60引发干旱防御反应的可能性较高,转录因子WRKY40和蛋白复合物WRKY40-40可以抑制干旱反应[126]。
4.2.4 WRKY转录因子与其他蛋白的相互作用除了前文提到的许多重要的调控蛋白,还存在其他一些蛋白与WRKY转录因子相互作用发挥调控作用。VQ蛋白是植物特异性转录调控的一种辅助因子。VQ蛋白与WRKY蛋白的相互作用可能导致构象的改变或翻译后的修饰,从而激活或抑制它们与靶基因启动子的结合[127]。在甜瓜中,共有24个WRKY基因与11个VQ家族基因共表达[128]。进一步研究发现,拟南芥VQ10和WRKY8可在植物细胞核中形成复合物。WRKY8的中间区域与VQ10之间的相互作用促进了WRKY8与DNA结合的活性,并正向调节植物对灰霉病菌的抗性[129]。最近研究表明,OsWRKY45的N端与细胞核中Pb1(穗芽抗性基因)蛋白的N端螺旋结构域(CC)相互作用对稻瘟菌产生抗性[114]。目前已经发现了大量与WRKY相互作用的蛋白质,未来在已鉴定出的WRKY相互作用蛋白的基础上,预计将会有更多的WRKY相互作用蛋白被传统的方法(如酵母双杂交)和最近发展的方法(如高密度蛋白质微阵列)所识别,并通过其相互作用蛋白在不同水平上的相互作用完善WRKY转录因子复杂调控功能网络。
5 展望WRKY转录因子在调控植物生长发育和衰老、非生物及生物胁迫中发挥着重要的作用。随着高通量转录组学、蛋白质组学、代谢组学等技术的发展,更深入地发掘WRKY转录因子生物学功能,更详细地确定WRKY靶基因、WRKY调控网络、WRKY转录因子相互作用的蛋白,为探索WRKY转录因子如何在启动子上发挥信号协同和信号拮抗的作用奠定生物化学基础,从而帮助人们深入了解WRKY转录因子之间的串扰机制,但要详细地明确这一机制还需要做更深入的研究。
WRKY转录因子在植物中是一个庞大的基因家族,WRKY转录因子种类繁多,其在调节重要的生物学功能的分子机制和功能多样性仍然还需进一步的研究和证明,其中一个重要的研究思路是将越来越多的测序数据与蛋白质-DNA和蛋白质-蛋白质相互作用的信息整合在WRKY介导的调控重要生物学功能的过程中,建立一个全面的WRKY信号和转录调控网络。在充分了解WRKY在分子水平上的作用机制后,通过分子辅助育种和生物技术工具,培育出达到经济目的的优良品种。
| [1] |
Xue GP. The DNA-binding activity of an Ap2 transcriptional activator Hvcbf2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature[J]. Plant J, 2003, 33(2): 373-383. DOI:10.1046/j.1365-313X.2003.01630.x |
| [2] |
Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors:genome-wide comparative analysis among eukaryotes[J]. Science, 2000, 290(5499): 2105-2110. DOI:10.1126/science.290.5499.2105 |
| [3] |
Yamasaki K, Kigawa T, Inoue M, et al. Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains[J]. Plant Physiology and Biochemistry, 2008, 46(3): 394-401. DOI:10.1016/j.plaphy.2007.12.015 |
| [4] |
Ishiguro S, Nakamura K. Characterization of a cdna encoding a novel DNA-binding protein, Spf1, that recognizes Sp8 sequences in the 5' upstream regions of genes coding for sporamin and beta-amylase from sweet potato[J]. Molecular and General Genetics, 1994, 244(6): 563-571. |
| [5] |
Rushton PJ, Macdonald H, Huttly AK, et al. Members of a new family of DNA-binding proteins bind to a conserved Cis-element in the promoters of Α-Amy2 genes[J]. Plant Mole Biol, 1995, 29(4): 691-702. DOI:10.1007/BF00041160 |
| [6] |
Wen F, Zhu H, Li P, et al. Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon[J]. DNA Research, 2014, 21(3): 327-339. DOI:10.1093/dnares/dst060 |
| [7] |
Rinerson CI, Rabara RC, Tripathi P, et al. The evolution of WRKY transcription factors[J]. BMC Plant Biology, 2015, 15(1): 66. |
| [8] |
Liu D, Lei BK, Zhao P, et al. Phylogenetic analysis of barley WRKY proteins and characterization of Hvwrky1 and-2 as repressors of the pathogen-inducible gene Hvger4c[J]. Molecular Genetics and Genomics, 2014, 289(6): 1331-1345. DOI:10.1007/s00438-014-0893-6 |
| [9] |
Li MY, Xu ZS, Tian C, et al. Genomic identification of WRKY transcription factors in carrot(Daucus carota)and analysis of evolution and homologous groups for plants[J]. Scientific Reports, 2016, 6: 23101. DOI:10.1038/srep23101 |
| [10] |
Liu JJ, Ekramoddoullah AKJ. Identification and characterization of the WRKY transcription factor family in Pinus monticola[J]. Genome, 2009, 52(1): 77-88. DOI:10.1139/G08-106 |
| [11] |
Guo C, Guo R, Xu X, et al. Evolution and expression analysis of the grape(Vitis vinifera L.)WRKY gene family[J]. Journal of Experimental Botany, 2014, 65(6): 1513-1528. DOI:10.1093/jxb/eru007 |
| [12] |
Da SEG, Ito TM, Souza SG. 'In silico'genome-wide identification and phylogenetic analysis of the WRKY transcription factor family in sweet orange (Citrus sinensis)[J]. Australian Journal of Crop Science, 2017, 11(6): 716. DOI:10.21475/ajcs.17.11.06.p471 |
| [13] |
Meng D, Li Y, Bai Y, et al. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress[J]. Plant Physiology, 2016, 103: 71-83. |
| [14] |
Yang Y, Zhou Y, Chi Y, et al. Characterization of soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode[J]. Scientific Reports, 2017, 7(1): 17804. DOI:10.1038/s41598-017-18235-8 |
| [15] |
Ülker B, Somssich IEJ. WRKY transcription factors:from DNA binding towards biological function[J]. Current Opinion in Plant Biology, 2004, 7(5): 491-498. |
| [16] |
Mangelsen E, Kilian J, Berendzen KW, et al. Phylogenetic and comparative gene expression analysis of barley(Hordeum vulgare)WRKY transcription factor family reveals putatively retained functions between monocots and dicots[J]. BMC Genomics, 2008, 9(1): 194. DOI:10.1186/1471-2164-9-194 |
| [17] |
Ning P, Liu C, Kang J, et al. Genome-wide analysis of WRKY transcription factors in wheat(Triticum aestivum L.)and differential expression under water deficit condition[J]. Peer J, 2017, 5: e3232. DOI:10.7717/peerj.3232 |
| [18] |
Xu H, Watanabe KA, Zhang L, et al. WRKY transcription factor genes in wild rice Oryza nivara[J]. DNA Research, 2016, 23(4): 311-323. DOI:10.1093/dnares/dsw025 |
| [19] |
Rice WRKY Working Group. Nomenclature report on rice WRKY's-conflict regarding gene names and its solution[J]. Rice, 2012, 5(1): 3. DOI:10.1186/1939-8433-5-3 |
| [20] |
Li L, Mu S, Cheng Z, et al. Characterization and expression analysis of the WRKY gene family in moso bamboo[J]. Scientific Reports, 2017, 7(1): 6675. DOI:10.1038/s41598-017-06701-2 |
| [21] |
Wei KF, Chen J, Chen YF, et al. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize[J]. DNA Research, 2012, 19(2): 153-164. DOI:10.1093/dnares/dsr048 |
| [22] |
Goel R, Pandey A, Trivedi PK, et al. Genome-wide analysis of the musa WRKY gene family:Evolution and differential expression during development and stress[J]. Front Plant Sci, 2016, 7: 299. |
| [23] |
Eulgem T, Rushton PJ, Robatzek S, et al. The WRKY superfamily of plant transcription factors[J]. Trends in Plant Science, 2000, 5(5): 199-206. DOI:10.1016/S1360-1385(00)01600-9 |
| [24] |
Rushton PJ, Somssich IE, Ringler P, et al. WRKY transcription factors[J]. Trends in Plant Science, 2010, 15(5): 247-258. DOI:10.1016/j.tplants.2010.02.006 |
| [25] |
Maeo K, Hayashi S, Kojima SH, et al. Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins[J]. Bioscience Biotechnology, 2001, 65(11): 2428-2436. |
| [26] |
Bakshi M, Oelmüller RJ. WRKY transcription factors:Jack of many trades in plants[J]. Plant Signaling Behavior, 2014, 9(2): e27700. DOI:10.4161/psb.27700 |
| [27] |
Yamasaki K, Kigawa T, Inoue M, et al. Solution structure of an Arabidopsis WRKY DNA binding domain[J]. The Plant Cell, 2005, 17(3): 944-956. DOI:10.1105/tpc.104.026435 |
| [28] |
Zhang Y, Wang LJ. The WRKY transcription factor superfamily:Its origin in eukaryotes and expansion in plants[J]. BMC Evolutionary Biology, 2005, 5(1): 1. |
| [29] |
Wan Y, Mao M, Wan D, et al. Identification of the WRKY gene family and functional analysis of two genes in Caragana intermedia[J]. BMC Plant Biology, 2018, 18(1): 31. |
| [30] |
Chen L, Song Y, Li S, et al. The role of WRKY transcription factors in plant abiotic stresses[J]. Biochimica et Biophysica Acta -Gene Regulatory Mechanisms, 2012, 1819(2): 120-128. DOI:10.1016/j.bbagrm.2011.09.002 |
| [31] |
Park CY, Lee JH, Yoo JH, et al. WRKY group iid transcription factors interact with calmodulin[J]. FEBS Letters, 2005, 579(6): 1545-1550. DOI:10.1016/j.febslet.2005.01.057 |
| [32] |
Bailey TL, Boden M, Buske FA, et al. Meme suite:tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(2): W202-W208. |
| [33] |
Vyas J, Gryk MR, Schiller MRJ. VENN, a tool for titrating sequence conservation onto protein structures[J]. Nucleic Acids Res, 2009, 37(18): e124. DOI:10.1093/nar/gkp616 |
| [34] |
Tamura K, Dudley J, Nei M, et al. Mega4:Molecular evolutionary genetics analysis(mega)software version 4. 0[J]. Molecular Biology Evolution, 2007, 24(8): 1596-1599. DOI:10.1093/molbev/msm092 |
| [35] |
Ou C, Jiang S, Wang F, et al. An RNA-Seq analysis of the pear(Pyrus communis L.)transcriptome, with a focus on genes associated with dwarf[J]. Plant Gene, 2015, 4: 69-77. DOI:10.1016/j.plgene.2015.08.003 |
| [36] |
Zhang D, Ren L, Yue Jh, et al. RNA-Seq-based transcriptome analysis of stem development and dwarfing regulation in Agapanthus praecox ssp. orientalis(leighton)leighton[J]. Gene, 2015, 565(2): 252-267. DOI:10.1016/j.gene.2015.04.013 |
| [37] |
Jiang J, Ma S, Ye N, et al. WRKY transcription factors in plant responses to stresses[J]. Journal of Integrative Plant Biology, 2017, 59(2): 86-101. DOI:10.1111/jipb.12513 |
| [38] |
Cai Y, Chen X, Xie K, et al. Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice[J]. PLoS One, 2014, 9(7): e102529. DOI:10.1371/journal.pone.0102529 |
| [39] |
Guo D, Zhang J, Wang X, et al. The WRKY transcription factor WRKY71/Exb1 controls shoot branching by transcriptionally regulating Rax genes in Arabidopsis[J]. The Plant Cell, 2015, 27(11): 3112-3127. DOI:10.1105/tpc.15.00829 |
| [40] |
Ma Y, Xue H, Zhang L, et al. Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple(Malus×domestica)[J]. Scientific Reports, 2016, 6: 26719. DOI:10.1038/srep26719 |
| [41] |
Soumelidou K, Morris D, Battey N, et al. Auxin transport capacity in relation to the dwarfing effect of apple rootstocks[J]. Journal of Horticultural Science, 1994, 69(4): 719-725. DOI:10.1080/14620316.1994.11516505 |
| [42] |
Michalczuk LJ. Indole-3-acetic acid level in wood, bark and cambial sap of apple rootstocks differing in growth vigour[J]. Acta Physiologiae Plantarum, 2002, 24(2): 131-136. |
| [43] |
Zhao B, Li JJ. Regulation of brassinosteroid biosynthesis and inactivation[J]. Journal of Integrative Plant Biology, 2012, 54(10): 746-759. DOI:10.1111/j.1744-7909.2012.01168.x |
| [44] |
Zheng X, Zhao Y, Shan D, et al. Mdwrky9 overexpression confers intensive dwarfing in the M26 rootstock of apple by directly inhibiting brassinosteroid synthetase Mddwf4 expression[J]. New Phytologist, 2018, 217(3): 1086-1098. DOI:10.1111/nph.14891 |
| [45] |
Xie Z, Zhang ZL, Zou X, et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells[J]. Plant Physiology, 2005, 137(1): 176-189. |
| [46] |
Qiu D, Xiao J, Ding X, et al. Oswrky13 mediates rice disease resistance by regulating defense-related genes in salicylate-and jasmonate-dependent signaling[J]. Molecular Plant-Microbe Interactions, 2007, 20(5): 492-499. DOI:10.1094/MPMI-20-5-0492 |
| [47] |
Zhang J, Peng Y, Guo ZJ. Constitutive expression of pathogen-inducible Oswrky31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants[J]. Cell Research, 2008, 18(4): 508. DOI:10.1038/cr.2007.104 |
| [48] |
Wang PT, Xu X, Tang Z, et al. Oswrky28 regulates phosphate and arsenate accumulation, root system architecture and fertility in rice[J]. Frontiers in Plant Science, 2018, 9: 1330. DOI:10.3389/fpls.2018.01330 |
| [49] |
Hu Z, Wang R, Zheng M, et al. Ta WRKY 51 promotes lateral root formation through negative regulation of ethylene biosynthesis in wheat(Triticum aestivum L.)[J]. Plant J, 2018, 96(2): 372-388. DOI:10.1111/tpj.14038 |
| [50] |
Cheng Y, Ahammed GJ, Yu J, et al. Putative WRKYs associated with regulation of fruit ripening revealed by detailed expression analysis of the WRKY gene family in pepper[J]. Scientific Reports, 2016, 6. DOI:10.1038/srep39000 |
| [51] |
Yang X, Li H, Yang Y, et al. Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon(Citrullus lanatus)[J]. PLoS One, 2018, 13(1): e0191308. DOI:10.1371/journal.pone.0191308 |
| [52] |
Li QL, Bo S, Deng wJ, et al. Avocado fruit pulp transcriptomes in the after-ripening process[J]. Notulae Botanicae Horti Agrobotanici Cluj-Napoca, 2019, 47(2): 308-319. |
| [53] |
Li D, Mou W, Xia R, et al. Integrated analysis of high-throughput sequencing data shows abscisic acid-responsive genes and mirnas in strawberry receptacle fruit ripening[J]. Horticulture Research, 2019, 6(1): 26. |
| [54] |
Guo Y, Cai Z, et al. Transcriptome of Arabidopsis Leaf senescence[J]. Plant, Cell Environment, 2004, 27(5): 521-549. DOI:10.1111/j.1365-3040.2003.01158.x |
| [55] |
Robatzek S, Somssich IEJ. A new member of the Arabidopsis WRKY transcription factor family, Atwrky6, is associated with both senescence-and defence-related processes[J]. Plant J, 2001, 28(2): 123-133. DOI:10.1046/j.1365-313X.2001.01131.x |
| [56] |
Robatzek S, Somssich IEJ. Targets of Atwrky6 regulation during plant senescence and pathogen defense[J]. Gene Development, 2002, 16(9): 1139-1149. DOI:10.1101/gad.222702 |
| [57] |
Gu L, Wei H, et al. Characterization and functional analysis of Ghwrky42, a group iid WRKY gene, in upland cotton(Gossypium hirsutum L.)[J]. BMC Genetics, 2018, 19(1): 48. DOI:10.1186/s12863-018-0653-4 |
| [58] |
Fan ZQ, Tan XL, Shan W, et al. Characterization of a transcriptional regulator, brwrky6, associated with gibberellin-suppressed leaf senescence of chinese flowering cabbage[J]. Journal of Agricultural Food Chemistry, 2018, 66(8): 1791-1799. DOI:10.1021/acs.jafc.7b06085 |
| [59] |
Gao H, Wang Y, Xu P, et al. Overexpression of a WRKY transcription factor Tawrky2 enhances drought stress tolerance in transgenic wheat[J]. Front Plant Sci, 2018, 9: 997. DOI:10.3389/fpls.2018.00997 |
| [60] |
Wang CT, Ru JN, Liu YW, et al. Maize WRKY transcription factor Zmwrky106 confers drought and heat tolerance in transgenic plants[J]. International Journal of Molecular Sciences, 2018, 19(10): 3046. DOI:10.3390/ijms19103046 |
| [61] |
Li DH, Wang CH, Liu XP, et al. Expression of Gmwrky35, a soybean WRKY gene, in transgenic tobacco confers drought stress tolerances[J]. Soybean Science, 2017, 36(5): 685-691. |
| [62] |
Lee H, Cha J, Choi C, et al. Rice WRKY11 plays a role in pathogen defense and drought tolerance[J]. Rice, 2018, 11(1): 5. DOI:10.1186/s12284-018-0199-0 |
| [63] |
Wang Y, Shu Z, Wang W, et al. Cswrky2, a novel WRKY gene from camellia sinensis, is involved in cold and drought stress responses[J]. Biologia Plantarum, 2016, 60(3): 443-451. DOI:10.1007/s10535-016-0618-2 |
| [64] |
Zhu D, Che YM, Xiao PL, et al. Functional analysis of a grape WRKY30 gene in drought resistance[J]. Plant Cell Tissue and Organ Culture, 2018, 132(3): 449-459. DOI:10.1007/s11240-017-1341-1 |
| [65] |
Yang G, Zhang W, Liu Z, et al. Both JrWRKY 2 and Jr WRKY 7 of Juglans regia mediate responses to abiotic stresses and abscisic acid through formation of homodimers and interaction[J]. Plant Biology, 2017, 19(2): 268-278. DOI:10.1111/plb.12524 |
| [66] |
Raineri J, Ribichich KF, Chan RLJ. The sunflower transcription factor Hawrky76 confers drought and flood tolerance to Arabidopsis thaliana plants without yield penalty[J]. Plant Cell Reports, 2015, 34(12): 2065-2080. DOI:10.1007/s00299-015-1852-3 |
| [67] |
He L, Wu YH, Zhao Q, et al. Chrysanthemum Dgwrky2 gene enha-nces tolerance to salt stress in transgenic chrysanthemum[J]. International Journal of Molecular Sciences, 2018, 19(7): 2062. DOI:10.3390/ijms19072062 |
| [68] |
Yang G, Zhang W, Sun Y, et al. Two novel WRKY genes from Juglans regia, Jrwrky6 and Jrwrky53, are involved in abscisic acid-dependent stress responses[J]. Biologia Plantarum, 2017, 61(4): 611-621. DOI:10.1007/s10535-017-0723-x |
| [69] |
Wu M, Liu H, Han G, et al. A Moso Bamboo WRKY gene Pewrky83 confers salinity tolerance in transgenic Arabidopsis plants[J]. Scientific Reports, 2017, 7(1): 11721. DOI:10.1038/s41598-017-10795-z |
| [70] |
Ullah A, Sun H, Hakim, et al. A novel cotton WRKY gene, Ghwrky6-like, improves salt tolerance by activating the aba signaling pathway and scavenging of reactive oxygen species[J]. Physiologia Plantarum, 2018, 162(4): 439-454. DOI:10.1111/ppl.12651 |
| [71] |
Pillai SE, Kumar C, Patel HK, et al. Overexpression of a cell wall damage induced transcription factor, Oswrky42, leads to enhanced callose deposition and tolerance to salt stress but does not enhance tolerance to bacterial infection[J]. BMC Plant Biology, 2018, 18(1): 177. |
| [72] |
Wang CT, Ru JN, Liu YW, et al. The maize WRKY transcription factor Zmwrky40 confers drought resistance in transgenic Arabidopsis[J]. International Journal of Molecular Sciences, 2018, 19(9): 2580. DOI:10.3390/ijms19092580 |
| [73] |
Hong C, Cheng D, Zhang G, et al. The role of Zmwrky4 in regulating maize antioxidant defense under cadmium stress[J]. Biochemical Biophysical Research Communications, 2017, 482(4): 1504-1510. DOI:10.1016/j.bbrc.2016.12.064 |
| [74] |
Sheng Y, Yan X, Huang Y, et al. The WRKY transcription factor, WRKY13, activates Pdr8 expression to positively regulate cadmium tolerance in Arabidopsis[J]. Plant, Cell and Environment, 2019, 42(3): 891-903. DOI:10.1111/pce.13457 |
| [75] |
Li GZ, Wang ZQ, Yokosho K, et al. Transcription factor WRKY 22 promotes aluminum tolerance via activation of Os Frdl 4 expression and enhancement of citrate secretion in rice(Oryza sativa)[J]. New Phytologist, 2018, 219(1): 149-162. DOI:10.1111/nph.15143 |
| [76] |
Dai X, Wang Y, Zhang WH. Oswrky74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice[J]. Journal of Experimental Botany, 2015, 67(3): 947-960. |
| [77] |
Jones JDG, Dangl JL. The plant immune system[J]. Nature, 2006, 444(7117): 323-329. DOI:10.1038/nature05286 |
| [78] |
Monaghan J, Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane[J]. Current Opinion in Plant Biology, 2012, 15(4): 349-357. DOI:10.1016/j.pbi.2012.05.006 |
| [79] |
Chisholm ST, Coaker G, Day B, et al. Host-microbe interactions:shaping the evolution of the plant immune response[J]. Cell, 2006, 124(4): 803-814. DOI:10.1016/j.cell.2006.02.008 |
| [80] |
Wang X, Guo R, Tu M, et al. Ectopic expression of the wild grape WRKY transcription factor Vqwrky52 in Arabidopsis Thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen botrytis cinerea[J]. Front Plant Sci, 2017, 8: 97. |
| [81] |
Arrano SP, Dominguez FJ, Herrera VA, et al. WRKY7, -11 and-17 transcription factors are modulators of the Bzip28 branch of the unfolded protein response during pamp-triggered immunity in Arabidopsis thaliana[J]. Plant Science, 2018, 277: 242-250. DOI:10.1016/j.plantsci.2018.09.019 |
| [82] |
Li J, Wang J, Wang N, et al. Ghwrky44, a WRKY transcription factor of cotton, mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana[J]. Plant Cell, Tissue Organ Culture, 2015, 121(1): 127-140. |
| [83] |
Wang L, Liu F, et al. Expression characteristics and functional analysis of the Scwrky3 gene from sugarcane[J]. International Journal of Molecular Sciences, 2018, 19(12): 4059. DOI:10.3390/ijms19124059 |
| [84] |
Wang H, Meng J, Peng X, et al. Rice WRKY4 acts as a transcrip-tional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight[J]. Plant Mole Biol, 2015, 89(1-2): 157-171. DOI:10.1007/s11103-015-0360-8 |
| [85] |
Shuo Y, Liang Z, Miao LY, et al. The expression and binding properties of the rice WRKY68 protein in the Xa21-Mediated resistance response to Xanthomonas Oryzae pv. Oryzae[J]. Journal of Integrative Agriculture, 2016, 15(11): 2451-2460. DOI:10.1016/S2095-3119(15)61265-5 |
| [86] |
Hwang SH, Kwon SI, Jang JY, et al. Oswrky51, a rice transcription factor, functions as a positive regulator in defense response against Xanthomonas Oryzae Pv. Oryzae[J]. Plant Cell Reports, 2016, 35(9): 1975-1985. DOI:10.1007/s00299-016-2012-0 |
| [87] |
Liu Z, Luan Y, Li JJ. Molecular cloning and expression analysis of Spwrky6 gene from Solanum Pimpinellifolium[J]. Biologia Plantarum, 2016, 60(2): 226-234. DOI:10.1007/s10535-016-0582-x |
| [88] |
Cui Q, Yan X, et al. Analysis of WRKY transcription factors and characterization of two botrytis cinerea-responsive Lrwrky genes from lilium regale[J]. Plant Physiology, 2018, 127: 525-536. |
| [89] |
Liu F, Li XX, Wang MR, et al. Interactions of Wrky15 and Wrky33 transcription factors and their roles in the resistance of Oilseed rape to Sclerotinia infection[J]. Plant Biotechnology Journal, 2018, 16(4): 911-925. DOI:10.1111/pbi.12838 |
| [90] |
Wang J, Tao F, Tian W, et al. The wheat WRKY transcription factors Tawrky49 and Tawrky62 confer differential high-temperature seedling-plant resistance to Puccinia striiformis F. sp. tritici[J]. PLoS One, 2017, 12(7): e0181963. DOI:10.1371/journal.pone.0181963 |
| [91] |
Cui J, Xu P, Meng J, et al. Transcriptome signatures of tomato leaf induced by phytophthora infestans and functional identification of transcription factor Spwrky3[J]. Theoretical and Applied Genetics, 2018, 131(4): 787-800. DOI:10.1007/s00122-017-3035-9 |
| [92] |
Wei W, Cui MY, Yang H, et al. Ectopic expression of Fvwrky42, a WRKY transcription factor from the diploid woodland strawberry(Fragaria Vesca), enhances resistance to powdery mildew, improves osmotic stress resistance, and increases abscisic acid sensitivity in Arabidopsis[J]. Plant Science, 2018, 275: 60-74. DOI:10.1016/j.plantsci.2018.07.010 |
| [93] |
Liu Q, Li X, Yan S, et al. Oswrky67 Positively regulates blast and bacteria blight resistance by direct activation of Pr genes in rice[J]. BMC Plant Biology, 2018, 18(1): 257. DOI:10.1186/s12870-018-1479-y |
| [94] |
Luan Q, Chen C, Liu M, et al. Cswrky50 mediates defense responses to pseudoperonospora cubensis infection in cucumis sativus[J]. Plant Science, 2019, 279: 59-69. DOI:10.1016/j.plantsci.2018.11.002 |
| [95] |
Shan W, Chen JY, Kuang JF, et al. Banana fruit nac transcription factor manac5 cooperates with mawrkys to enhance the expression of pathogenesis-related genes against Colletotrichum musae[J]. Molecular Plant Pathology, 2016, 17(3): 330-338. DOI:10.1111/mpp.12281 |
| [96] |
Hu L, Ye M, Li R, et al. Oswrky53, a versatile switch in regulating herbivore-induced defense responses in rice[J]. Plant Signaling, 2016, 11(4): e1169357. DOI:10.1080/15592324.2016.1169357 |
| [97] |
Ifnan KM, Zhang Y, Liu Z, et al. Cawrky40b in pepper acts as a negative regulator in response to ralstonia solanacearum by directly modulating defense genes including Cawrky40[J]. International journal of Molecular Sciences, 2018, 19(5): 1403. DOI:10.3390/ijms19051403 |
| [98] |
Wang Z, Zhu Y, Wang L, et al. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-Box elements of the galactinol synthase(Bhgols1)promoter[J]. Planta, 2009, 230(6): 1155-1166. DOI:10.1007/s00425-009-1014-3 |
| [99] |
Hussain RM, Sheikh AH, Haider I, et al. Arabidopsis WRKY50 and tga transcription factors synergistically activate expression of Pr1[J]. Front Plant Sci, 2018, 9: 930. DOI:10.3389/fpls.2018.00930 |
| [100] |
Miao Y, Laun TM, et al. Arabidopsis Mekk1 can take a short cut:It can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter[J]. Plant Mole Biol, 2007, 65(1-/2): 63-76. |
| [101] |
Sheikh AH, Eschen LL, et al. Regulation of WRKY46 transcription factor function by mitogen-activated protein kinases in Arabidopsis thaliana[J]. Front Plant Sci, 2016, 7: 61. |
| [102] |
Pandey SP, Somssich IEJ. The role of WRKY transcription factors in plant immunity[J]. Plant Physiology, 2009, 150(4): 1648-1655. DOI:10.1104/pp.109.138990 |
| [103] |
Arraño SP, Domínguez FJ, Herrera VA, et al. WRKY7, -11 and-17 transcription factors are modulators of the Bzip28 branch of the unfolded protein response during pamp-triggered immunity in Arabidopsis thaliana[J]. Plant Science, 2018, 277: 242-250. DOI:10.1016/j.plantsci.2018.09.019 |
| [104] |
Çelik Ö, Meriç S, Ayan A, 等. Epigenetic analysis of WRKY transcription factor genes in salt stressed rice(Oryza sativa L.)plants[J]. Environmental Experimental Botany, 2019, 159: 121-131. |
| [105] |
Pandey N, Pandey RSJ. Deciphering Uv-B-induced variation in DNA methylation pattern and its influence on regulation of Dbr2 expression in Artemisia annua L.[J]. Planta, 2015, 242(4): 869-879. DOI:10.1007/s00425-015-2323-3 |
| [106] |
Kim KC, Lai Z, Fan B, et al. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense[J]. The Plant Cell, 2008, 20(9): 2357-2371. DOI:10.1105/tpc.107.055566 |
| [107] |
Chakraborty J, Ghosh P, Sen S, et al. Epigenetic and transcriptional control of chickpea WRKY40 promoter activity under fusarium stress and its heterologous expression in Arabidopsis leads to enhanced resistance against bacterial pathogen[J]. Plant Science, 2018, 276: 250-267. DOI:10.1016/j.plantsci.2018.07.014 |
| [108] |
Eulgem T, Rushton PJ, Schmelzer E, et al. Early nuclear events in plant defence signalling:Rapid gene activation by WRKY transcription factors[J]. The EMBO Journal, 1999, 18(17): 4689-4699. DOI:10.1093/emboj/18.17.4689 |
| [109] |
Rushton PJ, Torres JT, Parniske M, et al. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley Pr1 genes[J]. EMBO Journal, 1996, 15(20): 5690-5700. DOI:10.1002/j.1460-2075.1996.tb00953.x |
| [110] |
Turck F, Zhou A, Somssich IEJ. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene Pcpr1-1 in parsley[J]. The Plant Cell, 2004, 16(10): 2573-2585. DOI:10.1105/tpc.104.024810 |
| [111] |
Maleck K, Levine A, Eulgem T, et al. The transcriptome of Arabidopsis thaliana during systemic acquired resistance[J]. Nat Genet, 2000, 26(4): 403-410. DOI:10.1038/82521 |
| [112] |
Mare C, Mazzucotelli E, Crosatti C, et al. Hv-WRKY38:A new transcription factor involved in cold-and drought-response in barley[J]. Plant Mole Biol, 2004, 55(3): 399-416. DOI:10.1007/s11103-004-0906-7 |
| [113] |
Cheng X, Zhao Y, Jiang Q, et al. Structural basis of dimerization and Dual W-Box DNA recognition by rice WRKY domain[J]. Nucleic Acids Res, 2019, 47(8): 4308-4318. DOI:10.1093/nar/gkz113 |
| [114] |
Colcombet J, Hirt HJ. Arabidopsis Mapks:A complex signalling network involved in multiple biological processes[J]. Biochemical Journal, 2008, 413(2): 217-226. |
| [115] |
Fiil BK, Petersen K, Petersen M, et al. Gene regulation by map kinase cascades[J]. Current Opinion In Plant Biology, 2009, 12(5): 615-621. DOI:10.1016/j.pbi.2009.07.017 |
| [116] |
Qiu JL, Fiil BK, et al. Arabidopsis map kinase 4 regulates gene expression through transcription factor release in the nucleus[J]. The EMBO Journal, 2008, 27(16): 2214-2221. DOI:10.1038/emboj.2008.147 |
| [117] |
Mao G, Meng X, Liu Y, et al. Phosphorylation of a WRKY transcription factor by two pathogen-responsive Mapks drives phytoalexin biosynthesis in Arabidopsis[J]. The Plant Cell, 2011, 23(4): 1639-1653. DOI:10.1105/tpc.111.084996 |
| [118] |
Guan Y, Meng X, Khanna R, et al. Phosphorylation of a WRKY transcription factor by Mapks is required for pollen development and function in Arabidopsis[J]. PLoS Genetics, 2014, 10(5): e1004384. DOI:10.1371/journal.pgen.1004384 |
| [119] |
Hu L, Ye M, Li R, et al. The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of mitogen-activated protein kinase activity[J]. Plant Physiology, 2015, 169(4): 2907-2921. |
| [120] |
Wang Y, Schuck S, Wu J, et al. A Mpk3/6-Wrky33-Ald1-pipecolic acid regulatory loop contributes to systemic acquired resistance[J]. The Plant Cell, 2018, 30(10): 2480-2494. DOI:10.1105/tpc.18.00547 |
| [121] |
Sarris PF, Duxbury Z, Huh SU, et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors[J]. Cell, 2015, 161(5): 1089-1100. DOI:10.1016/j.cell.2015.04.024 |
| [122] |
Shen QH, Saijo Y, Mauch S, et al. Nuclear activity of Mla immune receptors links isolate-specific and basal disease-resistance responses[J]. Science, 2007, 315(5815): 1098-1103. DOI:10.1126/science.1136372 |
| [123] |
Deslandes L, Olivier J, Theulières F, et al. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive Rrs1-R gene, a member of a novel family of resistance genes[J]. Proceedings of the National Academy of Sciences of the USA, 2002, 99(4): 2404-2409. DOI:10.1073/pnas.032485099 |
| [124] |
Deslandes L, Olivier J, Peeters N, et al. Physical interaction between Rrs1-R, a protein conferring resistance to bacterial Wilt, and Popp2, a type iii effector targeted to the plant nucleus[J]. Proceedings of the National Academy of Sciences of the USA, 2003, 100(13): 8024-8029. DOI:10.1073/pnas.1230660100 |
| [125] |
Vannozzi A, Wong DCJ, Höll J, et al. Combinatorial regulation of stilbene synthase genes by WRKY and Myb transcription factors in grapevine(Vitis vinifera L.)[J]. Plant Cell Physiology, 2018, 59(5): 1043-1059. DOI:10.1093/pcp/pcy045 |
| [126] |
Lahiri A, Venkatasubramani PS, Datta AJ. Bayesian modeling of plant drought resistance pathway[J]. BMC Plant Biology, 2019, 19(1): 96. |
| [127] |
Chi Y, Yang Y, Zhou Y, et al. Protein-protein interactions in the regulation of WRKY transcription factors[J]. Molecular Plant, 2013, 6(2): 287-300. DOI:10.1093/mp/sst026 |
| [128] |
Jiao Z, Sun J, Wang C, et al. Genome-wide characterization, evolutionary analysis of WRKY genes in cucurbitaceae species and assessment of its roles in resisting to powdery mildew disease[J]. PLoS One, 2018, 13(12): e0199851. DOI:10.1371/journal.pone.0199851 |
| [129] |
Chen J, Wang H, Li Y, et al. Arabidopsis Vq10 interacts with WRKY8 to Modulate basal defense against botrytis cinerea[J]. Journal of Integrative Plant Biology, 2018, 60(10): 956-969. DOI:10.1111/jipb.12664 |