赤霉素(gibberellins或gibberellic acid,GA)是一类属于双萜类化合物的植物激素,在植物整个生命周期中都起着重要作用,能促进细胞分裂和伸长、种子萌发、下胚轴和茎秆伸长、根的生长及开花等[1-3]。20世纪30年代,日本科学家发现GA能够促进植物生长。1926年,日本病理学家黑泽英一研究水稻“恶苗病”致病原因时,发现感染赤霉菌(Gibberella fujikuroi)的水稻植株会出现疯长现象。将赤霉菌培养基的滤液喷施到健康水稻幼苗上,发现幼苗虽然没有感染赤霉菌,但也会出现类似“恶苗病”的过度生长症状。1935年,日本薮田贞治郎和住木谕介从赤霉菌培养基的滤液中分离出这种活性物质,鉴定了它的化学结构,并将其命名为赤霉素。1956年,C.A.韦斯特和B.O.菲尼分别证明了高等植物中也普遍存在着类似的萜类化合物。迄今,已从不同维管植物、细菌及真菌中先后鉴定出了136种结构明确的GAs,并按照时间顺序将它们命名为GA1-GA136。但是,只有部分GAs具有调节植物生长的生理效应,如GA1、GA3、GA4和GA7等[4-5]。遗传学的证据表明,尽管植物中已分离鉴定出GA3,但是在许多植物中GA1和GA4是主要的活性GAs。此外,在拟南芥和水稻中,GA4的活性成分强于GA1[6-8]。赤霉素作为植物生长调节剂已被广泛应用于农业生产中,在促进种子萌发、茎秆伸长、果实发育以及提高植物耐逆性等方面发挥着重要作用。
植物需要产生和积累合适水平的活性GA以确保正常的生长发育。GA可以从合成位点转运至生长发育过程中需要GA的组织或器官中[9]。近来的研究表明,GA转运蛋白NPF(Nitrate transporter 1/peptide transporter family)家族及SWEET家族成员在GA的运输过程中起了重要作用[10]。NPF3.1已被证明是植物中独特的GA转运体[11]。此外,GTR1(Glucosinolate transporter 1,即NPF2.10)也能够转运GA3[12]。目前,SWEET家族成员中仅SWEET13及SWEET14被报道为GA转运蛋白[13]。有趣的是,SWEET13、SWEET14和NPF3的GA转运活性并不局限于活性GA或特定的中间形式,而是可以运输这两种类型[11, 13]。GA转运蛋白的发现进一步补充了GA可移动的证据,表明植物体内存在一种有效的调节机制来指导GA的分布和在植物中流动,以确保合适的GA信号。
自20世纪60年代起,“绿色革命”中半矮化育种的大规模推广大幅度地提高了世界主要粮食作物的产量。水稻和小麦的“绿色革命”都与赤霉素密切相关。水稻“绿色革命”基因sd1(semi-dwarf 1)编码赤霉素生物合成途径的一个关键酶GA20ox2;小麦“绿色革命”基因Rht1(Reduced height 1)编码赤霉素信号转导途径的关键调控元件DELLA蛋白[14-16]。近年来,随着植物分子生物学和功能基因组学的发展,有关赤霉素信号转导以及GA-DELLA与其它激素和环境因子互作调控植物生长发育等研究领域取得了突破性进展。
1 赤霉素信号转导途径植物体内存在GA信号转导的分子网络。当赤霉素受体感知GA信号后,激活信号传递通道,调控下游基因的表达,从而影响植物生长发育和形态建成。近年来,随着拟南芥和水稻功能基因组学、蛋白组学、代谢组学及系统生物学的发展,人们已经鉴定了多个参与GA信号转导途径的正/负调控因子,其中主要包括GA受体GID1蛋白、信号转导途径的F-box蛋白及DELLA蛋白等[17-20],为GA介导DELLA蛋白降解的分子模型的建立和赤霉素作用分子机理的解析奠定了基础。
1.1 GID1蛋白:可溶性的GA受体植物是如何感知GA信号并将其传递下去,从而引发一系列的生理学效应的?可溶性GA受体GID1蛋白的发现可谓是GA信号研究领域取得的突破性进展。2005年,日本科学家Ueguchi-Tanaka等[18]证实水稻GID1蛋白是一种可溶性的赤霉素受体,它能与活性GA结合,感知并传递GA信号,从而诱发一系列下游反应。GID1基因编码一种类似于激素敏感相关的脂肪酶HSL(Hormone sensitive lipase)蛋白。虽然它含有保守的HSL基序(motif):HGG和GXSXG,但研究发现GID1蛋白本身并不具有水解酶活性。GID1蛋白能与水稻的DELLA蛋白SLR1(SLENDER RICE1)直接互作,而这种互作依赖于活性GA的存在[18]。GID1蛋白主要定位于细胞核及细胞质中,目前并没有发现GID1在细胞膜上的定位。因此,是否存在质膜定位的GA受体还需进一步研究。
Nakajima等[21-23]从拟南芥中克隆了水稻GID1的3个同源基因:分别是AtGID1a,AtGID1b和AtGID1c。它们对活性GA都具有较强的结合能力,同时在GA存在的条件下均能够直接与DELLA蛋白相互作用。由于这3个基因在功能上存在冗余,因此单突变体没有明显的GA不敏感的表型,而三突变体则对GA信号完全丧失响应且生长发育受到抑制。但是在不同的生长发育阶段,3个功能基因的表达模式及作用不尽相同,例如,AtGID1a在座果形成中发挥重要作用,而AtGID1b和AtGID1c分别在种子发育和果荚伸长等方面具有重要功能[24]。
赤霉素受体GID1蛋白晶体结构的阐述对揭示GID1生物学功能和GA信号转导途径的分子机制提供了重要信息。2008年,日本两个研究小组成功解析了水稻GID1-GA-DELLA复合体以及拟南芥GID1a-GA-DELLA复合体的晶体结构[25-26]。对GID1蛋白晶体结构的解析发现GID1是一个球状的单体蛋白,其C端的核心结构形成GA结合的口袋,N端延伸结构形成了盖住口袋的盖子。GID1蛋白与活性GA结合后形成类似于HSL的结构:它们都是一种α/β水解酶折叠结构,由中心8条β折叠且β折叠两侧由α螺旋紧密连接组成,GA与GID1的结合位点与HSL的活性位点一致[27]。根据复合体的晶体结构,科学家们提出了GID1蛋白的“变构学说”。活性GA和GID1蛋白C端的核心结构结合后,促使GID1蛋白N端盖住GA结合的口袋,并将GA分子包在结合它的口袋中(类似于关闭盖子),进而诱导GID1蛋白构象转变,产生可与DELLA蛋白结合的疏水表面,促进GID1-GA-DELLA蛋白复合体的形成[2]。
近来的研究表明,GID1蛋白能够被E3泛素连接酶GARU(GA receptor RING E3 ubiquitin ligase)泛素化并经由蛋白酶体途径降解[28]。GID1能够与GARU在体内互作。染料木黄酮(Genistein)是酪氨酸激酶的抑制剂,染料木黄酮处理增强GID1A的泛素化,然而GA处理则抑制其泛素化。植物酪氨酸激酶TAGK2是染料木黄酮的靶标,能磷酸化GARU 321位的酪氨酸(Tyr321)进而抑制GARU-GID1A的相互作用。染料木黄酮诱导GID1的降解及DELLA蛋白的积累。相反,garu突变及过表达TAGK2则会促进GID1稳定及DELLA蛋白的降解。因此GA的响应受GARU-依赖的GID1泛素化负调控,而TAGK2磷酸化GARU的酪氨酸则能正调控GA的响应[28]。这些研究结果进一步阐明了GID1蛋白的降解途径。
1.2 DELLA蛋白:GA信号途径中的调节子通过遗传筛选和生化研究,科学家们分离鉴定了GA信号通路中的关键组分DELLA蛋白,它主要起阻遏作用,其N端都含有保守的DELLA结构域。已分离的DELLA蛋白主要有玉米的d8(dwarf 8)、小麦的Rht1、水稻的SLR1、大麦的SLN1(SLENDER1)、葡萄的VvGAI及番茄中的PROCERA[29-30]。拟南芥中含有5个DELLA蛋白:GAI(GA insensitive)、RGA(repressor of ga1-3)、RGLl(RGA-Like1)、RGL2和RGL3[31-35]。此外,GAI和RGA蛋白与拟南芥的SCR(SCARECROW)蛋白在C端同源性较高,属植物特异的GRAS(GAI、RGA和SCR)蛋白家族成员[36]。
DELLA蛋白的基本结构如图 1所示,其N端具有典型的DELLA和VHYNP结构域,中间是多聚Ser/Thr/Val结构域,C端则是GRAS结构域。进一步的研究表明,DELLA和VHYNP结构域是GA信号响应的功能域,主要介导DELLA与GID1蛋白的相互作用[31, 37-38]。多聚Ser/Thr/Val(多聚S/T/V),通常作为磷酸化和糖基化的靶位点,是调节结构域。C端的GRAS结构域主要有两个亮氨酸重复序列LHR(Leu heptad repeat)及一个核定位信号结构域(nuclear localization signal,NLS)和3个保守的VHIID、PFYRE及SAW等结构域。亮氨酸重复(LHR)介导蛋白-蛋白间相互作用;而VHIID、PFYRE及SAW等结构域是阻遏结构域,介导GID1与F-box蛋白的相互作用[38]。
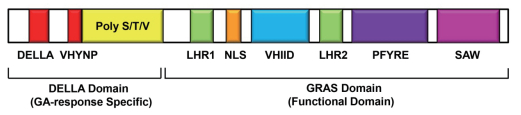
|
| 图 1 DELLA蛋白结构示意图 DELLA蛋白N端有两个非常保守的DELLA和VHYNP结构域,C端是GRAS结构域,含有一个核定位信号结构域(NLS)和两个亮氨酸重复序列LHR及3个保守的VHIID、PFYRE及SAW等结构域。Poly S/T/V,polymeric Ser and Thr and Val;LHR,Leu heptad repeat;NLS,nuclear localization signal |
DELLA结构域是GRAS蛋白家族中DELLA亚家族蛋白独有的,该结构域的功能与GA响应有关。拟南芥gai是一个对外源GA处理不敏感的半显性矮化突变体[39],它是由于GAI基因的一个51 bp碱基缺失突变,导致DELLA结构域中17个氨基酸丢失而造成。而RGA基因缺失相同的51 bp序列(rga-Δ17),其转基因拟南芥植株也会产生GA不敏感的严重矮化表型[31]。这些研究表明DELLA结构域的突变使gai(或者rga-Δ17)蛋白成为GA响应的组成型抑制子,阻遏GA介导的植物生长发育。GA促进植物的生长发育是通过解除DELLA蛋白的阻遏作用实现的,当DELLA蛋白感知GA信号后,DELLA蛋白的阻遏作用被解除,植株表现出正常生长发育。如果DELLA蛋白的GA响应功能域突变,使之不能感知GA信号,那么DELLA蛋白便组成性地阻遏植物生长发育,这类突变体表现为植株矮化、叶色深绿和晚花等类似GA缺失突变体表型,但对外源GA处理不敏感[34]。
1.3 DELLA蛋白降解依赖SCFSLY1/GID2/SNESCF(Skp1/cullin/F-box)复合体是26S蛋白酶体降解体系中的E3泛素连接酶,其中的F-box蛋白是SCF复合体的一个亚基,它决定了底物识别的特异性。在赤霉素信号转导中,DELLA蛋白的降解依赖泛素-蛋白酶体降解途径[40-41]。遗传学研究表明,拟南芥中的SLY1及水稻中的同源基因GID2编码F-box蛋白,是GA信号中的正调控因子[42]。
功能丧失的sly1突变体不能降解DELLA蛋白并且表现出GA不敏感的矮化表型[40]。DELLA蛋白能与SLY1蛋白在体内直接互作,表明DELLA蛋白是SLY1蛋白的直接靶标[42-43]。因此,SLY1蛋白作为SCFSLY1/GID2蛋白复合体组分之一,介导了GA诱导的DELLA蛋白的降解。当GA处理或有GA信号时,SLY1能特异地与DELLA蛋白发生亲和反应,通过26S蛋白酶体蛋白降解途径降解DELLA蛋白,从而去除DELLA蛋白的阻遏作用,实现GA的应答反应。
近来的研究表明,E3类泛素化(small ubiquitin-like modifier,SUMO)连接酶AtSIZ1能够与SLY1蛋白互作,并通过类泛素化SLY1进而正调控SLY1介导的GA信号[44]。与野生型相比,atsiz1-2突变体中的SLY1蛋白减少而DELLA蛋白增多。此外,GA促进SLY1的类泛素化,且类泛素化的SLY1与DELLA蛋白互作,调控DELLA蛋白的降解。这些研究表明SLY1的类泛素化对于GA介导的DELLA蛋白的降解也非常重要[44]。
拟南芥中SLY1的同源基因为SNEEZY(SNE)或SLEEPY2(SLY2),全长的SLY1与SNE基因在DNA及氨基酸序列上分别有55%及33%的相似性[45]。与sly1突变体相比,sly1 sne双突变体表现出严重矮化表型,且育性显著降低,表明SNE也是GA信号通路中功能冗余的正调控子[46]。此外,过量表达SNE(SLY2)能部分恢复sly1突变体的矮化表型,表明SNE在功能上能够替代SLY1[43, 47]。然而,在sly1突变体中过量表达SLY1能够导致RGA及RGL2蛋白的降解,但是在sly1突变体中过量表达SNE,只能导致RGA蛋白水平的降低,RGL2蛋白水平并不改变,表明SNE与SLY1的功能并不完成相同[45]。尽管AtSIZ1能与SLY1互作,但是AtSIZ1不能与SLY2互作,表明AtSIZ1以SLY1依赖的方式介导GA的响应[44]。
1.4 GID1介导DELLA蛋白降解的分子模型近年来,随着GID1、SLY1/GID2及DELLA蛋白功能研究的不断深入,人们对GA作用机理及其信号转导途径有了非常清晰的认识。在GA信号转导途径中,DELLA蛋白主要起阻遏作用并抑制植物生长发育,而GA促进植物生长发育是通过降解DELLA蛋白实现的。水稻GID2、拟南芥SLY1和SNE蛋白是SCFSLY1/GID2/SNE E3泛素连接酶蛋白复合体中的F-Box蛋白,而植物感知GA信号则依赖于GA受体GID1蛋白。当活性GA分子与受体GID1蛋白结合后,GA-GID1复合体与DELLA蛋白N端结合,导致DELLA蛋白构象改变并允许DELLA蛋白C端与SCFSLY1/GID2/SNE相互作用,GID1-GA-DELLA蛋白复合体的形成增强了DELLA与SCFSLY1/GID2/SNE间的互作,导致DELLA蛋白被泛素化,并经由26S蛋白酶复合体降解,进而解除DELLA蛋白的阻遏作用(图 2)[19-20, 48]。
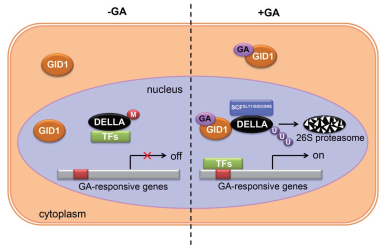
|
| 图 2 DELLA蛋白介导的GA信号转导模型 DELLA蛋白作为GA信号途径中的负调控因子,阻遏植物的生长发育。活性GA与GID1结合后,促进GID1与DELLA蛋白N端结合,导致DELLA蛋白构象改变,促进DELLA蛋白C端与SCFSLY1/GID2/SNE相互作用;GID1-GA-DELLA蛋白复合体形成,增强了DELLA与SCFSLY1/GID2/SNE间的互作,导致DELLA蛋白被泛素化,并经由26S蛋白酶复合体降解,进而解除DELLA蛋白的阻遏作用。U,ubiquitination of DELLA proteins。 |
尽管蛋白酶体依赖的DELLA蛋白降解途径能够解释大多数的GA响应,但是与GA合成缺陷的ga1-3突变体及gid1突变体相比,sly1及gid2突变体积累了更多的DELLA蛋白但并未表现出更加严重的矮化表型。研究表明,当存在有功能的DELLA结构域时,单独的GA和GID1蛋白就能去除DELLA蛋白的阻遏作用,而不需要SLY1/GID2介导的DELLA蛋白的降解[49]。过量表达GID1通过增加GA-GID1-DELLA复合体的形成进而解除了sly1-2突变体的种子休眠表型[50]。因此,可能存在另外一条不依赖于蛋白酶体降解的途径,这种途径可能是通过直接的蛋白-蛋白相互作用或者非直接地转录后修饰完成的[49, 51]。
近年来的研究表明,植物体内还存在磷酸化、糖基化和类泛素化等其他蛋白修饰作用影响DELLA蛋白活性。在水稻中的研究发现EL1(EARLY FLOWERING1),编码一种酪蛋白激酶,可以磷酸化SLR1,而SLR1中预测的磷酸化位点突变可以显著改变其对GA的响应,表明磷酸化的DELLA蛋白对于其活性和生物学功能具有重要的作用[52]。此外,磷酸酶TOPP4作为GA信号中的正调控因子,通过直接去磷酸化DELLA蛋白,进而促进GA介导的DELLA蛋白的降解[53]。糖基化修饰也在GA信号通路中起重要作用。SPY(SPINDLY)是GA信号途径负调控因子,SPY基因编码O-连N-乙酰基葡萄糖转移酶(O-linked N-acetylglucosamine(O-GlcNAc)transferases,OGTs),能O-GlcNAc糖基化修饰DELLA蛋白[54-57]。拟南芥的O-连N-乙酰基葡萄糖转移酶SECRET AGENT(SEC)也能够修饰DELLA蛋白。遗传学分析表明,SEC和SPY在调控GA信号中发挥的作用不同。SEC是GA响应的正调控因子,而SPY是负调控因子。在烟草细胞瞬时表达体系中,DELLA蛋白能够被SEC O-GlcNAc糖基化但不能被SPY O-GlcNAc糖基化。此外,O-GlcNAc糖基化的RGA抑制了其与互作蛋白PHYTOCHROME-INTERACTING FACTOR 3(PIF3)、PIF4、JA ZIM domain protein 1(JAZ1)及BRASSINAZOLE-RESISTANT 1(BZR1)的结合,表明O-GlcNAc糖基化修饰的DELLA蛋白能整合发育和环境信号精细调控GA响应[58]。最近研究表明,SPY作为新的O-岩藻糖转移酶(O-fucosyltransferase)可以O-岩藻糖基化修饰DELLA蛋白,这种O-岩藻糖基化修饰增强了DELLA蛋白的活性,促进了DELLA蛋白与PIF3、PIF4及BZR1互作,然而SEC O-GlcNAc糖基化修饰DELLA蛋白抑制了这种互作[59]。DELLA蛋白的类泛素化修饰(sumoylation)对于其活性和稳定性也起了重要作用。DELLA蛋白的羧基端能够被类泛素化修饰,GID1也含有类泛素化-互作基序。类泛素化的DELLA蛋白能够以GA非依赖的方式与GID1的类泛素化-互作基序互作,导致非类泛素化的DELLA蛋白积累,阻碍植物生长,以利于植物适应逆境环境[60]。
综上所述,磷酸化、糖基化和类泛素化等修饰作用对于DELLA蛋白的活性和稳定性具有重要的影响,不同修饰的DELLA蛋白整合发育和环境信号精细调控GA响应。
2 GA-DELLA整合其它激素和环境因子信号调控植物生长发育的机理越来越多的研究表明,DELLA蛋白作为GA信号途径中的调控因子,能够整合多种激素和环境信号调控植物的生长发育。DELLA蛋白缺少DNA结合结构域,且也没有直接的证据表明DELLA蛋白能够直接结合DNA。因此,DELLA蛋白调控靶基因的表达可能通过以下方式:一是,DELLA蛋白与转录因子互作,抑制转录因子的DNA结合活性,从而抑制靶基因的表达;二是,DELLA蛋白与其它的转录因子互作,作为转录激活子,调控下游靶基因的表达(图 3)[61-62]。此外,DELLA蛋白还可以作为非转录调控因子,调控植物生长发育。

|
| 图 3 DELLA整合其它激素和环境因子信号调控植物生长发育 DELLA蛋白作为转录激活子或者转录抑制子与其互作蛋白共同调控植物生长发育的多个进程 |
光和GA调控植物生长发育的多个过程。光诱导光形态建成,抑制下胚轴的伸长,然而GA促进黄化苗的生长及下胚轴的伸长。DELLA蛋白能够与bHLH转录因子家族的PIF3和PIF4互作,通过结合到这些转录因子的DNA结合区域,抑制PIFs靶基因的表达进而抑制下胚轴的伸长[63-64]。GA诱导DELLA蛋白的降解,释放出PIFs,促进PIFs激活基因的表达。因此,DELLA与PIFs互作整合GA和光信号进而调控植物的光形态建成(图 3)。
GA与油菜素内酯BR(Brassinosteroide)在调控植物的生长发育中也存在互作。在拟南芥中,转录因子BZR1及BRASSINOSTEROID-INSENSITIVE 1 EMS-SUPPRESSOR1(BES1)是BR信号途径中的正调控因子。DELLA蛋白与BZR1的直接互作阻碍了BZR1结合到BR调控基因的启动子区,从而抑制靶基因的表达,GA诱导DELLA蛋白降解后,释放出BZR1,解除了这种抑制[65-67]。此外,DELLA及BZR1能够与PIF4互作形成DELLA-BZR1-PIF4复合体,从而整合GA、BR及环境信号共同调控植物的生长发育[65, 68]。进一步研究表明,DELLA蛋白能够与生长素响应因子ARF6(Auxin response factor 6)互作,与BZR1及PIF4形成BZR1-ARF6-PIF-DELLA复合体,共同调控拟南芥下胚轴细胞的伸长[69](图 3)。
GA与乙烯信号在调控拟南芥顶端弯钩发育过程中也存在互作。乙烯通过促进下游转录因子ETHY-LENE INSENSITIVE3(EIN3)及EIN3-like(EIL)的积累,从而激活乙烯的信号响应。顶端弯钩形成的重要调控元件HLS1(HOOKLESS 1)是乙烯信号元件EIN3的直接靶基因。DELLA蛋白通过与EIN3/EIL1互作,抑制HLS1及RAP2.3(RELATED TO APETALA2.3)的表达及顶端弯钩的形成[70-71]。DE-LLA蛋白通过与EIN3/EIL1直接互作,抑制EIN3/EIL1的转录活性,从而拮抗乙烯的响应。DELLA-EIN3互作进一步揭示GA与乙烯共同调控顶端弯钩发育的作用机理(图 3)。
近来的研究表明,植物特异的转录因子TCP(TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR)参与独脚金内酯信号转导,能够抑制植物分枝[72-75]。DELLA蛋白能够与Ⅰ类的TCP转录因子(如TCP14和TCP15)互作,通过结合到TCP的DNA识别区域而阻碍TCP的功能[76-77](图 3)。在酵母中DELLA蛋白SLR1能够与独脚金内酯的受体D14(DWARF14)互作,表明GA信号与独脚金内酯也存在互作[78]。然而,DELLA-D14互作的功能还需要进一步研究。
茉莉酸(Jasmonic acid,JA)也是重要的植物激素,调控植物的生长发育及胁迫响应。研究表明GA和JA拮抗调控植物的生长发育及对生物及非生物胁迫的响应。JAZ是茉莉酸信号中的关键抑制子,JAZ1在JA信号途径中能与下游的关键正调控因子MYC2(JASMONATE INSENSITIVE1,JIN1/MYC2)相互作用,抑制MYC2对下游基因表达的调控作用。DELLA与JAZ互作抑制了JAZ的活性,增强了MYC2调控的JA介导的根的生长抑制及对病原菌Pst DC3000(Pseudomonas syringae pv tomato DC3000)的敏感性[79-81]。此外,DELLA和JAZ能够与WD-repeat/bHLH/MYB复合体中的成员GL3/EGL3及GL1互作,协同GA与JA信号调控毛状体的发育[82-84](图 3)。
在调控拟南芥开花方面,DELLA蛋白能够与转录因子SPL(SQUAMOSA PROMOTER BINDING-LIKE)互作,SPL通过激活miR172及MADS-box基因的表达进而促进开花。长日照条件下,DELLA蛋白与SPL蛋白互作干扰了SPL的转录活性,降低了叶中miR172的表达及顶端分生组织中MADS基因的表达,使得开花延迟[85](图 3)。在拟南芥中GA通过激活开花决定基因比如LEAFY(LFY)及SUPPRESSOR OF OVEREXPRESSION CONSTANS1(SOC1)进而促进开花。DELLA蛋白通过抑制LFY及SOC1的表达进而抑制短日照条件下的开花[86-89]。近来的研究表明,DELLA蛋白能够与重要的开花激活子CONSTANS(CO)互作,通过抑制CO的表达进而调控长日照条件下拟南芥的开花[90](图 3)。此外,DELLA蛋白能够与bHLH48及bHLH60互作,调控长日照条件下的开花[91]。DELLA蛋白也能够与WRKY12、WRKY13及WRKY75互作,调控拟南芥的开花时间[92-93]。其它DELLA互作蛋白包括BOTRYTIS SUSCEPTIBLE1 INTERACTOR(BOI),BOI-RELATED GENE1(BRG1),BRG2及BRG3,统称为BOIs,属于RING结构域蛋白家族。DELLA能与BOIs互作形成复合体,结合到GA响应基因的启动子区,抑制基因的响应,进而调控种子萌发、营养生长到生殖生长的转变及开花时间等[94]。
此外,在调控果实的发育方面,DELLA能够与bHLH家族成员ALCATRAZ(ALC)互作,抑制ALC靶基因的表达,调控离层的分化,进而调节果实的着生形态[95]。另有报道指出DELLA蛋白可以与同属于bHLH家族的SPT(SPATULA)相互作用,共同调控种子的萌发和子叶的扩张[96, 97](图 3)。
2.2 DELLA作为转录激活子整合激素和环境信号调控植物的生长发育除了作为转录抑制子,DELLA蛋白也能够作为转录激活子发挥作用。已有的研究表明,GA和ABA在调控种子的休眠和萌发中起拮抗作用。活性GA促进种子的萌发,而ABA却维持种子的休眠[98]。遗传学的研究表明,ABA信号途径中的转录因子ABSCISIC ACID INSENSITIVE 3(ABI3)及ABI5负调控种子的萌发[99]。近来研究表明,含有C3H锌指蛋白SOMNUS基因负调控种子的萌发,DELLA蛋白能与ABI3和ABI5蛋白互作形成复合体,结合在SOMNUS等基因的启动子区,激活这些基因的表达,从而调控逆境下种子的萌发[100]。DELLA蛋白与ABI3和ABI5互作揭示了GA与ABA互作调控种子萌发的机制(图 3)。
在拟南芥中SCL3(SCARECROW-LIKE 3)是GA信号中的正调控因子,SCL3的表达受DELLA的诱导但受GA的抑制。SCL3与DELLA蛋白互作通过拮抗DELLA蛋白的活性,进而调控根的生长和种子的萌发[101-102]。拟南芥的基因组中含有16个含锌指结构域的INDETERMINATE DOMAIN(IDD)蛋白[103],其中IDD1、IDD3、IDD5、IDD9、IDD10及IDD的同源蛋白GAF1能够与DELLA蛋白互作,调控GA响应[104-107]。IDD1与DELLA蛋白互作调控种子的休眠[104];IDD3、IDD5、IDD9及IDD10不仅与DELLA蛋白互作,也与SCL3互作。DELLA、SCL3及IDDs三个蛋白共同组成“共激活子/共抑制子调控系统”精细调控GA响应[61, 106](图 3)。在该系统中,DELLA蛋白作为共激活子,DELLA与IDD互作增强了SCL3的表达。此外,积累的SCL3作为共抑制子,SCL3-IDD蛋白复合体的增加抑制了SCL3自身的表达[61, 106]。
GA与细胞分裂素在调控植物的多个发育进程中发挥拮抗作用[108]。ARR1(Arabidopsis response regulator 1)是细胞分裂素信号通路中重要的转录因子,DELLA蛋白能够与ARR1互作,激活响应基因的表达,调控根的生长及光形态建成[109-110](图 3)。DELLA/ARR异二聚体揭示了GA与细胞分裂素互作的新调控机制,这与DELLA/IDD复合体类似,能转录激活目标基因的表达[106]。
2.3 DELLA作为非转录调控因子调控植物生长发育除了作为转录激活子或者转录抑制子外,DELLA蛋白还可作为非转录调控因子调控细胞的增殖。GA通过PFD(Prefoldin)复合体调控皮质微管Cortical Microtubule(cMT)的排列。PFD3及PFD5是微管蛋白伴侣分子,作用在胞质中协调细胞增殖过程中微管蛋白的组织及聚合[111]。PFD复合体在细胞核中不能发挥功能,但是可被转运至胞质中促进微管蛋白的折叠(tubulin folding)。DELLA蛋白能够与PFD3及PFD5互作,使PFD被限制在细胞核内并导致其不能在胞质中发挥功能。当有活性GA存在时,DELLA蛋白被降解,PFDs释放到胞质中促进微管蛋白二聚化[112]。因此,GA可以通过DELLA-PFD-tubulin folding-cMT聚合级联调控皮质微管的排列[113]。
除了调控微管蛋白的折叠外,DELLA蛋白作为非转录调控因子还参与到蛋白转运过程中。Salanenka等[114]的研究表明,GA能平衡蛋白质在液泡降解途径及返回细胞表面的转运过程。低GA水平促进转运物(包括PIN蛋白)向液泡传递和降解,而高GA水平促进它们再循环到质膜。GA的这种效应需要蛋白质逆向运输复合体(Retromer)组分,例如,Sorting Nexin 1(SNX1)及与它互作的微管相关蛋白CLASP1(the cytoplasmic linker-associated protein 1)。GA调控SNX1及CLASP1的亚细胞分布,且GA在蛋白质转运中的效应需要完整的微管细胞骨架。如前所述,DELLA蛋白能够与PFDs互作调控微管蛋白的折叠。因此,DELLA蛋白除了转录调控外,还可以通过其互作蛋白PFDs及下游的MT/CLASP1模块,调控蛋白质逆向运输复合体的活性及蛋白质从液泡途径到质膜的双向转运。通过这种机制,GA能够调控诸多转运蛋白和受体重新定位进出细胞表面,从而调控一系列细胞过程,包括生长过程中细胞的增殖。
综上所述,DELLA蛋白作为GA信号途径中的关键调控因子,通过与其它不同信号途径中的转录因子及非转录因子相互作用,影响下游基因的表达,从而实现对根的生长、下胚轴伸长、顶端弯钩的发育、开花、果实发育、植物光形态建成和防御反应等多种植物发育阶段和生理反应的调控。目前人们对这个复杂而精细的调控过程的认识还有待进一步阐明。
3 总结与展望GA-DELLA在调控植物的生长发育和胁迫反应中扮演了重要的作用。近年来的研究已从分子水平上揭示了GA通过GID1介导的DELLA蛋白的降解来促进植物的生长发育。尽管可溶性的GA受体GID1也存在细胞质中,但是GA-GID1-DELLA蛋白复合体定位在细胞核中。在谷物的糊粉层细胞中,GA诱导的α-淀粉酶的产生可能需要质膜定位的受体。因此,是否存在膜定位的依赖于GID1但是不依赖于DELLA蛋白的GA信号通路的共受体,还需进一步研究。去阻遏调控机制表明GA促进植物的生长发育是通过降解DELLA蛋白实现的。然而,对DELLA相关突变体的研究暗示,植物体内可能存在不依赖于DELLA蛋白的GA信号传递过程[115]。在Maymon等[116]的研究中,GA能够抵抗细胞分裂素对茎秆伸长的抑制作用,但是这个过程并不完全依赖于DELLA蛋白,因为DELLA缺失突变体global(缺失GAI、RGA、RGL1、RGL2及RGL3)对细胞分裂素并没有表现出完全不敏感的特点,暗示还有DELLA以外的因子参与该信号传递过程。此外,在研究global和ga1 global突变体时发现DELLA蛋白缺失并不等于GA作用完全饱和,global突变体仍然能够响应GA对单性结实果荚伸长的促进作用[117]。另外研究表明,GA诱导的细胞质中钙离子([Ca2+]cyt)的增加不依赖于DELLA蛋白的降解,在DELLA蛋白不能降解的转基因材料(RGAΔ17)及global突变体中,GA处理仍能快速增加细胞质中钙离子[118]。这些实验现象揭示,在已知的GA-DELLA信号调控模式之外,还可能存在未知的不依赖DELLA蛋白的信号调控模式参与GA响应,目前这种不依赖于DELLA蛋白的GA信号组分还需鉴定。DELLA蛋白的磷酸化、糖基化及类泛素化修饰对于DELLA蛋白的稳定性和活性至关重要,但是这些修饰的分子机制并不清楚。越来越多的证据表明,DELLA蛋白能够与各种转录因子互作,通过GA诱导的DELLA蛋白的降解,整合发育和环境信号调控细胞的分裂和分化。然而,GA动态调控DELLA相关复合体响应发育和环境信号的分子机制尚未可知,DELLA蛋白与其互作蛋白动态结合的调控机制仍需进一步阐述。水稻“绿色革命”基因sd1是赤霉素生物合成途径的一个关键酶,小麦“绿色革命”基因Rht1是赤霉素信号转导途径的关键成员——DELLA蛋白本身。今后利用遗传、生化及系统生物学的方法,进一步鉴定GA-DELLA响应的新组分及调控网络,加强对GA的生物学效应与其信号传递途径的分子机制研究,无论对于我们理解植物生长发育与GA的作用机理,还是对于赤霉素在农业生产中的应用,都具有非常重要的意义。
| [1] |
Daviere JM, de Lucas M, Prat S. Transcriptional factor interaction: a central step in DELLA function[J]. Curr Opin Genet Dev, 2008, 18: 295-303. DOI:10.1016/j.gde.2008.05.004 |
| [2] |
Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants[J]. Curr Biol, 2011, 21: R338-345. DOI:10.1016/j.cub.2011.02.036 |
| [3] |
Vera-Sirera F, Gomez MD, Perez-Amador MA. Chapter 20-DELLA proteins, a group of GRAS transcription regulators that mediate gibberellin signaling[J]. Plant Transcription Factors, 2016, 313-328. |
| [4] |
Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation[J]. Biochem J, 2012, 444: 11-25. DOI:10.1042/BJ20120245 |
| [5] |
Silverstone A, Sun T. Gibberellins and the green revolution[J]. Trends Plant Sci, 2000, 5: 1-2. DOI:10.1016/S1360-1385(99)01516-2 |
| [6] |
Magome H, Nomura T, Hanada A, et al. CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice[J]. Proc Natl Acad Sci USA, 2013, 110: 1947-1952. DOI:10.1073/pnas.1215788110 |
| [7] |
Nomura T, Magome H, Hanada A, et al. Functional analysis of Arabidopsis CYP714A1 and CYP714A2 reveals that they are distinct gibberellin modification enzymes[J]. Plant Cell Physiol, 2013, 54: 1837-1851. DOI:10.1093/pcp/pct125 |
| [8] |
Cowling RJ, Kamiya Y, Seto H, et al. Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis[J]. Plant Physiol, 1998, 117: 1195-1203. DOI:10.1104/pp.117.4.1195 |
| [9] |
Regnault T, Daviere JM, Achard P. Long-distance transport of endogenous gibberellins in Arabidopsis[J]. Plant Signal Behav, 2016, 11: e1110661. DOI:10.1080/15592324.2015.1110661 |
| [10] |
Binenbaum J, Weinstain R, Shani E. Gibberellin localization and transport in plants[J]. Trends Plant Sci, 2018, 23: 410-421. DOI:10.1016/j.tplants.2018.02.005 |
| [11] |
Tal I, Zhang Y, Jorgensen ME, et al. The Arabidopsis NPF3 protein is a GA transporter[J]. Nat Commun, 2016, 7: 11486. DOI:10.1038/ncomms11486 |
| [12] |
Saito H, Oikawa T, Hamamoto S, et al. The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis[J]. Nat Commun, 2015, 6: 6095. DOI:10.1038/ncomms7095 |
| [13] |
Kanno Y, Oikawa T, Chiba Y, et al. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes[J]. Nat Commun, 2016, 7: 13245. DOI:10.1038/ncomms13245 |
| [14] |
Peng J, Richards DE, Hartley NM, et al. 'Green revolution' genes encode mutant gibberellin response modulators[J]. Nature, 1999, 400: 256-261. DOI:10.1038/22307 |
| [15] |
Sasaki A, Ashikari M, Ueguchi-Tanaka M, et al. Green revolution: a mutant gibberellin-synthesis gene in rice[J]. Nature, 2002, 416: 701-702. DOI:10.1038/416701a |
| [16] |
Spielmeyer W, Ellis MH, Chandler PM. Semidwarf(sd-1), "green revolution" rice, contains a defective gibberellin 20-oxidase gene[J]. Proc Natl Acad Sci USA, 2002, 99: 9043-9048. DOI:10.1073/pnas.132266399 |
| [17] |
Sun T, Gubler F. Molecular mechanism of gibberellin signaling in plant[J]. Annual Review of Plant Biology, 2004, 55: 197-223. |
| [18] |
Ueguchi-Tanaka M, Ashikari M, Nakajima M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin[J]. Nature, 2005, 437: 693-698. DOI:10.1038/nature04028 |
| [19] |
Xu H, Liu Q, Yao T, et al. Shedding light on integrative GA signaling[J]. Curr Opin Plant Biol, 2014, 21: 89-95. DOI:10.1016/j.pbi.2014.06.010 |
| [20] |
Gao XH, Zhang YY, He ZH, et al. Chapter 4: Gibberellins[M]//Li JY, Li CY, Smith SM. Hormone Metabolism & Signaling in Plants. Elsevier, 2017: 107-160.
|
| [21] |
Griffiths J, Murase K, Rieu I, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis[J]. Plant Cell, 2006, 18: 3399-3414. |
| [22] |
Iuchi S, Suzuki H, Kim YC, et al. Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal[J]. Plant J, 2007, 50: 958-966. DOI:10.1111/j.1365-313X.2007.03098.x |
| [23] |
Nakajima M, Shimada A, Takashi Y, et al. Identification and characterization of Arabidopsis gibberellin receptors[J]. Plant J, 2006, 46: 880-889. DOI:10.1111/tpj.2006.46.issue-5 |
| [24] |
Gallego-Giraldo C, Hu J, Urbez C, et al. Role of the gibberellin receptors GID1 during fruit-set in Arabidopsis[J]. Plant J, 2014, 79: 1020-1032. DOI:10.1111/tpj.12603 |
| [25] |
Murase K, Hirano Y, Sun TP, et al. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1[J]. Nature, 2008, 456: 459-463. DOI:10.1038/nature07519 |
| [26] |
Shimada A, Ueguchi-Tanaka M, Nakatsu T, et al. Structural basis for gibberellin recognition by its receptor GID1[J]. Nature, 2008, 456: 520-523. DOI:10.1038/nature07546 |
| [27] |
Ueguchi-Tanaka M, Matsuoka M. The perception of gibberellins: clues from receptor structure[J]. Curr Opin Plant Biol, 2010, 503-508. |
| [28] |
Nemoto K, Ramadan A, Arimura GI, et al. Tyrosine phosphorylation of the GARU E3 ubiquitin ligase promotes gibberellin signalling by preventing GID1 degradation[J]. Nat Commun, 2017, 8: 1004. DOI:10.1038/s41467-017-01005-5 |
| [29] |
Boss PK, Thomas MR. Association of dwarfism and floral induction with a grape 'green revolution' mutation[J]. Nature, 2002, 416: 847-850. DOI:10.1038/416847a |
| [30] |
Olszewski N, Sun TP, Gubler F. Gibberellin signaling biosynthesis catabolism and response pathways[J]. The Plant Cell, 2002, 14: S61-S80. DOI:10.1105/tpc.010476 |
| [31] |
Dill A, Jung HS, Sun TP. The DELLA motif is essential for gibberellin-induced degradation of RGA[J]. Proc Natl Acad Sci USA, 2001, 98: 14162-14167. DOI:10.1073/pnas.251534098 |
| [32] |
Lee S, Cheng H, King KE, et al. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition[J]. Genes Dev, 2002, 16: 646-658. DOI:10.1101/gad.969002 |
| [33] |
Wen CK, Chang C. Arabidopsis RGL1 encodes a negative regulator of gibberellin responses[J]. Plant Cell, 2002, 14: 87-100. DOI:10.1105/tpc.010325 |
| [34] |
Peng J, Carol P, Richards DE, et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses[J]. Genes Dev, 1997, 11: 3194-3205. DOI:10.1101/gad.11.23.3194 |
| [35] |
Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway[J]. Plant Cell, 1998, 10: 155-169. DOI:10.1105/tpc.10.2.155 |
| [36] |
Pysh LD, Wysocka-Diller JW, Camilleri C, et al. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes[J]. Plant J, 1999, 18: 111-119. DOI:10.1046/j.1365-313X.1999.00431.x |
| [37] |
Asano K, Hirano K, Ueguchi-Tanaka M, et al. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice[J]. Mol Genet Genomics, 2009, 281: 223-231. DOI:10.1007/s00438-008-0406-6 |
| [38] |
Hirano K, Asano K, Tsuji H, et al. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice[J]. Plant Cell, 2010, 22: 2680-2696. DOI:10.1105/tpc.110.075549 |
| [39] |
Koornneef M, Elgersma A, Hanhart CJ, et al. A gibberellin insensitive mutant of Arabidopsis thaliana[J]. Physiol Plant, 1985, 65. |
| [40] |
McGinnis KM, Thomas SG, Soule JD, et al. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase[J]. Plant Cell, 2003, 15: 1120-1130. DOI:10.1105/tpc.010827 |
| [41] |
Wang F, Deng XW. Plant ubiquitin-proteasome pathway and its role in gibberellin signaling[J]. Cell Res, 2011, 21: 1286-1294. DOI:10.1038/cr.2011.118 |
| [42] |
Dill A, Thomas SG, Hu J, et al. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation[J]. Plant Cell, 2004, 16: 1392-1405. DOI:10.1105/tpc.020958 |
| [43] |
Fu X, Richards DE, Fleck B, et al. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates[J]. Plant Cell, 2004, 16: 1406-1418. DOI:10.1105/tpc.021386 |
| [44] |
Kim SI, Park BS, Kim do Y, et al. E3 SUMO ligase AtSIZ1 positively regulates SLY1-mediated GA signalling and plant development[J]. Biochem J, 2015, 469: 299-314. DOI:10.1042/BJ20141302 |
| [45] |
Ariizumi T, Lawrence PK, Steber CM. The role of two f-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol[J]. 2011, 155: 765-775.
|
| [46] |
Ariizumi T, Steber CM. Mutations in the F-box gene SNEEZY result in decreased Arabidopsis GA signaling[J]. Plant Signal Behav, 2011, 6: 831-833. DOI:10.4161/psb.6.6.15164 |
| [47] |
Strader LC, Ritchie S, Soule JD, et al. Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY[J]. Proc Natl Acad Sci USA, 2004, 101: 12771-12776. DOI:10.1073/pnas.0404287101 |
| [48] |
Daviere JM, Achard P. A pivotal role of DELLAs in regulating multiple hormone signals[J]. Mol Plant, 2015, 9: 10-20. |
| [49] |
Ariizumi T, Murase K, Sun TP, et al. Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1[J]. Plant Cell, 2008, 20: 2447-2459. DOI:10.1105/tpc.108.058487 |
| [50] |
Ariizumi T, Hauvermale AL, Nelson SK, et al. Lifting della repression of Arabidopsis seed germination by nonproteolytic gibberellin signaling[J]. Plant Physiol, 2013, 162: 2125-2139. DOI:10.1104/pp.113.219451 |
| [51] |
Ueguchi-tanaka M, Hirano K, Hasegawa Y, et al. Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant[J]. Plant Cell, 2008, 20: 2437-2446. DOI:10.1105/tpc.108.061648 |
| [52] |
Dai C, Xue HW. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling[J]. EMBO J, 2010, 29: 1916-1927. DOI:10.1038/emboj.2010.75 |
| [53] |
Qin Q, Wang W, Guo X, et al. Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4[J]. PLoS Genet, 2014, 10: e1004464. DOI:10.1371/journal.pgen.1004464 |
| [54] |
Filardo F, Robertson M, Singh DP, et al. Functional analysis of HvSPY, a negative regulator of GA response, in barley aleurone cells and Arabidopsis[J]. Planta, 2009, 229: 523-537. DOI:10.1007/s00425-008-0843-9 |
| [55] |
Shimada A, Ueguchi-Tanaka M, Sakamoto T, et al. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis[J]. Plant J, 2006, 48: 390-402. DOI:10.1111/tpj.2006.48.issue-3 |
| [56] |
Swain SM, Tseng TS, Olszewski NE. Altered expression of SPINDLY affects gibberellin response and plant development[J]. Plant Physiol, 2001, 126: 1174-1185. |
| [57] |
Swain SM, Tseng TS, Thornton TM, et al. SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant[J]. Plant Physiol, 2002, 129: 605-615. DOI:10.1104/pp.020002 |
| [58] |
Zentella R, Hu J, Hsieh WP, et al. O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis[J]. Genes Dev, 2016, 30: 164-176. DOI:10.1101/gad.270587.115 |
| [59] |
Zentella R, Sui N, Barnhill B, et al. The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA[J]. Nat Chem Biol, 2017, 13: 479-485. DOI:10.1038/nchembio.2320 |
| [60] |
Conti L, Nelis S, Zhang C, et al. Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin[J]. Dev Cell, 2014, 28: 102-110. DOI:10.1016/j.devcel.2013.12.004 |
| [61] |
Yoshida H, Ueguchi-Tanaka M. DELLA and SCL3 balance gibberellin feedback regulation by utilizing INDETERMINATE DOMAIN proteins as transcriptional scaffolds[J]. Plant Signal Behav, 2014, 9: e29726. DOI:10.4161/psb.29726 |
| [62] |
Van De Velde K, Ruelens P, Geuten K, et al. Exploiting DELLA signaling in cereals[J]. Trends Plant Sci, 2017, 22: 880-893. |
| [63] |
de Lucas M, Daviere JM, Rodriguez-Falcon M, et al. A molecular framework for light and gibberellin control of cell elongation[J]. Nature, 2008, 451: 480-484. DOI:10.1038/nature06520 |
| [64] |
Feng S, Martinez C, Gusmaroli G, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins[J]. Nature, 2008, 451: 475-479. |
| [65] |
Bai MY, Shang JX, Oh E, et al. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis[J]. Nat Cell Biol, 2012, 14: 810-817. DOI:10.1038/ncb2546 |
| [66] |
Gallego-Bartolome J, Minguet EG, Grau-Enguix F, et al. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis[J]. Proc Natl Acad Sci USA, 2012, 109: 13446-13451. DOI:10.1073/pnas.1119992109 |
| [67] |
Li QF, Wang C, Jiang L, et al. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis[J]. Sci Signal, 2012, 5: ra72. |
| [68] |
Bernardo-Garcia S, de Lucas M, Martinez C, et al. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth[J]. Genes Dev, 2014, 28: 1681-1694. DOI:10.1101/gad.243675.114 |
| [69] |
Oh E, Zhu JY, Bai MY, et al. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl[J]. Elife, 2014, 3. |
| [70] |
An F, Zhang X, Zhu Z, et al. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings[J]. Cell Res, 2012, 22: 915-927. DOI:10.1038/cr.2012.29 |
| [71] |
Marín-de la Rosa N, Sotillo B, Miskolczi P, et al. Large-scale identification of gibberellin-related transcription factors defines group Ⅶ ETHYLENE RESPONSE FACTORS as functional DELLA partners[J]. Plant Physiol, 2014, 166: 1022-1032. DOI:10.1104/pp.114.244723 |
| [72] |
Braun N, de Saint Germain A, Pillot JP, et al. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching[J]. Plant Physiol, 2012, 158: 225-238. DOI:10.1104/pp.111.182725 |
| [73] |
Drummond RS, Janssen BJ, Luo Z, et al. Environmental control of branching in petunia[J]. Plant Physiol, 2015, 168: 735-751. DOI:10.1104/pp.15.00486 |
| [74] |
Guan JC, Koch KE, Suzuki M, et al. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork[J]. Plant Physiol, 2012, 160: 1303-1317. DOI:10.1104/pp.112.204503 |
| [75] |
Rameau C, Bertheloot J, Leduc N, et al. Multiple pathways regulate shoot branching[J]. Front Plant Sci, 2015, 5: 741. |
| [76] |
Daviere JM, Wild M, Regnault T, et al. Class Ⅰ TCP-DELLA interactions in inflorescence shoot apex determine plant height[J]. Curr Biol, 2014, 24: 1923-1928. DOI:10.1016/j.cub.2014.07.012 |
| [77] |
Resentini F, Felipo-Benavent A, Colombo L, et al. TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana[J]. Mol Plant, 2015, 8: 482-485. DOI:10.1016/j.molp.2014.11.018 |
| [78] |
Nakamura H, Xue YL, Miyakawa T, et al. Molecular mechanism of strigolactone perception by DWARF14[J]. Nat Commun, 2013, 4: 2613. DOI:10.1038/ncomms3613 |
| [79] |
Hou X, Lee LY, Xia K, et al. DELLAs modulate jasmonate signaling via competitive binding to JAZs[J]. Dev Cell, 2010, 19: 884-894. DOI:10.1016/j.devcel.2010.10.024 |
| [80] |
Wild M, Daviere JM, Cheminant S, et al. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses[J]. Plant Cell, 2012, 24: 3307-3319. DOI:10.1105/tpc.112.101428 |
| [81] |
Yang DL, Yao J, Mei CS, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade[J]. PNAS, 2012, E1192-E1200. |
| [82] |
Grebe M. The patterning of epidermal hairs in Arabidopsis--updated[J]. Curr Opin Plant Biol, 2012, 15: 31-37. DOI:10.1016/j.pbi.2011.10.010 |
| [83] |
Qi T, Song S, Ren Q, et al. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana[J]. Plant Cell, 2011, 23: 1795-1814. DOI:10.1105/tpc.111.083261 |
| [84] |
Qi T, Huang H, Wu D, et al. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy[J]. Plant Cell, 2014, 26: 1118-1133. DOI:10.1105/tpc.113.121731 |
| [85] |
Yu S, Galvao VC, Zhang YC, et al. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors[J]. Plant Cell, 2012, 24: 3320-3332. DOI:10.1105/tpc.112.101014 |
| [86] |
Achard P, Herr A, Baulcombe DC, et al. Modulation of floral development by a gibberellin-regulatedmicroRNA[J]. Development, 2004, 131: 3357-3365. DOI:10.1242/dev.01206 |
| [87] |
Achard P, Liao L, Jiang C, et al. DELLAs contribute to plant photomorphogenesis[J]. Plant Physiol, 2007, 143: 1163-1172. DOI:10.1104/pp.106.092254 |
| [88] |
Moon J, Suh SS, Lee H, et al. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis[J]. Plant J, 2003, 35: 613-623. DOI:10.1046/j.1365-313X.2003.01833.x |
| [89] |
Mutasa-Göttgens E, Hedden P. Gibberellin as a factor in floral regulatory networks[J]. J Exp Bot, 2009, 60: 1979-1989. DOI:10.1093/jxb/erp040 |
| [90] |
Xu F, Li T, Xu PB, et al. DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis[J]. FEBS Lett, 2016, 590: 541-549. DOI:10.1002/1873-3468.12076 |
| [91] |
Li Y, Wang H, Li X, et al. Two DELLA-interacting proteins bHLH48 and bHLH60 regulate flowering under long-day conditions in Arabidopsis thaliana[J]. J Exp Bot, 2017, 68: 2757-2767. DOI:10.1093/jxb/erx143 |
| [92] |
Li W, Wang H, Yu D. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions[J]. Mol Plant, 2016, 9: 1492-1503. DOI:10.1016/j.molp.2016.08.003 |
| [93] |
Zhang L, Chen L, Yu D. Transcription Factor WRKY75 Interacts with DELLA proteins to affect flowering[J]. Plant Physiol, 2018, 176: 790-803. DOI:10.1104/pp.17.00657 |
| [94] |
Park J, Nguyen KT, Park E, et al. DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis[J]. Plant Cell, 2013, 25: 927-943. DOI:10.1105/tpc.112.108951 |
| [95] |
Arnaud N, Girin T, Sorefan K, et al. Gibberellins control fruit patterning in Arabidopsis thaliana[J]. Genes Dev, 2010, 24: 2127-2132. DOI:10.1101/gad.593410 |
| [96] |
Gallego-Bartolomé J, Minguet EG, Marin JA, et al. Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis[J]. Mol Biol Evol, 2010, 27: 1247-1256. DOI:10.1093/molbev/msq012 |
| [97] |
Josse EM, Gan Y, Bou-Torrent J, et al. A DELLA in Disguise: SPATULA restrains the growth of the developing Arabidopsis seedling[J]. Plant Cell, 2011, 23: 1337-1351. DOI:10.1105/tpc.110.082594 |
| [98] |
Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination[J]. Curr Opin Plant Biol, 2002, 5: 33-36. DOI:10.1016/S1369-5266(01)00219-9 |
| [99] |
Piskurewicz U, Jikumaru Y, Kinoshita N, et al. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity[J]. Plant Cell, 2008, 20: 2729-2745. DOI:10.1105/tpc.108.061515 |
| [100] |
Lim S, Park J, Lee N, et al. ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis[J]. Plant Cell, 2013, 25: 4863-4878. DOI:10.1105/tpc.113.118604 |
| [101] |
Heo JO, Chang KS, Kim IA, et al. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root[J]. Proc Natl Acad Sci USA, 2011, 108: 2166-2171. DOI:10.1073/pnas.1012215108 |
| [102] |
Zhang ZL, Ogawa M, Fleet CM, et al. Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis[J]. Proc Natl Acad Sci USA, 2011, 108: 2160-2165. DOI:10.1073/pnas.1012232108 |
| [103] |
Colasanti J, Tremblay R, Wong AY, et al. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants[J]. BMC Genomics, 2006, 7: 158. DOI:10.1186/1471-2164-7-158 |
| [104] |
Feurtado JA, Huang D, Wicki-Stordeur L, et al. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation[J]. Plant Cell, 2011, 23: 1772-1794. DOI:10.1105/tpc.111.085134 |
| [105] |
Fukazawa J, Teramura H, Murakoshi S, et al. DELLAs Function as coactivators of GAI-ASSOCIATED FACTOR1 in regulation of gibberellin homeostasis and signaling in Arabidopsis[J]. The Plant Cell, 2014, 26: 2920-2938. DOI:10.1105/tpc.114.125690 |
| [106] |
Yoshida H, Hirano K, Sato T, et al. DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins[J]. Proc Natl Acad Sci USA, 2014, 111: 7861-7866. DOI:10.1073/pnas.1321669111 |
| [107] |
Fukazawa J, Mori M, Watanabe S, et al. DELLA-GAF1 complex is a main component in gibberellin feedback regulation of GA20 oxidase 2[J]. Plant Physiol, 2017, 175: 1395-1406. DOI:10.1104/pp.17.00282 |
| [108] |
Weiss D, Ori N. Mechanisms of cross talk between gibberellin and other hormones[J]. Plant Physiol, 2007, 144: 1240-1246. DOI:10.1104/pp.107.100370 |
| [109] |
Marín-de la Rosa N, Pfeiffer A, Hill K, et al. Genome wide binding site analysis reveals transcriptional coactivation of Cytokinin-responsive genes by DELLA proteins[J]. PLoS Genet, 2015, 11: e1005337. DOI:10.1371/journal.pgen.1005337 |
| [110] |
Moubayidin L, Perilli S, Dello Ioio R, et al. The rate of cell differentiation controls the Arabidopsis root meristem growth phase[J]. Curr Biol, 2010, 20: 1138-1143. DOI:10.1016/j.cub.2010.05.035 |
| [111] |
Rodriguez-Milla MA, Salinas J. Prefoldins 3 and 5 play an essential role in Arabidopsis tolerance to salt stress[J]. Mol Plant, 2009, 2: 526-534. DOI:10.1093/mp/ssp016 |
| [112] |
Locascio A, Blazquez MA, Alabadi D. Dynamic regulation of cortical microtubule organization through prefoldin-DELLA interaction[J]. Curr Biol, 2013, 23: 804-809. DOI:10.1016/j.cub.2013.03.053 |
| [113] |
Chen X, Wu S, Liu Z, et al. Environmental and endogenous control of cortical microtubule orientation[J]. Trends Cell Biol, 2016, 26: 409-419. DOI:10.1016/j.tcb.2016.02.003 |
| [114] |
Salanenka Y, Verstraeten I, Lofke C, et al. Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane[J]. Proc Natl Acad Sci USA, 2018, 115: 3716-3721. DOI:10.1073/pnas.1721760115 |
| [115] |
Ito T, Okada K, Fukazawa J, et al. DELLA-dependent and -independent gibberellin signaling[J]. Plant Signal Behav, 2018, e1445933. |
| [116] |
Maymon I, Greenboim-Wainberg Y, Sagiv S, et al. Cytosolic activity of SPINDLY implies the existence of a DELLA-independent gibberellin-response pathway[J]. Plant J, 2009, 58: 979-988. DOI:10.1111/tpj.2009.58.issue-6 |
| [117] |
Fuentes S, Ljung K, Sorefan K, et al. Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses[J]. Plant Cell, 2012, 24: 3982-3996. DOI:10.1105/tpc.112.103192 |
| [118] |
Okada K, Ito T, Fukazawa J, et al. Gibberellin induces an increase in cytosolic Ca2+ via a DELLA-independent signaling pathway[J]. Plant Physiol, 2017, 175: 1536-1542. DOI:10.1104/pp.17.01433 |




