2. 中国烟草中南农业试验站,长沙 410004;
3. 湖南农业大学生物科学技术学院,长沙 410128
2. Central-South Agricultural Experiment Station of China Tobacco, Changsha 410004;
3. College of Biological Science and Technology, Hunan Agricultural University, Changsha 410128
植物受多种非生物胁迫影响,低温是影响植物生长发育以及地理分布的主要限制因素。低温刺激导致植物体内的各生理代谢活动紊乱,表型上产生一系列的生理变化如苗期早花,植株矮小等。相关研究表明转录因子在植物信号调控网络中具有重要作用,转录因子可以通过识别下游基因启动子上的功能元件来激活或者抑制目标基因表达,引起植物体内的低温相关代谢途径产生变化,从而改变植物对于外界低温胁迫刺激的适应能力[1-3]。本文着重从转录因子调控低温胁迫相关基因的转录表达过程来分析植物体内的抗低温转导途径,进一步揭示在低温逆境条件下的相关基因的表达调控机理,为今后的逆境研究工作提供了众多线索与依据。
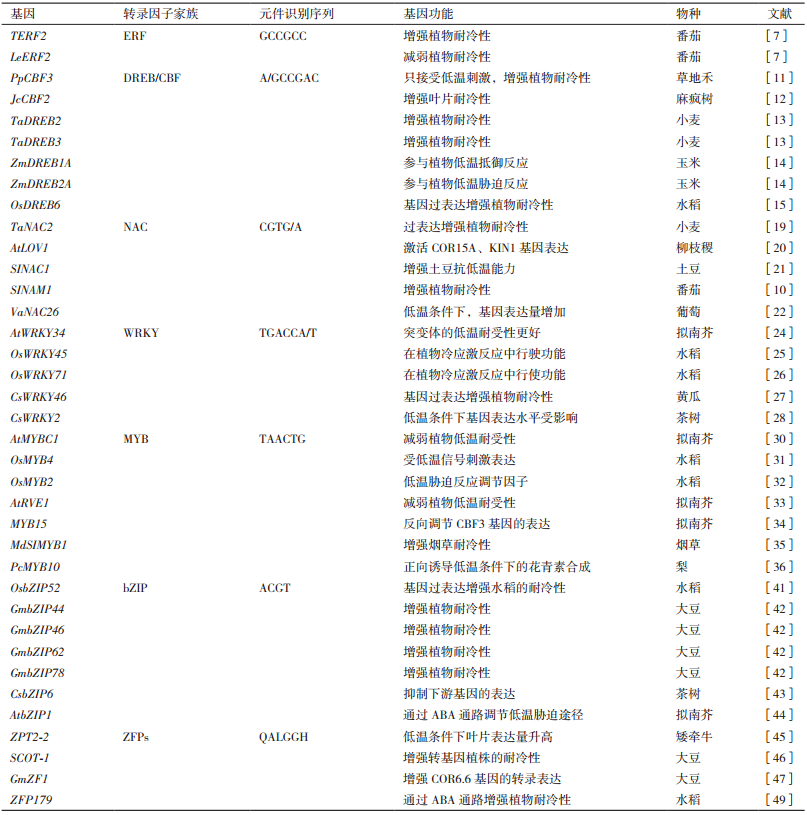
1 转录因子在低温胁迫中的研究转录因子的转录调控是植物对低温胁迫反应的关键部分。目前,参与低温信号响应的转录因子有AP2/ERF、NAC、WRKY、MYB、bZIP和ZFPs等[4]。每个转录因子基本包含4个独立的结构域,即DNA结合域、转录功能域、核定位信号和寡聚化位点[5],转录因子在表达调控网络中的主要功能由其转录功能域决定。
1.1 AP2/ERF家族转录因子在低温胁迫中的研究 1.1.1 AP2/ERF转录因子家族低温诱导AP2/ERF家族转录因子表达,激活或抑制下游低温响应基因表达[6]。AP2/ERF家族成员分为AP2、RAV、ERF和DREB四类,DREBs和ERFs转录因子家族是AP2/ERF转录因子家族最重要的亚家族。ERF转录因子可以直接结合下游基因启动子上的GCC盒(GCCGCC)调控其表达。在番茄中发现了ERF家族基因TERF2和LeERF2,过表达或敲除TERF2/LeERF2可以增强或减弱蕃茄低温耐受力[7]。有研究表明,DREB/CBF途径是植物中公认的关键的低温信号调节通路,DREB转录因子通过识别与结合低温响应基因的启动子上顺式作用元件(A/GCCGAC)激活其表达[8]。同时ICE-CBF通路又是DREB/CBF反应途径中关键的调节通路,ICE基因可以结合CBF基因启动子上的MYC元件(CACATG)调控下游基因的转录表达[9]。相关研究表明,当植物接收到低温信号时,CBF基因受到ICE的刺激表达上调,直接结合低温调节基因启动子上的CRT/DRE顺式元件,引起低温响应基因的转录产物和后续合成的蛋白质产量的增多[10]。
在草地禾中发现的PpCBF3只接收低温刺激,在拟南芥中过表达PpCBF3增强了自身的耐低温性[11]。在麻风树基因组克隆得到JcCBF2,RT-PCR结果表明,JcCBF2在低温胁迫早期快速调节低温响应基因,过表达植株的叶片对于低温耐受力比野生型更好[12]。小麦TaDREB2和TaDREB3在大麦中表达,明显增强了大麦的耐低温能力,LEA/COR/DHN的表达量也明显上调[13]。在玉米中发现的2个同源基因ZmDREB1A和ZmDREB2A也参与了植物体内的低温信号转导途径[14]。在水稻日本晴中发现了DREB基因OsDREB6,酵母单杂交分析表明,OsDREB6特异结合CRT/DRE顺式结合元件激活转录调控,过表达OsDREB6增强转基因水稻低温耐性,发现RNA干扰型比野生型对于低温更敏感[15]。
1.1.2 NAC转录因子家族NAC参与调节植物的发育过程和环境胁迫应答的转录因子[16]。NAC家族基因包括了N末端高度保守的DNA结合域和C末端可变的转录调控区[16-17]。在拟南芥中已被证实NAC蛋白通过识别并结合核心序列(CGTG/A)调控下游低温胁迫相关基因的表达[18]。转基因拟南芥过表达TaNAC2提高植物自身的耐低温能力[19]。AtLOV1通过激活COR15A和KIN1的表达增强拟南芥的耐低温性[20]。过表达SINAC1提高马铃薯的耐低温能力,同时检测到体内的CBF基因表达量上调,ROS积累量减少[21]。番茄NAC转录因子SINAM1转入烟草,可以提高其自身耐低温能力,说明SINAM1的表达增强了植物的耐低温性[10]。低温时,葡萄体VaNAC26表达上调[22]。以上研究表明,NAC转录因子参与低温代谢途径的调控,而且多数基因具有正向调节作用。
1.1.3 WRKY转录因子家族WRKY转录因子家族是最大的高等植物转录因子家族。它可以结合目标基因启动子上的作用元件W-BOX(TGACCA/T),调控下游目的基因表达,从而调节植物低温应激反应。WRKY转录因子编码的蛋白质通常表现为抑制作用,低温处理后下游靶基因表达量下调[23]。AtWRKY34在花粉中特异性表达,在低温处理后基因表达量上调并且反向调节拟南芥成熟花粉低温敏感性[24]。水稻OsWRKY45和OsWRKY76被证明在植物低温应激反应过程中行使功能[25-26]。过表达黄瓜CsWRKY46的拟南芥低温耐受性增强,体内脯氨酸含量增加,丙二醛含量减少,电解质渗漏水平降低,说明CsWRKY46是植物提高低温耐受能力的正向调节因子[27]。有研究表明茶树CsWRKY2表达水平受ABA和低温调控。低温刺激下,外源ABA引起CsWRKY2表达量上调,表明CsWRKY2通过参与ABA信号转导途径响应低温胁迫[28]。
1.1.4 MYB转录因子家族MYB转录因子家族是目前基因数最为庞大的植物转录因子家族。根据其结构内保守的DNA结合域(MYB结构域)命名。MYB结合元件(TAACTG)是MYB转录因子结合目的基因的核心序列[29]。AtMYBC1在CBF通路中减弱植物低温耐受力[30]。OsMYB4受低温信号刺激表达,转OsMYB4拟南芥对低温胁迫具有较强耐受力[31]。HOS10编码重要的胁迫反应协调因子OsMYB2蛋白[32]。拟南芥MYB基因AtRVE1负向调节植物的低温耐受能力[33]。拟南芥MYB15可以结合CBF3基因启动子上的作用元件并且抑制其表达[34]。过表达烟草MdSIMYB1可以增强烟草的耐低温性[35]。梨PcMYB10正向诱导低温状态下花青素的合成[36]。以上结果表明,该转录因子家族成员能够较为明显提高植物低温耐能力。
1.1.5 bZIP转录因子家族bZIP转录因子在低温胁迫中扮演重要的角色。bZIP蛋白的保守序列(PYC-GTGG)可以特异性结合目标基因启动子上的ABRE元件(ACGT)调控下游低温响应基因表达[37-38]。水稻OsbZIP52被证实参与低温胁迫调控,酵母双杂交试验证明OsbZIP52通过识别并结合下游目标基因启动子上的G-BOX激活其转录表达,同时证明过表达OsbZIP52可以增强水稻自身低温敏感性[39-41]。在大豆中发现的bZIP基因被命名为GmbZIPs,在拟南芥中分别过表达GmbZIP44、GmbZIP46、GmbZIP62和GmbZIP78,拟南芥的耐低温能力均被增强[42]。CsbZIP6是茶树低温应激反应的负调控因子,导致下游低温响应基因表达量下调并且影响茶树对ABA的敏感性[43]。拟南芥基因AtbZIP1结合ABRE作用元件,通过ABA依赖的信号传导通路调节植物的低温胁迫应答[44],表明bZIP转录因子通过ABA依赖途径调控植物低温胁迫应激。
1.1.6 ZFPs转录因子家族ZFPs构成真核生物转录调节因子的最大家族之一,家族成员均包含保守的锌指结构域(QALGGH)。C2H2锌指蛋白是植物体内基因对抵御和适应胁迫环境的关键转录阻遏物。在矮牵牛中发现的转录因子ZPT2-2,包含了2个C2H2型锌指结构域和一个转录抑制结构域,低温处理后该基因在叶片的表达量明显上调[45]。大豆基因SCOF-1具有提高转基因植物的抗低温能力的积极作用[46]。在大豆中新发现的GmZF1蛋白,拟南芥中过表达GmZF1激活下游COR6.6的表达,提高转基因植株的耐低温能力[47],由此证明GmZF1是通过激活COR6.6在拟南芥响应低温信号通路中具有调控作用。
ZFP179通过参与ABA依赖和ABA独立的途径,来增强植物本身抗非生物逆境胁迫能力,其中耐低温能力增强效果最为显著[49]。说明锌指蛋白这一类转录因子对植物抗低温胁迫也具有一定作用。
1.2 低温胁迫下第二信使和激素分子的调控表达Ca2+是重要的第二信使分子之一,当植物受到外界低温胁迫的刺激后,COLD1与G蛋白相互作用感知低温并激活Ca2+通道,使细胞内钙离子浓度增加。钙离子通过钙调蛋白结合转录因子CAMTA家族的成员与CM2序列结合,其中CAMTA3转录因子是CBF2表达的正调控因子[50-51]。
低温胁迫条件下,乙烯在水稻低温信号转导通路中与AP2 / ERF家族成员相互作用,APETALA2/乙烯响应因子(AP2 / ERF)是认为在水稻胁迫应答途径中起作用的转录因子。在乙烯信号传导途径中,乙烯通过与初级乙烯反应元件(PERE)结合后,PERE再与EIN3和EIL1结合。EIN3和EIL1是介导乙烯信号的中心转录因子,能调节乙烯反应蛋白的表达,其中包括转录因子乙烯反应因子1(ERF1)[52]。
此外,EIN3可以结合CBF以ARR5/7/15的启动子区域,乙烯负向调控低温信号通过EIN3直接转录控制低温调控CBF基因的表达[53]。有研究表明JAZ蛋白是茉莉酸信号传导的抑制因子,与ICE1和ICE2转录因子发生物理相互作用,其中JAZ1和JAZ4抑制ICE1的转录功能,从而减弱调节表达。茉莉酸还可以通过EIN3和EIL1负调节CBF/DREB1基因的表达[54-55]。
2 低温响应相关启动子及其功能元件的分析与鉴定核心启动子含有多个顺式作用元件,转录因子的作用是特异性结合目的基因核心启动子上顺式作用元件,调控下游基因表达[61-63]。在转录起始前通用转录因子TFIID、TFIIB与RNA聚合酶Ⅱ形成蛋白复合物,转录过程中,转录因子特异性结合作用元件,同时与转录复合体相互作用,调节基因的转录水平[64]。表 1中列举一些受转录因子调控的参与低温胁迫的靶基因及其相关功能。
目前,常用的查找和鉴定启动子的数据库有Gene MANIA、Plant CARE、PLACE、Plant PAN、Soft Berry、Genomatix和Matinspector[65]。使用Plant CARE和Gene MANIA网站可以预测和建立可能存在的分子调控网络。植物体内的顺式作用元件数量较多,目前被确定参与植物低温胁迫响应的顺式作用元件有ERF、ABRE、DRE、CRT、W-BOX和LTRE等[64]。拟南芥中CBF基因可以结合COR和DRE基因启动子上的CRT/DRE的顺式作用元件[66]。缺失分析LRK6(亮氨酸受体激酶)启动子的差分序列DSLP2,酵母单杂交表明与其互作的ERF蛋白是OsERF3。通过序列分析和致突变试验表明在DSLP2中发现了OsERF3的结合序列(TAA(A)GT)[67]。缺失分析ABA诱导启动子,确定了ABA响应元件(ACGT)并命名为ABRE[68]。
3 响应低温胁迫的基因表达调控网络在的正常生长发育过程中,植物难免遭受多种外界胁迫伤害,其中以低温伤害最为严重。在逆境胁迫条件下,为了保障自身的正常生长发育,它们进化出一个稳定且灵活基因调控网络[4]。接收到胁迫信号刺激后,植物体内的信号分子如激素、次级信使和酶等物质激活各转录因子表达,激活的转录因子激活或者抑制下游低温相关基因的表达,从而调节植物自身的低温耐受能力。
已有研究表明植物低温信号转导途径由各类转录因子家族成员共同参与、相互作用。钙调蛋白是不仅是CBF转录因子的正向调节因子,同时也能结合转录激活因子CAMTA3[50]。研究表明MYB15是CBF调控网络的重要调控因子,MYB15蛋白是CBF基因的负调节因子,它识别结合CBF3基因启动子上的MYC元件,并且与ICE1基因相互作用抑制CBF3基因表达。相关研究表明,R2R3-MYB蛋白作为乙烯响应因子诱导低温胁迫早期植物体内的低温信号应激反应[69],乙烯分子参与了植物的低温信号通路。
在低温胁迫条件下,ICE1基因的突变几乎对CBF1和CBF2基因表达没有影响,这说明了在植物的低温信号转导通路中除了CBF转录因子还有其他家族转录因子的参与。另有研究表明AtWRKY34是通过激活CBF调控因子的转录表达反向调节拟南芥成熟花粉的低温敏感性[24]。除了CBF与MYB转录因子的相互作用,植物体内的低温调控网络也有NAC、WRKY、bZIP等转录因子的参与。已被证明拟南芥AtNAC019参与了AtMYB88基因调控的低温信号途径[35]。
另有研究证明,在发现的101个BdNAC中至少有一个激素和低温胁迫有关的顺式作用元件,其中44个基因有超过5个以上的作用元件,说明BdNAC基因参与植物低温胁迫与激素相互作用的过程[70-71]。同时,在BdNAC启动子上检测到135个ABA应答元件(ABREs),107个茉莉酸应答元件,99个MYB转录因子结合位点,66个SA应答元件,表明NAC基因在脱落酸、茉莉酸、水杨酸信号通路中扮演重要角色[18]。DREB1A、DREB1C和DEAR3蛋白可以结合目的基因启动子上的ABRE、ERF作用元件来响应低温胁迫,表明DREB转录因子不仅参与植物低温应激反应,并且参与脱落酸、乙烯等植物激素信号通路[65]。
在构树中发现82个AP2/ERF、39个NAC、71个WRKY、45个MYB、89个bZIP、41个ZFPs基因参与低温胁迫响应反应[72]。在拟南芥成熟花粉中,有93个来自多个转录因子家族的基因如AP2/ERF、NAC、WRKY、MYB、bZIP参与植物低温信号转导途径[73]。在木薯顶端芽,鉴定出了32个在低温早期反应中产生作用的低温响应转录因子。其中有6个AP2/ERF、5个MYB、和5个GRAS转录因子[74]。这些研究结果说明,植物体内的低温调控网络由AP2/ERF、NAC、WRKY、MYB、bZIP和ZFPs等多种转录因子共同调控而实现其功能的。
同时,将参与低温信号转导途径的6类转录因子及其相关功能列举出来,如表 2所示。
在ABA依赖途径中,bZIP转录因子结合基因启动子上的ABER元件,激活冷响应基因表达;在ABA独立信号途径中,MYB与MYC转录因子相互作用识别MYB作用元件,激活冷响应基因表达。其他CBF、NAC、WRKY几类转录因子家族,通过识别下游冷响应基因上的CRT/DRE、ERF、W-BOX、GCC-BOX等元件激活下游基因表达,从而引起转基因植株的相关表型发生改变,如图 1所示。
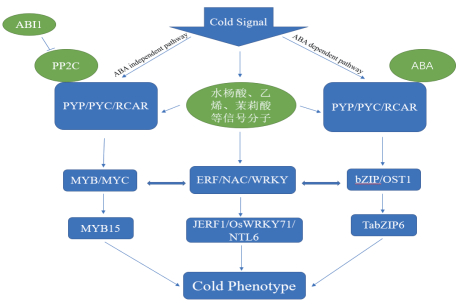
|
| 图 1 低温状态下植物体内转录因子相互作用的调控网络 |
转录因子在植物低温信号途径中扮演十分重要的角色。NAC、WRKY、MYB、b ZIP、ZFPs等转录因子家族激活低温相关基因的表达从而调控植物的低温耐受能力。同时参与ABA(脱落酸)、JAs(茉莉酸)、SA(水杨酸)、乙烯等途径进一步增强植物对低温胁迫的调节能力。虽然在现代生物技术手段的帮助下,转录因子在低温信号转导通路中的调控机理的研究取得了突破,但是由于低温信号途径中参与的调控因子数量过于巨大,其间相互关系也十分复杂,目前仍然存在着很多问题,如同一转录因子在不同信号通路中是否具有相同功能;各转录因子家族是否同时参与低温应激反应等,这些问题仍然是人们未来研究工作的重点内容。进一步了解转录因子对逆境胁迫调控的作用机理,为今后的育种选育与改良提供了非常坚实的理论依据和研究背景。
| [1] |
Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants[J]. Trends in Plant Science, 2007, 12(10): 444. DOI:10.1016/j.tplants.2007.07.002 |
| [2] |
Zhang X, Fowler SG, Cheng H, et al. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis[J]. The Plant Journal, 2004, 39(6): 905. DOI:10.1111/tpj.2004.39.issue-6 |
| [3] |
Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and cis-acting regulatory elements[J]. Plant Science, 2014, 217-218(1): 109-119. |
| [4] |
Tweneboah S, Oh SK. Biological roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in solanaceous crops[J]. Journal of Plant Biotechnology, 2017, 44(1): 1-11. DOI:10.5010/JPB.2017.44.1.001 |
| [5] |
Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors:genome-wide comparative analysis among eukaryotes[J]. Science, 2000, 290(5499): 2105-2110. DOI:10.1126/science.290.5499.2105 |
| [6] |
Bai B, Wu J, Sheng WT, et al. Comparative analysis of anther transcriptome profiles of two different rice male sterile lines genotypes under cold stress[J]. International Journal of Molecular Sciences, 2015, 16(5): 11398. |
| [7] |
Zhang ZJ, Huang RF. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis[J]. Plant Molecular Biology, 2010, 73(3): 241-249. DOI:10.1007/s11103-010-9609-4 |
| [8] |
Xu ZS, Chen M, Li LC, et al. Functions and application of the AP2/ERF transcription factor family in crop improvement[J]. Bulletin of Botany, 2011, 53(7): 570-585. |
| [9] |
李瑞梅, 惠杜娟, 刘姣, 等. 植物抗寒转录因子CBF和ICE研究进展[J]. 广东农业科学, 2012, 39(23): 132-135. DOI:10.3969/j.issn.1004-874X.2012.23.043 |
| [10] |
Li XD, Zhuang KY, Liu ZM, et al. Overexpression of a novel NAC-type tomato transcription factor, SlNAM1, enhances the chilling stress tolerance of transgenic tobacco[J]. Journal of Plant Physiology, 2016, 204: 54-65. DOI:10.1016/j.jplph.2016.06.024 |
| [11] |
Zhuang L, Yuan X, Chen Y, et al. PpCBF3 from cold-tolerant kentucky bluegrass involved in freezing tolerance associated with up-regulation of cold-related genes in transgenic Arabidopsis thaliana[J]. PLoS One, 2015, 10(7): e0132928. DOI:10.1371/journal.pone.0132928 |
| [12] |
Wang L, Gao J, Qin X, et al. JcCBF2 gene from Jatropha curcas improves freezing tolerance of Arabidopsis thaliana during the early stage of stress[J]. Molecular Biology Reports, 2015, 42(5): 937-945. DOI:10.1007/s11033-014-3831-0 |
| [13] |
Morran S, Eini O, Pyvovarenko T, et al. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors[J]. Plant Biotechnology Journal, 2011, 9(2): 230. DOI:10.1111/pbi.2010.9.issue-2 |
| [14] |
Sobkowiak A, Jończyk M, Adamczyk J, et al. Molecular foundations of chilling-tolerance of modern maize[J]. BMC Genomics, 2016, 17(1): 125. DOI:10.1186/s12864-016-2453-4 |
| [15] |
Ke YG, Yang ZJ, Yu SW, et al. Characterization of OsDREB6 responsive to osmotic and cold stresses in rice[J]. Journal of Plant Biology, 2016, 42(7): 9264-9269. |
| [16] |
Puranik S, Sahu PP, Srivastava PS, et al. NAC proteins:regulation and role in stress tolerance[J]. Trends in Plant Science, 2012, 17(6): 369-381. DOI:10.1016/j.tplants.2012.02.004 |
| [17] |
Ooka H, Satoh K, Doi K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana[J]. Genes & Genomes, 2003, 10(6): 239. |
| [18] |
Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses:progress and prospects[J]. Frontiers in Plant Science, 2015, 6(902): 81. |
| [19] |
Mao X, Zhang H, Qian X, et al. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis[J]. Journal of Experimental Botany, 2012, 63(8): 2933-2946. DOI:10.1093/jxb/err462 |
| [20] |
Yoo SY, Kim Y, Kim SY, et al. Control of flowering time and cold response by a NAC-domain protein in Arabidopsis[J]. PLoS One, 2007, 2(7): e642. DOI:10.1371/journal.pone.0000642 |
| [21] |
Ma N, Zuo Y, Liang X, et al. The multiple stress-responsive transcription factor SlNAC1 improves the chilling tolerance of tomato[J]. Physiologia Plantarum, 2013, 149(4): 474-486. DOI:10.1111/ppl.2013.149.issue-4 |
| [22] |
Fang L, Su L, Sun X, et al. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis[J]. Journal of Experimental Botany, 2016, 67(9): 2829-2845. DOI:10.1093/jxb/erw122 |
| [23] |
Tripathi P, Rabara RC, Rushton PJ. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants[J]. Planta, 2014, 239(2): 255-266. DOI:10.1007/s00425-013-1985-y |
| [24] |
Zou C, Jiang W, Yu D. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis[J]. Journal of Experimental Botany, 2010, 61(14): 3901-3914. DOI:10.1093/jxb/erq204 |
| [25] |
Zeng T, Kou Y, Liu H, et al. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice[J]. Journal of Experimental Botany, 2011, 62(14): 4863. DOI:10.1093/jxb/err144 |
| [26] |
Kim CY, Vo KTX, Cong DN, et al. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71[J]. Plant Biotechnology Reports, 2016, 10(1): 13-23. DOI:10.1007/s11816-015-0383-2 |
| [27] |
Zhang Y, Yu H, Yang X, et al. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner[J]. Plant Physiology & Biochemistry, 2016, 108: 478-487. |
| [28] |
Wang Y, Shu Z, Wang W, et al. CsWRKY2, a novel WRKY gene from Camellia sinensis, is involved in cold and drought stress responses[J]. Biologia Plantarum, 2016, 60(3): 1-9. |
| [29] |
李濯雪, 陈信波. 植物诱导型启动子及相关顺式作用元件研究进展[J]. 生物技术通报, 2015, 31(10): 8-15. |
| [30] |
Zhai H, Bai X, Zhu Y, et al. A single-repeat R3-MYB transcription factor MYBC1 negatively regulates freezing tolerance in Arabidopsis[J]. Biochemical & Biophysical Research Communications, 2010, 394(4): 1018. |
| [31] |
Pasquali G, Biricolti S, Locatelli F, et al. Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples[J]. Plant Cell Reports, 2008, 27(10): 1677. DOI:10.1007/s00299-008-0587-9 |
| [32] |
Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice[J]. Journal of Experimental Botany, 2012, 63(7): 2541. DOI:10.1093/jxb/err431 |
| [33] |
Meissner M, Orsini E, Ruschhaupt M, et al. Mapping quantitative trait loci for freezing tolerance in a recombinant inbred line population of Arabidopsis thaliana accessions Tenela and C24 reveals REVEILLE1 as negative regulator of cold acclimation[J]. Plant Cell & Environment, 2013, 36(7): 1256-1267. |
| [34] |
Ding Z, Li S, An X, et al. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana[J]. Hereditas, 2009, 36(1): 17-29. |
| [35] |
Baldoni E, Genga A, Cominelli E. Plant MYB transcription factors:their role in drought response mechanisms[J]. International Journal of Molecular Sciences, 2015, 16(7): 15811-15851. DOI:10.3390/ijms160715811 |
| [36] |
Yang YN, Zhao G, Yue WQ, et al. Molecular cloning and gene expression differences of the anthocyanin biosynthesis-related genes in the red/green skin color mutant of pear(Pyrus communis L.)[J]. Tree Genetics & Genomes, 2013, 9(5): 1351-1360. |
| [37] |
Indeok H, Kumar MR, Kang JG, et al. Genome-wide identification and characterization of bZIP transcription factors inbrassica oleraceaunder cold stress[J]. BioMed Research International, 2016, 2016(2016): 1-18. |
| [38] |
李田, 孙景宽, 刘京涛. 植物转录因子家族在耐盐抗旱调控网络中的作用[J]. 生命科学, 2015, 27(2): 217-227. |
| [39] |
Ma Q, Dai X, Xu Y, et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes[J]. Plant Physiology, 2009, 150(1): 244-256. DOI:10.1104/pp.108.133454 |
| [40] |
Hossain MA, Jungil C, Han M, et al. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice[J]. Journal of Plant Physiology, 2010, 167(17): 1512. DOI:10.1016/j.jplph.2010.05.008 |
| [41] |
Liu C, Wu Y, Wang X. bZIP transcription factor OsbZIP52/RISBZ5:a potential negative regulator of cold and drought stress response in rice[J]. Planta, 2012, 235(6): 1157-1169. DOI:10.1007/s00425-011-1564-z |
| [42] |
曹红利, 岳川, 王新超, 杨亚军. bZIP转录因子与植物抗逆性研究进展[J]. 南方农业学报, 2012, 43(8): 1094-1100. DOI:10.3969/j:issn.2095-1191.2012.08.1094 |
| [43] |
Wang L, Cao H, Qian W, et al. Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic arabidopsis[J]. Annals of Botany, 2017, 119(7): 1195-1209. DOI:10.1093/aob/mcx011 |
| [44] |
Sun XL, Li Y, Cai H, et al. Arabidopsis bZIP1 transcription factor binding to ABRE cis -element regulates abscisic acid signal transduction[J]. Acta Agronomica Sinica, 2011, 37(4): 612-619. |
| [45] |
Liu DC, Qi WU, Wang YC, et al. Cloning and expression analysis of PtrZPT2-2 from trifoliate orange(Poncirus trifoliata)[J]. Acta Horticulturae Sinica, 2014, 41(1): 9-16. |
| [46] |
Kim JC, Lee SH, Cheong YH, et al. A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants[J]. Plant J, 2001, 25(3): 247-259. DOI:10.1046/j.1365-313x.2001.00947.x |
| [47] |
Yu GH, Jiang LL, Ma XF, et al. A soybean C2H2-type zinc finger gene GmZF1 enhanced cold tolerance in transgenic Arabidopsis[J]. PLoS One, 2014, 9(10): e109399. DOI:10.1371/journal.pone.0109399 |
| [48] |
Ciftci-Yilmaz S, Mittler R. The zinc finger network of plants[J]. Cellular & Molecular Life Sciences, 2008, 65(7/8): 1150-1160. |
| [49] |
Sun SJ, Guo SQ, Yang X, et al. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice[J]. Journal of Experimental Botany, 2010, 61(10): 2807. DOI:10.1093/jxb/erq120 |
| [50] |
Doherty CJ, Buskirk HAV, Myers SJ, et al. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance[J]. The Plant Cell, 2009, 21(3): 972. DOI:10.1105/tpc.108.063958 |
| [51] |
Ma Y, Dai X, Xu Y, et al. COLD1 confers chilling tolerance in rice[J]. Cell, 2015, 160(6): 1209. DOI:10.1016/j.cell.2015.01.046 |
| [52] |
Abiri R, Shaharuddin NA, Maziah M, et al. Role of ethylene and the APETALA 2/ethylene response factor superfamily in rice under various abiotic and biotic stress conditions[J]. Environmental & Experimental Botany, 2017, 134: 33-44. |
| [53] |
Shi Y, Tian S, Hou L, et al. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis[J]. The Plant Cell, 2012, 24(6): 2578-2595. DOI:10.1105/tpc.112.098640 |
| [54] |
Niu YJ, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis[J]. Journal of Experimental Botany, 2011, 62(6): 2143. DOI:10.1093/jxb/erq408 |
| [55] |
Hu Y, Jiang L, Wang F, et al. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis[J]. The Plant Cell, 2013, 25(8): 2907-2924. DOI:10.1105/tpc.113.112631 |
| [56] |
Wu LJ, Chen XL, Ren HY, et al. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco[J]. Planta, 2007, 226(4). |
| [57] |
Pil JS, Mi JK, Park JY, et al. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis[J]. The Plant Journal, 2010, 61(4): 661-671. DOI:10.1111/tpj.2010.61.issue-4 |
| [58] |
Agarwal M, Hao YJ, Kapoor A, et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance[J]. J Biol Chem, 2006, 281(49): 37636-37645. DOI:10.1074/jbc.M605895200 |
| [59] |
Cai WT, Yang YL, Wang WW, et al. Overexpression of a wheat(Triticum aestivum L.)bZIP transcription factor gene, TabZIP6, decreased the freezing tolerance of transgenic Arabidopsis seedlings by down-regulating the expression of CBFs[J]. Plant Physiology and Biochemistry, 2018, 124: 100-111. DOI:10.1016/j.plaphy.2018.01.008 |
| [60] |
Luo X, Bai X, Zhu D, et al. GsZFP1, a new Cys2/His2-type zinc-finger protein, is a positive regulator of plant tolerance to cold and drought stress[J]. Planta, 2012, 235(6): 1141-1155. DOI:10.1007/s00425-011-1563-0 |
| [61] |
Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and cis-acting regulatory elements[J]. Plant Science An International Journal of Experimental Plant Biology, 2014, 217-218(1): 109-119. |
| [62] |
Vysotskii DA, Ij VVL, Souer E, et al. ABF transcription factors of Thellungiella salsuginea:Structure, expression profiles and interaction with 14-3-3 regulatory proteins[J]. Plant Signaling & Behavior, 2013, 8(1): e22672. |
| [63] |
Ma NN, Zuo YQ, Liang XQ, et al. The multiple stress-responsive transcription factor SlNAC1 improves the chilling tolerance of tomato[J]. Physiologia Plantarum, 2013, 149(4): 474-486. DOI:10.1111/ppl.2013.149.issue-4 |
| [64] |
Zou C, Sun K, Mackaluso JD, et al. Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana[J]. Proc Natl Acad Sci USA, 2011, 108(36): 14992-14997. DOI:10.1073/pnas.1103202108 |
| [65] |
Sazegari S, Niazi A, Ahmadi FS. A study on the regulatory network with promoter analysis for Arabidopsis DREB-genes[J]. Bioinformation, 2015, 11(2): 101-106. DOI:10.6026/bioinformation |
| [66] |
Ha NT, Leipner J, Guerra-Peraza O, et al. Article 3:Characterization of the stress-induced gene ZmCOI6.1 in maize:Expression and promoter sequences[J]. Tap Chi Sinh Hoc, 2014, 31(3): 71-80. |
| [67] |
Wang Q, Qi W, Wang Y, et al. Isolation and identification of an AP2/ERF factor that binds an allelic cis-element of rice gene LRK6[J]. Genetics Research, 2011, 93(5): 319-332. DOI:10.1017/S0016672311000218 |
| [68] |
Mishra S, Shukla A, Upadhyay S, et al. Identification, occurrence, and validation of DRE and ABRE Cis-regulatory motifs in the promoter regions of genes of Arabidopsis thaliana[J]. Bulletin of Botany, 2014, 56(4): 388-399. |
| [69] |
Yun KY, Park MR, Mohanty B, et al. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress[J]. BMC Plant Biology, 2010, 10(1): 16. DOI:10.1186/1471-2229-10-16 |
| [70] |
You J, Zhang L, Song B, et al. Systematic analysis and identification of stress-responsive genes of the NAC gene family in Brachypodium distachyon[J]. PLoS One, 2015, 10(3): e0122027. DOI:10.1371/journal.pone.0122027 |
| [71] |
Lv X, Lan S, Guy KM, et al. Global expressions landscape of NAC transcription factor family and their responses to abiotic stresses in Citrullus lanatus[J]. Scientific Reports, 2016, 6: 30574. DOI:10.1038/srep30574 |
| [72] |
Peng X, Wu Q, Teng L, et al. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors[J]. BMC Plant Biology, 2015, 15(1): 108. DOI:10.1186/s12870-015-0489-2 |
| [73] |
Zou C, Yu D. Analysis of the cold-responsive transcriptome in the mature pollen of Arabidopsis[J]. Journal of Plant Biology, 2010, 53(6): 400-416. DOI:10.1007/s12374-010-9129-4 |
| [74] |
An D, Yang J, Zhang P. Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress[J]. BMC Genomics, 2012, 13(1): 64. DOI:10.1186/1471-2164-13-64 |