2. 四川省旺苍县畜牧食品局,广元 628200;
3. 成都农业科技职业学院,成都 6111305;
4. 四川省南江县农业局,巴中 635600
2. Sichuan Food and Animal Husbandry Bureaus of Wangcang County, Guangyuan 628200;
3. Chengdu Vocational College of Agriculture and Technology, Chengdu 6111305;
4. Sichuan Agricultural Bureau of Nanjiang County, Bazhong 635600
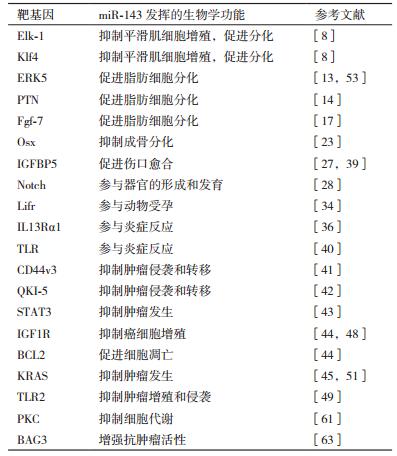
microRNA是一类在分子进化中十分保守的非编码RNA,长度约22个核苷酸,是基因表达的重要调节分子,广泛存在于真核细胞中,在动植物生长、发育、繁殖等诸多生理过程中发挥着重要作用[1]。自20世纪90年代科学家在线虫中发现第一个microRNA以来,相关microRNA的研究成为了生物学领域热点,据预测microRNA的种类有上千个,而单个的microRNA可以调控多个目标基因,涉及多条生物学通路。miR-143位于人类第5号染色体长臂3区2带,miR-143的功能发挥是建立在调控其靶基因和相关信号通路上,通过转录后水平抑制靶基因的表达来调控相关信号通路,对细胞的增殖、分化、凋亡产生影响,继而调控机体生长发育和疾病的发生。近年来的研究表明,miR-143具有高度的保守性,且广泛存在于动物和人体组织中,是一个多功能microRNA,参与了动物心血管、运动、神经、免疫和生殖等方面的调控,并与肥胖、炎症和肿瘤的发生有密切关系。本文综述了miR-143在动物正常生长发育和疾病发生方面已有的研究,以期为进一步挖掘miR-143的生物学功能、对miR-143调控网络的构建,以及相关疾病的诊断和治疗等方面提供参考(表 1)。
心脏和血管组成的心血管系统在机体内是一个相对封闭的管道系统,其最重要的生理机能就是维持血液循环,保证机体内环境的相对恒定和新陈代谢的正常进行。血液循环一旦停止,生命活动就不能正常进行,最后将导致机体的死亡。
心脏是哺乳动物胚胎发育时期最早形成的器官。Dicer基因参与了microRNA的形成[2],有研究发现Dicer敲除小鼠心脏特异性microRNA表达普遍下降,出生后4 d内全部死于扩张型心肌病和心衰[3]。同时,在心脏中存在的一些特异性并且高表达micro-RNA的异常表达也将导致心脏发育异常[4]。miR-143虽然不是心脏特异性表达的miRNA,但它也对心脏发育产生重要影响。ADD3(Adducin3) 是一种细胞膜骨架蛋白,参与细胞膜网状结构的构建和维持,还具有细胞信号转导、细胞膜离子转运等功能[5],Deacon等[6]发现miR-143-add3基因通路通过调节心肌细胞的形态影响心腔的形成和功能。敲除miR-143或中断miR-143-add3交互作用抑制了心室心肌细胞f -肌动蛋白(F-actin capping protein)重塑,阻碍了心室肌细胞的正常生长和伸长,导致心室坍塌和收缩下降。Miyasaka等[7]在模式动物斑马鱼上发现miR-143的表达可以影响心跳节律,敲除miR-143引起维甲酸(Retinoic acid)信号通路脱抑制,造成心室流出道产生异常。
血管平滑肌细胞是血管的重要组成部分,在血管生成期间具有增殖和迁移能力,在成熟血管中则具有收缩功能,可调节动脉张力和血压。同时平滑肌细胞还具有高度的可塑性,在血管发生损伤时可以从收缩表型转换便于增殖、迁移和胶原生成的状态,而这种转换可导致动脉粥样硬化、高血压等疾病的发生。miR-143常与miR-145共同影响血管平滑肌。Cordes等[8]的研究证实了miR-143和miR-145调控着平滑肌细胞的宿命和可塑性。miR-143和miR-145通过靶向结合Elk-1(Ets-like protein 1)、Klf4(Kruppel-like factor4) 等基因,抑制平滑肌细胞的增殖,促进平滑肌细胞分化,可以定向诱导心脏祖细胞分化为心脏平滑肌细胞。Boettger等[9]研究发现miR-143/145基因簇控制着平滑肌细胞的合成和收缩,并直接影响血管平滑肌的收缩。Elia等[10]在急性血管损伤的大鼠模型中过表达miR-143和mir-145发现,新生内膜的形成会减少,同时miR-143和mir-145缺失会引起血管平滑肌细胞分化不完全,从而导致主动脉的结构发生改变。与Cordes等的研究结果不同,Xin等[11]的研究发现缺乏miR-143和mir-145的老鼠并不表现明显的平滑肌分化异常,生活力表现正常,但是由于血管张力的降低,缺乏miR-143和mir-145的老鼠血压显著降低,同时由于肌动蛋白纤维的混乱和平滑肌细胞迁移活性的降低严重阻碍了血管损伤后的内膜增生。这可能与一个靶基因可以受多个microRNA调控,同时一个microRNA也可以调控多个靶基因发挥不同的生物学功能的特性有关[12]。Bhattachariya等[5]发现在门静脉中miR-143/145基因簇对维持拉伸诱导的收缩分化和钙信号中也发挥着重要作用。
1.2 miR-143对脂肪细胞分化和能量代谢的影响Esau等[13]的研究发现miR-143可以促进脂肪细胞分化。他们将反义核苷酸转染到3T3细胞中发现,抑制miR-143可以抑制脂肪细胞的分化,同时miR-143的靶基因细胞外信号调节激酶5(Extracellular signal regulated kinase,ERK5) 低表达,因此他们推测miR-143的作用途径可能是通过靶向结合ERK5进而对脂肪细胞的分化产生影响。Yi等[14]发现miR-143可以通过靶向结合多效生长因子(Pleiotrophin,PTN)促进3T3脂肪细胞分化。Li[15]等通过测序技术在猪上也发现miR-143参与脂肪细胞的分化。Takanabe等[16]在高脂诱导肥胖的小鼠脂肪组织中并没有发现miR-143与ERK5基因的显著负相关关系。而后来He等[17]沉默小鼠miR-143后没有发现脂肪细胞任何的异常分化,一些脂肪形成标志基因PPARγ(Peroxisome proliferator-activated receptorγ)和C/EBPα(CCAAT/enhancer binding proteinα)的表达也没有受到明显的影响,抑制miR-143后ERK5基因的转录未发生明显的抑制。在小鼠脂肪形成过程中,他们通过双荧光素酶报告系统确定成纤维细胞生长因子7(fibroblast growth factor 7,Fgf-7) 为miR-143的靶基因,猜测Fgf-7可能通过激活酪氨酸激酶(Tyrosine kinase)信号系统促进脂肪细胞的增殖和分化。造成miR-143靶基因差异的原因可能是因为实验对象不同,Esau等的实验材料是人类细胞,而He等使用的是小鼠。
目前的研究表明miR-143可以影响甘油三酯(Triacylglyceride,TAG)的积累和糖酵解过程,影响机体的能量代谢。Xie等[18]发现在前脂肪细胞中,miR-143通过调节脂肪生成标记基因和增加甘油三酯积累加速脂肪在早期阶段的形成。Wang等[19]在猪脂肪细胞上的研究也表明miR-143可以通过增加脂肪细胞中甘油三酯的积累促进脂肪形成。Zhao等[20]发现miR-143可以抑制糖酵解并降低胶质瘤样干细胞的多能性。Peschiaroli等[21]关于肿瘤细胞的研究表明miR-143可以调控己糖激酶2(hexokinase 2,HK 2) 的表达。
1.3 miR-143对运动系统和神经系统的影响骨骼和骨骼肌是运动系统的主要组成部分,miR-143对骨细胞和骨骼肌细胞的分化都有重要影响。成骨相关转录因子(Osterix,Osx)是由Nakashima[22]等发现的成骨细胞特异性转录因子,Osx对维持骨骼的生长和动态平衡具有重要的调控作用。Li等[23]发现miR-143在分化的成骨细胞中表达下降,同时他们证实miR-143可以靶向结合成骨形成相关转录因子抑制成骨分化。Zuo等[24]在猪骨骼肌卫星细胞中过表达(或抑制)ssc-miR-143-3p会诱导增加(或减少)慢肌纤维标志性基因和蛋白的表达,证实了miR-143可以调控骨骼肌细胞肌纤维的分化,其作用途径可能是通过HDAC4-MEF2(Histone deacetylase 4-myocyte enhancer factor-2) 通路调节MYH7基因(Beta-myosin heavy chin gene)。Chen等[25]利用鳜鱼研究发现抑制miR-143之后MyoD(Myogenic differentiation)基因和快速肌球蛋白重链(Fast myosin heavy chain)基因的表达显著上调,研究提示miR-143参与调控了脊椎动物肌纤维类型的分化。Li等[26]发现miR-135和miR-143抑制剂可能诱发牙髓干细胞肌原性的分化。Soriano-Arroquia等[27]发现miR-143在卫星细胞中的表达随年龄的增长而下降,通过进一步研究发现miR-143可能通过靶向结合IGFBP5(Insulin-like growth factor-binding protein 5) 参与了卫星细胞与年龄相关的功能调节。
目前有关miR-143在神经发育方面的研究较少,但近期Rani等[28]在灵长类的狭鼻猴类分支中发现了一个有趣的lncRNA,命名为IncND(Neurodeveloment,ND),该研究表明:在灵长类动物神经前体细胞中LncND通过介导miRNA调控Notch信号通路从而影响大脑皮质的扩张。lncND有16个miR-143-3p的miRNA反应元件,通过LncND结合和释放miR-143-3p调控Notch受体的表达。他们发现在发育的人类脑的脑室和脑室下带中,LncND在放射状胶质细胞(Radial glia cells,RGCs)中大量富集。在神经母细胞瘤细胞中下调能够降低细胞的增殖,诱导神经分化。
1.4 miR-143对免疫系统和生殖系统的影响脾脏是机体最大的免疫器官,Lagos-Qaintana等[29]通过测序发现miR-143在小鼠脾中高度表达,约占所有miRNA总丰度的30%,推测miR-143在脾脏发育和分化过程中发挥着重要作用。Trakooljul等[30]在禽类的研究中发现14 d的小鸡脾脏中miR-143显著高表达。Yuan等[31]的研究报道miR-143通过调节细胞凋亡和自噬的相互作用而调控小胶质细胞存活,miR-143和促凋亡蛋白(P53 up-regulated modulator of apoptosis,PUMA)共同调控小胶质细胞的功能发挥。PUMA的表达在转录后水平受miR-143调节,anti-miR-143能够通过上调PUMA调节细胞的自噬和凋亡,减轻甲基苯丙胺引起的小胶质细胞的死亡。在整体的动物模型上,anti-miR-143脑微注射进海马脑区,能够缓解小鼠脑内甲基苯丙胺导致的小胶质细胞减少,在miR-143基因敲除小鼠体内也得到了进一步的证实。
Huang等[32]通过测序发现miR-143在奶牛卵巢和睾丸中均高表达,预测miR-143的靶基因可能参与了GnRH(Gonadotropin-releasing hormone)细胞信号通路。Zhang等[33]发现miR-143可以抑制前颗粒细胞和下调相关基因的表达抑制原始卵泡,参与卵巢发育和调节卵巢功能。Shi等[34]发现大鼠胚胎移植后子宫miR-143在5-7 d的表达水平高于3-4 d,通过进一步的研究发现子宫miR-143的表达可能参与了动物成功受孕,其机制可能是通过调节白血病抑制因子受体(Leukemia inhibitory factor receptor,Lifr)影响胚泡植入的过程。
2 miR-143对疾病发生的影响 2.1 miR-143与炎症反应Yu等[35]利用测序技术发现miR-143在过敏性鼻炎患者鼻黏膜中表达水平降低。Teng等[36]也发现miR-143在变应性鼻炎中表达下降,miR-143通过靶向结合IL-13Rα1(Interleukin 13 recepor α1),诱导变应性鼻炎患者的鼻上皮细胞炎性细胞因子和黏液产生。Tam等[37]发现完全弗氏佐剂(Complete freunds adjuvant,CFA)炎性疼痛能够引起miR-143表达显著下调。Pekow等[38]发现,与正常结肠黏膜相比,慢性结肠炎患者miR-143和miR-145表达量下调明显。Chivukula等[39]利用葡聚糖硫酸钠(Dextran sulfate sodium salt)诱导急性溃疡性结肠炎模型小鼠,结果发现,野生型小鼠结肠上皮细胞可快速增殖,填补损伤伤口,而敲除miR-143和miR-145的小鼠,肠道细胞无法切换到这种修复模式,伤口不能愈合,因此小鼠无法生存。miR-143和miR-145可能作用于IGFBP5促进伤口的愈合,敲除miR-143和miR-145后则可抵消这种效应。Xia等[40]发现在表皮葡萄球菌(Staphylococcus epidermidis)的脂磷壁酸(Lipoteichoic acid,LTA)可以诱导miR-143抑制丙酸菌属(Propionibacterium acnes)介导的炎症反应。
2.2 miR-143与肿瘤 2.2.1 对消化系统肿瘤的影响近年的研究报道发现,miR-143几乎对从口腔到结直肠的整个消化道以及消化腺肿瘤的发生都产生了影响。
Xu等[41]在口腔鳞状上皮细胞癌中发现miR-143表达量明显减少,miR-143可以显著抑制口腔鳞状上皮细胞癌的迁移和入侵能力,miR-143可能作用于黏附分子CD44变异型-3(CD44 v3) 信号通路影响原癌基因c-Met磷酸化抑制肿瘤。He和Jia等[42, 43]分别发现miR-143可以通过靶向结合QKI-5(Quaking 5) 和STAT3(Signal transducers and activators of transcription 3) 抑制食管鳞状细胞癌的增殖和入侵能力。Zhuang等[44]发现在胃癌组织和胃癌细胞系中miR-143水平显著下降,同时miR-143可以通过与其目标基因IGF1R(Insulin-like growth factor1receptor)和BCL2(B-cell lymphoma-2) 的结合,影响人类胃癌细胞株对顺铂的耐药性。Chen等[45]在结肠癌的研究表明miR-143可以通过抑制KRAS(Kirsten rat sarcoma viral oncogene)的翻译从而抑制结肠癌。Borralho等[46]发现miR-143可以抑制裸鼠异体移植肿瘤的生长。Akao等[47]也发现miR-143对结直肠肿瘤有抑制作用。Su等[48]发现miR-143和mirR-145可以通过调控IGF1R从而抑制大肠癌细胞增殖。
肝脏是机体最大的消化腺,Liu等[49]发现miR-143可以下调TLR2(Toll like receptor 2) 在肝癌细胞中表达,抑制肝癌细胞增殖和入侵。Pham等[50]研究结果表明miR-143通过MAPK(Mitogen-activated protein kinase)通路抑制COX-2(Cyclooxygenase 2) 的表达,抑制胰腺癌细胞的增殖。Hu等[51]的研究发现miR-143可以抑制胰腺癌的转移并下调GEF1(grain extended filling 1)、GEF40(Grain extended filling 40) 和KRAS基因的表达。
2.2.2 miR-143对生殖和泌尿系统肿瘤的影响Yan等[52]的研究表明miR-143和miR-145可以协同作用于ERBB3(Epidermal growth factor receptor 3),抑制乳腺癌细胞的增殖和入侵。Clape等[53]发现miR-143的表达水平与晚期前列腺癌之间存在负相关关系,miR-143可能通过抑制ERK5信号抑制肿瘤生长。Xu等[54]发现miR-143可以通过抑制KRAS提高前列腺癌细胞对紫杉醇的敏感性,抑制前列腺癌的增殖和迁移。Chu等[55]发现miR-143可以通过KLK2(Kallikrein 2) 抑制前列腺肿瘤。Zhang等[56]研究发现miR-143和mir-145表达下调可能导致DNA甲基转移酶3 B(DNAmethyltransferase 3B,DnMT3B)的超表达,从而导致子宫内膜癌的预后变得更差。Chen等[57]发现miR-143可以影响宫颈鳞状上皮细胞瘤的大小,但miR-143不参与紫杉醇敏感性反应,这与Xu等[54]的研究结果不同,可能是肿瘤所处微环境不同的缘故[58]。Puerta-cril等[59]的研究结果表明miR-143可结合miR-222和miR-452作为膀胱癌分级和无创诊断的生物标记。
2.2.3 miR-143对其他部位肿瘤的影响Xu等[60]发现miR-143可以抑制鼻咽癌;Zhang等[61]发现miR-143通过靶向结合PKC(Protein kinase C)调节肺癌细胞凋亡过程;Wang等[62]发现miR-143可以抑制表皮生长因子受体(Epithelial growth factor receptor,EGFR)信号抑制骨肉瘤的入侵。Liu等[63]在人类恶性胶质瘤干细胞中发现miR-143可以通过BAG3(Bcl-2-associated athanogene 3) 提高紫草素的抗肿瘤活性。
上述研究表明:miR-143可以抑制肿瘤的生长,其作用途径可以是直接与生长、凋亡相关的基因靶向结合来抑制肿瘤的生长,或者是影响能量代谢抑制肿瘤生长,也可以通过相关途径提高肿瘤对抗肿瘤药物的敏感性或降低自身的耐药性抑制肿瘤生长,还可以通过肿瘤微环境影响肿瘤的生长。
3 结语miR-143是一个典型的多功能microRNA,参与了机体的生长发育和疾病发生过程。其调控网络也十分复杂,不仅可以作用于多个靶基因,还可以联合其他microRNA共同发挥作用,但是目前的研究都是针对单个或少数几个基因进行功能验证,miR-143与其他非编码RNA之间、miR-143各靶基因之间的相互作用还不甚了解,仍需进一步深入探讨。对其调控网络的深入挖掘可以为调控生长发育以及疾病治疗和诊断提供新的思路。miR-143在肿瘤组织中呈现低表达,过表达miR-143可以抑制肿瘤生长,因此miR-143有可能用于癌症的治疗和肿瘤标志物研究;但是过表达miR-143可能破坏机体正常功能,带来严重的副作用,如要利用其治疗癌症则需要找到既不严重影响机体正常机能又对抑制肿瘤有效的平衡点;由于miR-143并不是肿瘤特异表达miRNA,因此单独作为肿瘤标志物诊断癌症不太可靠,需要联合其他肿瘤标志物来进行诊断。对这些问题的深入探讨将为我们更深入了解microRNA的作用机制和利用microRNA提供帮助。
| [1] | Ambros V. microRNAs:tiny regulators with great potential[J]. Cell, 2001, 107 (7): 823–826. DOI:10.1016/S0092-8674(01)00616-X |
| [2] | Pham JW, Pellino JL, Lee YS, et al. A Dicer-2-dependent 80S complex cleaves targeted mRNAs during RNAi in Drosophila[J]. Cell, 2004, 117 (1): 83–94. DOI:10.1016/S0092-8674(04)00258-2 |
| [3] | Chen JF, Murchison EP, Tang R, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure[J]. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105 (6): 2111–2116. DOI:10.1073/pnas.0710228105 |
| [4] | Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2[J]. Cell, 2007, 129 (2): 303–317. DOI:10.1016/j.cell.2007.03.030 |
| [5] | Bhattachariya A, Dahan D, Ekman M, et al. Spontaneous activity and stretch-induced contractile differentiation are reduced in vascular smooth muscle of miR-143/145 knockout mice[J]. Acta Physiologica, 2015, 215 (3): 133–143. DOI:10.1111/apha.2015.215.issue-3 |
| [6] | Deacon DC, Nevis KR, Cashman TJ, et al. The miR-143-adducin3 pathway is essential for cardiac chamber morphogenesis[J]. Development, 2010, 137 (11): 1887–1896. DOI:10.1242/dev.050526 |
| [7] | Miyasaka KY, Kida YS, Banjo T, et al. Heartbeat regulates cardiogenesis by suppressing retinoic acid signaling via expression of miR-143[J]. Mechanisms of Development, 2011, 128 (1-2): 18–28. DOI:10.1016/j.mod.2010.09.002 |
| [8] | Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate decisions[J]. Nature, 2009, 460 (7256): 705–710. |
| [9] | Boettger T, Beetz N, Kostin S, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster[J]. Journal of Clinical Investigation, 2009, 119 (9): 2634–2647. DOI:10.1172/JCI38864 |
| [10] | Elia L, Quintavalle M, Zhang J, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice:correlates with human disease[J]. Cell Death & Differentiation, 2009, 16 (12): 1590–1598. |
| [11] | Xin M, Small EM, Sutherland LB, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury[J]. Genes & Development, 2009, 23 (18): 2166–2178. |
| [12] | Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA[J]. Science, 2012, 337 (6099): 1159–1161. DOI:10.1126/science.337.6099.1159 |
| [13] | Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation[J]. Journal of Biological Chemistry, 2004, 279 (50): 52361–52365. DOI:10.1074/jbc.C400438200 |
| [14] | Yi C, Xie WD, Li F, et al. MiR-143 enhances adipogenic differentiation of 3T3-L1 cells through targeting the coding region of mouse pleiotrophin[J]. Febs Letters, 2011, 585 (20): 3303–3309. DOI:10.1016/j.febslet.2011.09.015 |
| [15] | Li G, Li Y, Li X, et al. MicroRNA identity and abundance in developing swine adipose tissue as determined by solexa sequencing[J]. Journal of Cellular Biochemistry, 2011, 112 (5): 1318–1328. DOI:10.1002/jcb.v112.5 |
| [16] | Takanabe R, Ono K, Abe Y, et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet[J]. Biochemical & Biophysical Research Communications, 2008, 376 (4): 728–732. |
| [17] | He Z, Yu J, Zhou C, et al. MiR-143 is not essential for adipose development as revealed by in vivo antisense targeting[J]. Biotechnology Letters, 2013, 35 (4): 499–507. DOI:10.1007/s10529-012-1112-3 |
| [18] | Xie H, Bing L, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity[J]. Diabetes, 2009, 58 (5): 1050–1057. DOI:10.2337/db08-1299 |
| [19] | Wang T, Li M, Guan J, et al. MicroRNAs miR-27a and miR-143 regulate porcine adipocyte lipid metabolism[J]. International Journal of Molecular Sciences, 2011, 12 (11): 7950–7959. |
| [20] | Zhao S, Liu H, Liu Y, et al. miR-143 inhibits glycolysis and depletes stemness of glioblastoma stem-like cells[J]. Cancer Letters, 2013, 333 (2): 253–260. DOI:10.1016/j.canlet.2013.01.039 |
| [21] | Peschiaroli A, Giacobbe A, Formosa A, et al. miR-143 regulates hexokinase 2 expression in cancer cells[J]. Oncogene, 2013, 32 (6): 797–802. DOI:10.1038/onc.2012.100 |
| [22] | Nakashima K, Xin Z, Kunkel G, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation[J]. Cell, 2002, 108 (1): 17–29. DOI:10.1016/S0092-8674(01)00622-5 |
| [23] | Li E, Zhang J, Yuan T, et al. MiR-143 suppresses osteogenic differentiation by targeting Osterix[J]. Molecular & Cellular Biochemistry, 2014, 390 (1-2): 69–74. |
| [24] | Zuo J, Wu F, Liu Y, et al. MicroRNA transcriptome profile analysis in porcine muscle and the effect of miR-143 on the MYH7 gene and protein[J]. PLoS One, 2014, 10 (4): 1–21. |
| [25] | Chen L, Wu P, Guo XH, et al. miR-143:a novel regulator of MyoD expression in fast and slow muscles of Siniperca chuatsi[J]. Current Molecular Medicine, 2014, 14 (3): 370–375. DOI:10.2174/1566524014666140228100250 |
| [26] | Li D, Deng T, Li H, et al. MiR-143 and miR-135 inhibitors treatment induces skeletal myogenic differentiation of human adult dental pulp stem cells[J]. Arch Oral Biol, 2015, 60 (11): 1613–1617. DOI:10.1016/j.archoralbio.2015.08.010 |
| [27] | Soriano-Arroquia A, Mccormick R, Molloy AP, et al. Age-related changes in miR-143-3p:Igfbp5 interactions affect muscle regeneration[J]. Aging Cell, 2016, 15 (2): 361–369. DOI:10.1111/acel.2016.15.issue-2 |
| [28] | Rani N, Nowakowski T, Zhou H, et al. A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA[J]. Neuron, 2016, 90 (6): 1174–1188. DOI:10.1016/j.neuron.2016.05.005 |
| [29] | Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse[J]. Current Biology Cb, 2002, 12 (9): 735–739. DOI:10.1016/S0960-9822(02)00809-6 |
| [30] | Trakooljul N, Hicks JA, Liu HC. Identification of target genes and pathways associated with chicken microRNA miR-143[J]. Animal Genetics, 2010, 41 (4): 357–364. |
| [31] | Yuan Z, Kai S, Ying B, et al. Mir-143/BBC3 cascade reduces microglial survival via interplay between apoptosis and autophagy:Implications for methamphetamine-mediated neurotoxicity[J]. Autophagy, 2016 : 1–22. |
| [32] | Huang J, Ju Z, Li Q, et al. Solexa sequencing of novel and differentially expressed microRNAs in testicular and ovarian tissues in Holstein cattle[J]. International Journal of Biological Sciences, 2011, 7 (7): 1016–1026. DOI:10.7150/ijbs.7.1016 |
| [33] | Zhang J, Ji X, Zhou D, et al. miR-143 is critical for the formation of primordial follicles in mice[J]. Frontiers in Bioscience, 2013, 18 (2): 588–597. DOI:10.2741/4122 |
| [34] | Shi T, Xing S, Lu Q, et al. MiR-143, and rat embryo implantation[J]. Biochimica et Biophysica Acta(BBA)-General Subjects, 2015, 1850 (4): 708–721. DOI:10.1016/j.bbagen.2014.11.023 |
| [35] | Yu SQ, Zhang RX, Liu GJ, et al. Microarray analysis of differentially expressed microRNAs in allergic rhinitis[J]. American Journal of Rhinology & Allergy, 2011, 25 (6): 242–246. |
| [36] | Teng Y, Zhang R, Liu C, et al. miR-143 inhibits interleukin-13-induced inflammatory cytokine and mucus production in nasal epithelial cells from allergic rhinitis patients by targeting IL13Rα1[J]. Biochemical & Biophysical Research Communications, 2014, 457 (1): 58–64. |
| [37] | Tam TS, Bastian I, Zhou XF, et al. MicroRNA-143 expression in dorsal root ganglion neurons[J]. Cell & Tissue Research, 2011, 346 (2): 163–173. |
| [38] | Pekow JR, Dougherty U, Mustafi R, et al. miR-143 and miR-145 are downregulated in ulcerative colitis:Putative regulators of inflammation and protooncogenes[J]. Inflammatory Bowel Diseases, 2012, 18 (1): 94–100. DOI:10.1002/ibd.21742 |
| [39] | Chivukula R, Shi G, Acharya A, et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration[J]. Cell, 2014, 157 (5): 1104–1116. DOI:10.1016/j.cell.2014.03.055 |
| [40] | Xia X, Li Z, Liu K, et al. Staphylococcal LTA-induced miR-143 inhibits propionibacterium acnes-mediated inflammatory response in skin[J]. Journal of Investigative Dermatology, 2015, 136 (3): 621–630. |
| [41] | Xu P, Li Y, Yang S, et al. Micro-ribonucleic acid 143(MiR-143) inhibits oral squamous cell carcinoma(OSCC)cell migration and invasion by downregulation of phospho-c-Met through targeting CD44 v3[J]. Oral Surg Oral Med Oral Pathol Oral Radiol, 2015, 120 (1): 43–51. DOI:10.1016/j.oooo.2015.02.486 |
| [42] | He Z, Yi J, Liu X, et al. MiR-143-3p functions as a tumor suppressor by regulating cell proliferation, invasion and epithelial-mesenchymal transition by targeting QKI-5 in esophageal squamous cell carcinoma[J]. Molecular Cancer, 2016, 15 (1): 1–17. DOI:10.1186/s12943-015-0489-8 |
| [43] | Jia L, Yu M, Zhang D, et al. MiR-143 inhibits tumor cell proliferation and invasion by targeting STAT3, in esophageal squamous cell carcinoma[J]. Cancer Letters, 2016, 373 (1): 97–108. DOI:10.1016/j.canlet.2016.01.023 |
| [44] | Zhuang M, Shi Q, Zhang X, et al. Involvement of miR-143 in cisplatin resistance of gastric cancer cells via targeting IGF1R and BCL2[J]. Tumour Biology the Journal of the International Society for Oncodevelopmental Biology & Medicine, 2015, 36 (4): 2737–2745. |
| [45] | Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis[J]. Oncogene, 2009, 28 (10): 1385–1392. DOI:10.1038/onc.2008.474 |
| [46] | Borralho PM, Gomes S, Lima RT, et al. miR-143 over-expression reduces the growth of xenograft tumors from colon carcinoma cells[J]. Bmc Proceedings, 2010, 4 (Suppl 2): 59. DOI:10.1186/1753-6561-4-s2-p59 |
| [47] | Akao Y, Nakagawa Y, Hirata I, et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors[J]. Cancer Gene Therapy, 2010, 17 (6): 398–408. DOI:10.1038/cgt.2009.88 |
| [48] | Su J, Liang H, Yao W, et al. MiR-143 and MiR-145 regulate IGF1R to suppress cell proliferation in colorectal cancer[J]. PLoS One, 2014, 9 (12): e114420–e114420. DOI:10.1371/journal.pone.0114420 |
| [49] | Liu X, Gong J, Xu B. miR-143 down-regulates TLR2 expression in hepatoma cells and inhibits hepatoma cell proliferation and invasion[J]. International Journal of Clinical & Experimental Pathology, 2015, 8 (10): 12738–12747. |
| [50] | Pham H. miR-143 decreases COX-2 mRNA stability and expression in pancreatic cancer cells[J]. Biochemical & Biophysical Research Communications, 2013, 439 (1): 6–11. |
| [51] | Hu Y, Ou Y, Wu K, et al. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway[J]. Tumor Biology, 2012, 33 (6): 1863–1870. DOI:10.1007/s13277-012-0446-8 |
| [52] | Yan X, Chen X, Liang H, et al. miR-143 and miR-145 synergistically regulate ERBB3 to suppress cell proliferation and invasion in breast cancer[J]. Molecular Cancer, 2014, 13 (1): 126–136. DOI:10.1186/1476-4598-13-126 |
| [53] | Clape C, Fritz V, Henriquet C, et al. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice[J]. PLoS One, 2009, 4 (10): e7542. DOI:10.1371/journal.pone.0007542 |
| [54] | Xu B, Niu X, Zhang X, et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS[J]. Molecular & Cellular Biochemistry, 2011, 350 (1-2): 207–213. |
| [55] | Chu HY, Zhong DY, Tang JL, et al. A functional variant in miR-143 promoter contributes to prostate cancer risk[J]. Archives of Toxicology, 2014, 90 (2): 403–414. |
| [56] | Zhang XM, Dong Y, Ti HJ, et al. Down-regulation of miR-145 and miR-143 might be associated with DNA methyltransferase 3B overexpression and worse prognosis in endometrioid carcinomas[J]. Human Pathology, 2013, 44 (11): 2571–2580. DOI:10.1016/j.humpath.2013.07.002 |
| [57] | Chen Y, Ma C, Zhang W, et al. Down regulation of miR-143 is related with tumor size, lymph node metastasis and HPV16 infection in cervical squamous cancer[J]. Diagnostic Pathology, 2014, 9 (6): 901–906. |
| [58] | Almeida MI, Calin GA. The miR-143/miR-145 cluster and the tumor microenvironment:unexpected roles[J]. Genome Medicine, 2016, 8 (1): 1–3. DOI:10.1186/s13073-015-0257-9 |
| [59] | Puerta-Gil P, García-Baquero R, Jia AY, et al. miR-143, miR-222, and miR-452 are useful as tumor stratification and noninvasive diagnostic biomarkers for bladder cancer[J]. American Journal of Pathology, 2012, 180 (5): 1808–1815. DOI:10.1016/j.ajpath.2012.01.034 |
| [60] | Xu YF, Li YQ, Guo R, et al. Identification of miR-143 as a tumour suppressor in nasopharyngeal carcinoma based on microRNA expression profiling[J]. International Journal of Biochemistry & Cell Biology, 2015, 61 : 120–128. |
| [61] | Zhang N, Su Y, Xu L. Targeting PKCε by miR-143 regulates cell apoptosis in lung cancer[J]. Febs Letters, 2013, 587 (22): 3661–3667. DOI:10.1016/j.febslet.2013.09.018 |
| [62] | Wang Q, Cai J, Wang J, et al. MiR-143 inhibits EGFR-signaling-dependent osteosarcoma invasion[J]. Tumour Biology the Journal of the International Society for Oncodevelopmental Biology & Medicine, 2014, 35 (12): 12743–12748. |
| [63] | Liu J, Qu CB, Xue YX, et al. MiR-143 enhances the antitumor activity of shikonin by targeting BAG3 expression in human glioblastoma stem cells[J]. Biochemical & Biophysical Research Communications, 2015, 468 (1-2): 105–112. |