细菌的非编码小RNA(small non-coding RNA,sRNA),其长度通常在50-500个核苷酸之间,由于缺乏编码蛋白的开放阅读框(open reading frame,ORF),故在基因组中只被转录而不编码蛋白质[1]。大部分sRNA位于两个编码蛋白的基因之间,这类sRNA的转录起始于一段能折叠成稳定茎环结构的序列,终止于不依赖Rho因子的转录终止子,由于茎环结构的存在,sRNA相对比较稳定;还有少数几个sRNA是从mRNA的5'或3'非翻译区(untranslated region,UTR)剪切而来[2, 3]。细菌sRNA已被证明是众多调控网络中的重要组成部分,尤其在细菌转录后水平的调控中发挥重要作用,其分类鉴定、作用机制以及对细菌毒力调控已成为当前研究的热点[4, 5]。
黄单胞菌属细菌属于薄壁菌门、变形纲中γ-亚纲、假单胞菌科。菌体单端极生鞭毛,直杆状,专性好氧,能产生一种不溶于水的黄色素,属于革兰氏阴性细菌。第八版的伯杰氏手册将黄单胞菌属分为6个种,其中的大部分成员都是植物病原菌,能引起如十字花科黑腐病、水稻白叶枯病、水稻条斑病和柑橘溃疡病等[6, 7]。统计发现,黄单胞菌中的140个致病变种至少能引起124种单子叶植物和268种双子叶植物发病,作为重要的模式病原菌,一直是科学家们重要的研究对象[8, 9]。近十几年来,随着测序技术的革新,黄单胞菌的sRNA逐渐被鉴定和研究,本文重点就细菌sRNA的分类、生物学功能以及黄单胞菌中已知sRNA参与的生物学功能进行综述,以期为黄单胞菌引起的作物病害防控提供新的思路。
1 细菌sRNA的研究进展 1.1 细菌sRNA的分类根据生物学功能及作用机制可将sRNA分为3类[10-12]:(1)具有持家功能的sRNA,如核糖核酸酶P RNA(RNase P RNA)、转移-信使RNA(transfer-messenger RNA,tmRNA)、4.5S RNA,该类sRNA有的具有催化功能,有的作为其他蛋白的一部分参与代谢调控[13]。(2)与目标mRNA互作调控基因表达的sRNA;这类sRNA与mRNA通过碱基互补配对调节基因的表达,这种作用机制的sRNA可分为两类:顺式—编码的反义sRNA和反式—编码的反义sRNA[14, 15],前者与mRNA形成完全的互补配对,抑制靶mRNA翻译起始或者结合在靶标mRNA的3'端,起到稳定靶标mRNA的作用[16];后者与mRNA的5' UTR不完全配对,影响靶标基因的表达[17, 18]。反式—编码的反义sRNA在伴侣蛋白Hfq(host factor required for phage Qβ RNA replication)的协助下促进其与其靶标mRNA互补配对,调节靶标基因的翻译或稳定性[19]。(3)与蛋白质相互作用影响蛋白质功能的sRNA,如6S RNA、CsrB[20, 21]。另外,也有研究发现SgrS和RNAⅢ具有sRNA和mRNA两种功能[22, 23]。
1.2 细菌sRNA的功能随着分子生物学技术的不断进步,越来越多的sRNA被鉴定出来,人们对sRNA的功能研究也越来越深入。细菌sRNA已被证明是众多调控网络中的重要组成部分,已知的大部分细菌sRNA主要在蛋白调节、代谢调控、转录后调节和毒力调控等生理过程中起重要作用。
1.2.1 细菌sRNA在蛋白调节中的作用 1.2.1.1 调控外膜蛋白的表达革兰氏阴性细菌的外膜蛋白(outer membrane proteins,OMP)是寄主与病原物互作的关键因子,sRNA通过与mRNA互作调控OMP的表达,进而影响病原菌的黏附、浸染和毒素分泌[24, 25]。大肠杆菌(Escherichia coli)中的MicC sRNA能与Omp C的mRNA结合,抑制其mRNA翻译,控制Omp C的表达[26];沙门菌属(Salmonella)中,高度保守的InvR sRNA在RNA伴侣蛋白Hfq的作用下,抑制外膜孔蛋白Omp D的合成,帮助T3SS锚定在膜上[27, 28]。
1.2.1.2 调节蛋白活性1967年,6S RNA在大肠杆菌中作为一类特殊sRNA被发现,它能够与σ70-RNA聚合酶相互作用并改变该聚合酶对启动子识别的特异性[29]。CsrB家族的sRNA CsrB和CsrC能与转录后调节因子CsrA蛋白互作形成拮抗反馈通路,并调节该蛋白的活性[21, 30, 31]。
1.2.2 sRNA调节细菌代谢研究发现,sRNA能调节细菌的糖代谢和铁代谢,从而使细菌能更好的适应外界环境。大肠杆菌中当外界葡萄糖含量增高时,体内的sRNA SgrS能负调控运输葡萄糖载体ptsG的mRNA的翻译,最终使得胞内葡萄糖含量恢复正常水平[32, 33]。大肠杆菌中铁代谢同时受到铁吸收调节蛋白(ferric uptake regulator,Fur)和sRNA RyhB的调控,在两者的协同调控下,使细胞内Fe2+维持正常水平[34, 35]。
1.2.3 细菌sRNA调节转录关键因子细菌转录因子的编码基因rpoS在遇到生存压力时可转录出大量应答基因,作出应激反应,从而适应环境变化[36, 37]。目前发现有4种sRNA(DsrA、RprA、ArcZ和OxyS)能与rpoS基因的mRNA碱基互补配对,参与rpoS的转录后调控过程[38],DsrA、RprA和ArcZ能够激活RNA聚合酶σ因子(RNA polymerase sigma factor,RpoS)的翻译,而OxyS则抑制其表达[39, 40]。
1.2.4 细菌sRNA在毒力调控中的作用sRNA能调控病原菌毒力基因的表达。RNAⅢ可作为mRNA翻译溶血素蛋白,同时也是发现的第一个在金黄色葡萄球菌(Staphylococcus aureus)的致病性方面具有调控功能的sRNA[41]。金黄色葡萄球菌A蛋白(staphylococal protein A,Spa)和α溶血素是该菌中重要的毒素因子,Spa是细胞壁抗原的主要成分,能刺激抗体的产生,它与金黄色葡萄球菌的抗吞噬作用有关[42];α溶血素是对人类致病起主要作用的溶血素,它能攻击血红细胞,破坏细胞膜,使细胞裂解。Spa和α溶血素都受到RNAⅢ的调控,并影响该菌的毒性。
2 黄单胞菌sRNA研究进展黄单胞菌属的细菌大多数属于植物病原菌,作为模式病原物,主要研究集中在:引起辣椒-番茄细菌性疮痂病的疮痂病病原菌(Xanthomonas euvesicatoria、X. vesicatoria、X. perforans和X. gard-neri)、引起水稻细菌性条斑病的水稻细菌性条斑病菌(X. oryzae pv. Oryzicola、Xooc)、引起水稻白叶枯病的水稻白叶枯病菌(X. oryzae pv. Oryzae、Xoo)、引起十字花科植物病害的十字花科黑腐病菌(X. campestris pv. Campestris、Xcc)、引起柑橘溃疡病的柑橘溃疡病菌(X. citri subsp. Citri、Xcc)[43]等。
黄单胞菌属的病原菌依赖T3SS使植物发病,该分泌系统由hrp基因簇编码,该基因簇包括两部分:一部分由hrpX和hrpG组成;另一部分包含23个基因簇,是黄单胞菌入侵寄主的关键通道[44, 45]。随着深度测序技术的不断进步,人们对黄单胞菌中sRNA的认识也逐渐深入。目前,对于辣椒-番茄细菌性疮痂病菌、水稻白叶枯病菌及十字花科黑腐病菌3个致病种的黄单胞菌属的sRNA研究较多[46],发现部分sRNA与黄单胞菌的致病性相关,且受HrpG和HrpX蛋白调控。
2.1 辣椒-番茄细菌性疮痂病菌(Xcv85-10)中的sRNA除了细菌中广泛存在的tRNA和rRNA,2010年Findeiss等[47]在Xcv85-10菌株中发现和报道了质粒转运反义RNA(plasmid transferred anti-sense RNA,PtaRNA1),2011年其利用dRNA-seq和Northern blot在Xcv85-10中鉴定出23个sRNA[48],包括16个基因间隔区的sRNA即sX1-15和6S,7个顺式—编码反义RNA,即asX1-7,如表 1。
Ⅰ型毒素-抗毒素系统(toxin-antitoxin system,TA)中毒素是蛋白,抗毒素为反义RNA,是由编码毒素的基因反向转录而来。在Xcv85-10菌株中发现Xcv2162/ptaRNA1属于Ⅰ型TA,Xcv2162在跨膜区域编码小蛋白,并具有毒素典型特征;PtaRNA1作为抗毒素,Xcv2162/ptaRNA1两者在细胞中协同发挥作用,以保持细胞能更好的适应外界环境以及维持代谢平衡。PtaRNA1具有70个核苷酸,是Xcv85-10菌株中组成型表达sRNA,由5'茎环结构以及长的3'发卡结构构成其二级结构,具有水平基因转移(horizontal gene transfer,HGT)特征,以质粒为载体,通过频繁的横向转移扩散[47]。该类sRNA的发现丰富了sRNA种类、对sRNA有了更深的认识。
sX13长度为115个核苷酸,存在于基因间隔区,同源性分析发现,该sRNA仅存在于黄单胞科,且具有高度保守性。功能研究发现,在温度、盐离子浓度骤变的情况下,sX13能大量积累,以适应不同的环境。缺失sX13的菌株在培养基上生长缓慢,hrp基因的表达降低,这影响了细菌的T3SS中蛋白的活性,进而使得突变株发病迟缓。基因芯片分析还表明sX13能调控63个基因,包括信号转导、运动、转录与转录后调控以及毒力基因的表达,其功能的发挥不依赖Hfq蛋白[49]。
另外,sX3和asX5为辣椒-番茄细菌性疮痂病菌特有,sX6有341个核苷酸能编码80个氨基酸。sX12具有78个核苷酸,受到HrpX的诱导表达,在细胞平台期积累,通过获得sX12缺失突变菌株,发现与野生型相比其在平板上生长缓慢,且在植物上发病延迟,说明它与sX13一样能影响Xcv85-10菌株的毒力[48]。
2.2 水稻白叶枯病菌(XooPX099)中的sRNALiang等[50]在XooPX099中发现了8个sRNA,命名为:sRNA-Xoo1到sRNA-Xoo8,如表 2。其中sRNA-Xoo1、sRNA-Xoo2、sRNA-Xoo3发挥作用依赖于Hfq蛋白,构建hfq基因缺失突变体,通过双向电泳(two-dimensional electrophoresis,2-DE)将突变体与野生型相关蛋白分离,分析与之相关蛋白的表达量,hfq基因的缺失使得与sRNA-Xoo1、sRNA-Xoo2相关的很多代谢蛋白表达量显著降低。
sRNA-Xoo1来自基因间隔区,长度为103个核苷酸。缺失该sRNA使得6个蛋白表达上调,包括外膜蛋白、伴侣蛋白以及其他酶类物质,改变氨基酸代谢、蛋白合成、物质运输、基因转录等;16个蛋白的表达下调,对细菌的代谢、氨基酸合成等产生影响。
sRNA-Xoo3来自基因间隔区,长度为93个核苷酸。缺失sRNA-Xoo3可以降低参与氧化还原过程中的还原酶和过氧化物歧化酶,而参与ATP合成相关的酶以及调节细胞代谢、氨基酸合成相关的蛋白都增加。
sRNA-Xoo4来自基因间隔区,长度为145个核苷酸。sRNA-Xoo4缺失突变体中有9个与基因转录、rRNA合成等过程中相关蛋白下调,参与物质运输的外膜蛋白表达上调。
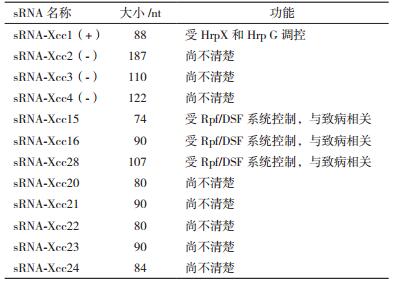
2.3 十字花科黑腐病菌(Xcc8004)中的sRNAJiang等[51]于2010年报道了Xcc8004中4个sRNA,命名为sRNA-Xcc1到sRNA-Xcc4;An等[52]通过Northern blot技术发现了8个sRNA,命名为:sRNA-Xcc15、sRNA-Xcc16、sRNA-Xcc18、sRNA-Xcc28、sRNA-Xcc20到sRNA-Xcc24,如表 3。
可扩散信号分子(diffusible signal factor,DSF)作为新发现的群体感应信号,该家族的所有信号分子的主要化学成分都是顺式-2-不饱和脂肪酸。rpf家族的基因与DSF紧密相关,RpfF参与DSF的合成,RpfC/RpfG组成双组分感应系统参与DSF信号转导。DSF对十字花科黑腐病菌胞外酶、胞外多糖等毒素因子的调节有重要作用[53]。
sRNA-Xcc1来源于基因间隔区,长度为88个核苷酸。在十字花科黑腐病菌中sRNA-Xcc1与辣椒-番茄细菌性疮痂病菌中PtaRNA1一样以质粒为载体通过HGT转运,sRNA-Xcc1具有独立的启动子,其转录方向与其他基因间隔区的启动子方向相反。sRNA-Xcc1,受到HrpX和HrpG调控,缺失HrpX和HrpG引起sRNA-Xcc1的表达下调,并推测该sRNA可能与十字花科黑腐病菌的毒力调控有关[54]
sRNA-Xcc15/16/28受到DSF信号转导途径中RpfF和RpfG的系统控制[52],该系统又与细菌的致病性密切相关,单独缺失某一个sRNA都不影响Xcc8004的毒力,同时缺失sRNA-Xcc15/16/28则会大大降低该菌的毒力。
3 展望近年来,通过深度测序技术在黄单胞菌属少数几个致病变种中发现了一定数量的sRNA,这些sRNA有的与致病相关,如sX12和sX13,还有少数发挥特殊作用的sRNA,如PtaRNA1行使抗毒素功能,但大部分已发现的黄单胞菌属中的sRNA功能并不清楚。为深入解析黄单胞菌属病原中sRNA的种类及其功能,今后在该研究领域有必要开展以下研究:(1)利用深度测序、交联免疫沉淀等技术进一步开展黄单胞菌中sRNA的鉴定,尤其是针对引起重要经济危害的该属病原开展相关研究。如柑橘溃疡病是我国检疫性病害,该病对我国乃至全球柑橘产业造成了巨大的危害,但目前尚未见柑橘溃疡病菌sRNA的研究报道。(2)开展黄单胞菌属内及相近属间比较转录组分析,更好的发掘进化保守性的转录起始位点及功能性的sRNA[55, 56]。(3)加快sRNA功能研究步伐,解析其在黄单胞菌生理与环境适应响应,以及病原-寄主互作中的作用机制;同时在深入了解sRNA作用机制基础上,开展RNA调控研究,以期为该类病害防控提供新的思路。
| [1] | Livny J, Waldor MK. Identification of small RNAs in diverse bacter-ial species. Current Opinion in Microbiology, 2007, 10 (2): 96–101. DOI:10.1016/j.mib.2007.03.005 |
| [2] | Chao Y, Kai P, Reinhardt R, et al. An atlas of Hfq-bound transcripts reveals 3' UTRs as a genomic reservoir of regulatory small RNAs. EMBO Journal, 2012, 31 : 4005–4019. DOI:10.1038/emboj.2012.229 |
| [3] | Oliva G, Sahr T, Buchrieser C. Small RNAs, 5' UTR elements and RNA-binding proteins in intracellular bacteria:impact on metabolism and virulence. FEMS Microbiology Reviews, 2015, 39 (3): 331–349. DOI:10.1093/femsre/fuv022 |
| [4] | Gottesman S, Storz G. Bacterial small RNA regulators:versatile roles and rapidly evolving variations. Cold Spring Harbor Perspectives in Biology, 2011, 3 (12): 723–729. |
| [5] | Sievers S, Lillebæk EMS, Jacobsen K, et al. A multicopy sRNA of Listeria monocytogenes regulates expression of the virulence adhesin LapB. Nucleic Acids Research, 2014, 42 (14): 9383–9398. DOI:10.1093/nar/gku630 |
| [6] | Da Silva AR, Ferro JA, Reinach F, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature, 2002, 417 (6887): 459–463. DOI:10.1038/417459a |
| [7] | Afolabi O, Milan B, Amoussa R, et al. First report of Xanthomonas oryzae pv. oryzicola causing bacterial leaf streak of rice in Burundi. Applied and Environmental Microbiology, 2015, 81 : 688–698. DOI:10.1128/AEM.02768-14 |
| [8] | Leyns F, Cleene MD, Swings JG, et al. The host range of the genus Xanthomonas. Botanical Review, 1984, 50 (3): 308–356. DOI:10.1007/BF02862635 |
| [9] | 龙海, 李一农, 李芳荣, 等. 植物病原菌黄单胞菌的分类研究进展. 植物保护, 2010, 36(5): 11–16. |
| [10] | Mikulík K, Palečková P, Felsberg J, et al. SsrA genes of streptomycetes and association of proteins to the tmRNA during development and cellular differentiation. Proteomics, 2008, 8 (7): 1429–1441. DOI:10.1002/(ISSN)1615-9861 |
| [11] | Eddy SR. Non-coding RNA genes and the modern RNA world. Nature Reviews Genetics, 2001, 2 (12): 919–929. DOI:10.1038/35103511 |
| [12] | Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Molecular Microbiology, 1998, 29 (6): 1321–1330. DOI:10.1046/j.1365-2958.1998.01021.x |
| [13] | Kazantsev AV, Pace NR. Bacterial RNase P:a new view of an ancient enzyme. Nature Reviews Microbiology, 2006, 4 (10): 729–740. DOI:10.1038/nrmicro1491 |
| [14] | Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Current Opinion in Microbiology, 2007, 10 (2): 102–109. DOI:10.1016/j.mib.2007.03.012 |
| [15] | Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Current Opinion in Microbiology, 2007, 10 (2): 134–139. DOI:10.1016/j.mib.2007.03.010 |
| [16] | Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiology and Molecular Biology Reviews, 2011, 75 (2): 286–300. DOI:10.1128/MMBR.00032-10 |
| [17] | Chambers JR, Sauer K. Small RNAs and their role in biofilm formation. Trends in Microbiology, 2013, 21 (1): 39–49. DOI:10.1016/j.tim.2012.10.008 |
| [18] | Prévost K, Desnoyers G, Jacques JF, et al. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes & Development, 2011, 25 (4): 385–396. |
| [19] | Brennan RG, Link TM. Hfq structure, function and ligand binding. Current Opinion in Microbiology, 2007, 10 (2): 125–133. DOI:10.1016/j.mib.2007.03.015 |
| [20] | Warrier I, Hicks LD, Battisti JM, et al. Identification of novel small RNAs and characterization of the 6S RNA of Coxiella burnetii. PLoS One, 2014, 9 (6): e100147. DOI:10.1371/journal.pone.0100147 |
| [21] | Kulkarni PR, Cui X, Williams JW, et al. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Research, 2006, 34 (11): 3361–3369. DOI:10.1093/nar/gkl439 |
| [22] | Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA:SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proceedings of the National Academy of Sciences, 2007, 104 (51): 20454–20459. DOI:10.1073/pnas.0708102104 |
| [23] | Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-function RNA regulators in bacteria. Biochimie, 2011, 93 (11): 1943–1949. DOI:10.1016/j.biochi.2011.07.016 |
| [24] | Grabowicz M, Koren D, Silhavy TJ. The CpxQ sRNA negatively regulates skp to prevent mistargeting of β-Barrel outer membrane proteins into the cytoplasmic membrane. Microbiology, 2016, 7 (2): 312–328. |
| [25] | Song T, Mika F, Lindmark B, et al. A new Vibrio cholerae, sRNA modulates colonization and affects release of outer membrane vesicles. Molecular Microbiology, 2008, 70 (1): 100–111. DOI:10.1111/j.1365-2958.2008.06392.x |
| [26] | Hoe CH, Raabe CA, Rozhdestvensky TS, et al. Bacterial sRNAs:regulation in stress. International Journal of Medical Microbiology, 2013, 303 (5): 217–229. DOI:10.1016/j.ijmm.2013.04.002 |
| [27] | Pfeiffer V, Papenfort K, Lucchini S, et al. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nature Structural & Molecular Biology, 2009, 16 (8): 840–846. |
| [28] | Pfeiffer V, Sittka A, Tomer R, et al. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Molecular Microbiology, 2007, 66 (5): 1174–1191. DOI:10.1111/mmi.2007.66.issue-5 |
| [29] | Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell, 2000, 101 (6): 613–623. DOI:10.1016/S0092-8674(00)80873-9 |
| [30] | Leng Y, Vakulskas CA, Zere TR, et al. Regulation of CsrB/C sRNA decay by EIIAGlc of the phosphoenolpyruvate:carbohydrate phosphotransferase system. Molecular Microbiology, 2015, 99 (4): 627–639. |
| [31] | Holmqvist E, Vogel J. A small RNA serving both the Hfq and CsrA regulons. Genes & Development, 2013, 27 (10): 1073–1081. |
| [32] | Kimata K, Tanaka Y, Inada T, et al. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. The EMBO Journal, 2001, 20 (13): 3587–3595. DOI:10.1093/emboj/20.13.3587 |
| [33] | Maki K, Otaka MH, Aiba H. A minimal base-pairing region of a bacterial small RNA SgrS required for translational repression of ptsG mRNA. Molecular Microbiology, 2010, 76 (3): 782–792. DOI:10.1111/mmi.2010.76.issue-3 |
| [34] | Semsey S, Andersson AM, Krishna S, et al. Genetic regulation of fluxes:iron homeostasis of Escherichia coli. Nucleic Acids Research, 2006, 34 (17): 4960–4967. DOI:10.1093/nar/gkl627 |
| [35] | Jacques JF, Jang S, Prévost K, et al. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Molecular Microbiology, 2006, 62 (4): 1181–1190. DOI:10.1111/mmi.2006.62.issue-4 |
| [36] | Schellhorn HE. Elucidating the function of the RpoS regulon. Future Microbiology, 2014, 9 (4): 497–507. DOI:10.2217/fmb.14.9 |
| [37] | Dalia AB. RpoS is required for natural transformation of Vibrio cholerae, through regulation of chitinases. Environmental Microbiology, 2016 . DOI:10.1111/1462-2920.13302 |
| [38] | Mccullen CA, Benhammou JN, Majdalani N, et al. Mechanism of positive regulation by DsrA and RprA small noncoding RNAs:pairing increases translation and protects rpoS mRNA from degradation. Journal of Bacteriology, 2010, 192 (21): 5559–5571. DOI:10.1128/JB.00464-10 |
| [39] | Wilf NM, Salmond GP. The stationary phase sigma factor, RpoS, regulates the production of a carbapenem antibiotic, a bioactive prodigiosin and virulence in the enterobacterial pathogen Serratia sp. ATCC 39006. Microbiology, 2012, 158 (Pt 3): 648–658. |
| [40] | Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. The EMBO Journal, 2010, 29 (18): 3094–3107. DOI:10.1038/emboj.2010.179 |
| [41] | Chevalier C, Boisset S, Romilly C, et al. Staphylococcus aureus RNAⅢ binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathogens, 2010, 6 (3): e1000809. DOI:10.1371/journal.ppat.1000809 |
| [42] | Huntzinger E, Boisset S, Saveanu C, et al. Staphylococcus aureus RNAⅢ and the endoribonuclease Ⅲ coordinately regulate spa gene expression. The EMBO Journal, 2005, 24 (4): 824–835. DOI:10.1038/sj.emboj.7600572 |
| [43] | Jones JB, Lacy GH, Bouzar H, et al. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Systematic & Applied Microbiology, 2004, 27 (6): 755–762. |
| [44] | Zimaro T, Thomas L, Marondedze C, et al. The type Ⅲ protein secretion system contributes to Xanthomonas citri subsp. citri biofilm formation. BMC Microbiology, 2014, 14 (1): 50–57. DOI:10.1186/1471-2180-14-50 |
| [45] | Ryan RP, Vorhölter FJ, Potnis N, et al. Pathogenomics of Xanthomonas:understanding bacterium-plant interactions. Nature Reviews Microbiology, 2011, 9 (5): 344–355. DOI:10.1038/nrmicro2558 |
| [46] | Abendroth U, Schmidtke C, Bonas U. Small non-coding RNAs in plant-pathogenic Xanthomonas spp.. RNA Biology, 2014, 11 (5): 457–463. DOI:10.4161/rna.28240 |
| [47] | Findeiss S, Schmidtke C, Stadler PF, et al. A novel family of plasmid-transferred anti-sense ncRNAs. RNA Biology, 2010, 7 (2): 120–124. DOI:10.4161/rna.7.2.11184 |
| [48] | Schmidtke C, Findeiss S, Sharma CM, et al. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Research, 2012, 40 (5): 2020–2031. DOI:10.1093/nar/gkr904 |
| [49] | Schmidtke C, Abendroth U, Brock J, et al. Small RNA sX13:a multifaceted regulator of virulence in the plant pathogen Xanthomonas. PLoS Pathogens, 2013, 9 (9): e1003626. DOI:10.1371/journal.ppat.1003626 |
| [50] | Liang H, Zhao YT, Zhang JQ, et al. Identification and functional characterization of small non-coding RNAs in Xanthomonas oryzae pathovar oryzae. BMC Genomics, 2011, 12 (1): 87–101. DOI:10.1186/1471-2164-12-87 |
| [51] | Jiang RP, Tang DJ, Chen XL, et al. Identification of four novel small non-coding RNAs from Xanthomonas campestris pathovar campestris. BMC Genomics, 2010, 11 (1): 316–325. DOI:10.1186/1471-2164-11-316 |
| [52] | An SQ, Febrer M, Mccarthy Y, et al. High-resolution transcriptional analysis of the regulatory influence of cell-to-cell signalling reveals novel genes that contribute to Xanthomonas phytopathogenesis. Molecular Microbiology, 2013, 88 (6): 1058–1069. DOI:10.1111/mmi.2013.88.issue-6 |
| [53] | 周莲, 王杏雨, 何亚文. 植物病原黄单胞菌DSF信号依赖的群体感应机制及调控网络. 中国农业科学, 2013, 46(14): 2910–2922. |
| [54] | Chen XL, Tang DJ, Jiang RP, et al. sRNA-Xcc1, an integron-encoded transposon-and plasmid-transferred trans-acting sRNA, is under the positive control of the key virulence regulators HrpG and HrpX of Xanthomonas campestris pathovar campestris. RNA Biology, 2011, 8 (6): 947–953. DOI:10.4161/rna.8.6.16690 |
| [55] | Shao W, Price MN, Deutschbauer AM, et al. Conservation of transcription start sites within genes across a bacterial genus. Microbiology, 2014, 5 (4): 01398–01412. |
| [56] | Cohen O, Doron S, Wurtzel O, et al. Comparative transcriptomics across the prokaryotic tree of life. Nucleic Acids Research, 2016 . DOI:10.1093/nar/gkw394 |