2. 北京农学院生物科学与工程学院,北京 102206;
3. 北京林果业生态环境功能提升协同创新中心,北京 102206
2. College of Biological Science and Engineering, Beijing University of Agriculture, Beijing 102206;
3. Beijing Collaborative Innovation Center for Eco-environmental Improvement with Forestry and Fruit Trees, Beijing 102206
生长素(Auxin)是第一个被发现的植物激素,其能促进植物细胞的膨大,引起植物向性生长,参与植物组织器官的形成及果实的发育成熟等,在植物响应逆境胁迫过程中发挥重要的调控作用[1]。
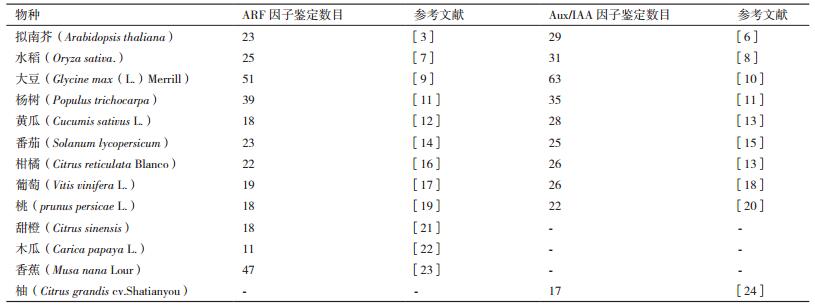
目前研究表明,生长素引起植物生理反应的细胞信号转导途径既有直接作用机制,也存在分子上的调控。参与生长素调节转录过程的一系列基因叫做原初生长素反应基因,已知的这类基因包括3个基因家族Aux/IAA(Auxin/indole acetic acid),GH3(Gretchen Hagen3)和SAUR(Small auxin up RNA),它们的启动子上游均含有的保守序列“TGTCTC”,该序列被称之为生长素响应原件(AuxREs)[2-3]。生长素响应因子(Auxin response factors,ARF)是一类能够识别并与AuxREs结合,调控生长素响应基因表达的转录因子[3]。ARF调节基因的表达决定其结构存在的状态,其可与其它的ARF或生长素阻遏蛋白(Auxin/Indole-3-Acetic Acid,Aux/IAA)形成二聚体,阻遏ARF的转录活性[4]。在生长素信号转导过程中,当内源生长素浓度增高时,生长素与受体TIR1/AFBs(Transport inhibitor resistant/Auxin signaling Fbox)结合后,使TIR1易于结合AUX/IAA蛋白并诱导发生泛素化反应,在26S蛋白酶降解作用下ARF被释放,开启对下游基因的调控作用(图 1)[5]。ARF与Aux/IAA是生长素的信号转导途径中的重要因子,其在植物体内广泛存在(表 1),并对一些生理过程发挥着重要的调控作用。
本文将对生长素信号转导相关因子ARF和Aux/IAA在果实成熟发育过程中的调控作用以及因子间相互作用的研究进展进行概述,以期为生长素调控果实发育成熟的分子机制研究提供参考。
1 生长素响应因子ARF 1.1 ARFs因子的结构一个典型的ARF因子由N-末端保守的DNA结合域(DNA-binding domain,DBD),C-末端保守的二聚体结合域(Dimerization domain,CTD)以及中间非保守序列(MR)组成(图 1)。其蛋白分子量一般在67-129 kD。其中,DBD结合于生长素响应基因上游的生长素响应元件[3],CTD结构域在由于组成上与Aux/IAA结构域Ⅲ、Ⅳ的相似性很高因此可与之结合[26]。MR的氨基酸序列可以决定ARFs转录因子对下游基因的激活或者抑制活性,如果富含谷氨酰胺,其ARF因子具有激活功能,如拟南芥AtARF5、6、7、8、9和19;如果富含脯氨酸、苏氨酸和丝氨酸,其ARF因子则为转录抑制子,如拟南芥AtARF1、2、4和9[27]。此外,有小部分ARF因子结构不包含CTD结构域,如拟南芥AtARF3、13和17[3],这类因子的功能尚未深入研究。
1.2 ARFs因子在果实发育成熟过程中的功能ARF因子在植物组织内普遍表达,且具有时空特异性[25]。研究表明拟南芥AtARF2、3、5-8、17和19分别参与调控植物体形态生长,如顶芽形成、花粉壁合成、维管束发育、下胚轴向性运动、不定根形态建成等[28-33]。在植物激素信号转导方面,AtARF2和AtARF19被认为是生长素和乙烯信号的关键位点,JAZ/TAFY10A的表达在被茉莉酸(Jasmonic acid,JA)诱导的同时受生长素响应因子AtARF6和AtARF8控制[29]。在维管束发育方面,AtARF5等位基因monopteros(mp)的强mp突变体与弱mp突变体均导致植株维管束发育紊乱,无效mp基因具有致死效应[34-35]。有研究表明,AtARF5可直接调节AtARR7和AtARR15参与维持顶端分生组织发育,可调控AtATHB8影响叶片维管束组织的结构[36-37]。刘振华等[25]总结了大量的研究成果,发现ARF sister pair基因双突变的表型往往比单突变要强,这证明了ARF因子在功能上的冗余性。
ARF对果实成熟发育调控的研究多集中于番茄。迄今为止,番茄中共发现有21个ARF因子[38],其中SlARF3、5、6、13、16和17在花、果实发育的绿熟期与红熟期表达较高,SlARF1、2、4、7、8、11和14在绿熟期时期表达较高[14]。在果实形态发育方面,Goetz等[39-40]发现抑制SlARF8表达的单性结实植株突变体果实比普通单性结实植株果实直径明显增加,推测SlARF8因子可能是番茄果实膨大的重要负调控因子,且该研究结果在拟南芥与茄子中也得到了验证。此外,SlARF7被证明也是果实发育初期的负调控子[41]。在果实糖分积累方面,Sagar等[42-43]发现抑制SlARF4表达可释放SlGLK1基因表达并诱导叶绿素大量合成最终促进果实的糖分积累,且在SlGLK1基因启动子区域发现AuxREs结构,推测SlARF4为该因子的负调控因子。此外,有研究推测SlARF4还与番茄细胞壁结构建成相关。在花器官发育中,拟南芥AtARF6、AtARF8参与调控JA合成及花器官成熟,AtARF17参与调控果实发育并且与胚授粉密切相关[44]。此外,在乙烯信号转导途径中,ARF因子同样参与调控。乙烯(Ethylene)被认为是调控果实成熟的主要植物激素,木瓜中乙烯信号转导因子CpETR1和CpETR2以及乙烯合成因子CpACS1、CpACS2和CpACO1的启动子区域均发现了AuxREs结构[22]。
ARF因子在其它果实发育中的研究也取得了一定进展。甜橙(Citrus sinensis)基因组中筛选出19个ARF因子,通过不同组织部位转录水平的检测,CsARF8和12在发育后期果实中表达显著[21]。木瓜(Carica papaya L.)中共筛选出11个ARF成员,其中CpARF1在果实发育过程中的表达显著增加[22]。苹果(Malus domestica Borkh)中也检测到共计29个ARF编码基因[45]。桃(Prunus persicae L.)基因组中筛选出了18个ARFs编码基因,其中大部分ARF因子在桃的组织部位广泛存在,个别ARF因子的表达部位具有局限性,如PpARF13和16在根与茎中没有表达,PpARF5仅在果实中表达[19]。我们实验室发现‘晚24号’桃在硬核期PpARF1表达量明显上调。由于在果实硬核期中果皮生长基本停滞,养分大量向种子中的胚和胚乳集中[46],因此推测该因子可能与桃胚形态建成以及内果皮木质化密切相关,此外有研究推测PpARF6因子参与调控果实花色素苷积累[20, 47]。
2 生长素/吲哚乙酸蛋白Aux/IAA 2.1 Aux/IAAs因子的结构Aux/IAAs因子是一类半衰期较短的核蛋白,普遍包含4个保守的结构域(图 1)[4, 48]。结构域Ⅰ中含有保守的亮氨酸序列(LxLxLx),乙烯信号阻遏蛋白(EAR)中也存在类似结构,该序列赋予Aux/IAA转录抑制子的特性,但抑制效力与LxLxLx序列的成分无明确规律[15, 49]。结构域Ⅱ包含13个氨基酸,序列高度保守。在信号转导过程中生长素受体TIR1与结构域Ⅱ结合引起Aux/IAA因子泛素化降解,从而调控下游基因的表达[3, 5, 26]。结构域Ⅲ、Ⅳ具有ARFs蛋白CTD的同源结构域,主要负责Aux/IAA蛋白自身的二聚化和多聚化以及与ARFs的二聚化,从而抑制生长素反应基因的转录。然而,有个别非典型性Aux/IAA因子缺失部分结构域,这类因子普遍表达量较低且受束性较强。如番茄SlIAA33同时缺失结构域Ⅰ与Ⅱ,目前在各个组织部位均未能有效检测到该因子的mRNA;SlIAA32缺失转录抑制功能的结构域Ⅱ,仅在番茄“转色期”发现该因子的生长素信号抑制活性,目前该因子尚未研究透彻[15];拟南芥AtIAA20和AtIAA31被发现缺失结构域Ⅱ,经检测这两个因子的半衰期长于其他结构完整的拟南芥Aux/IAA因子[50]。在功能方面,目前人们普遍认为Aux/IAA具有转录调控和参与组成生长素信号转导复合体两种作用。
2.2 Aux/IAAs在果实发育成熟过程中的功能Aux/IAA因子在植物发育过程中起重要调控作用,其作用主要包括向性生长,细胞伸长、分裂,根毛、维管组织的发育,以及花、果和种子的形态建成等[2, 51]。拟南芥AtIAA12突变体iaa12/bdl-1的根系发育不良[52]。AtIAA-28表达抑制突变体IAA28-1表现出侧根生长旺盛,顶端优势缺失等与生长素关联表型的变异[53]。AtIAA-1抑制型突变体axr5-1、番茄突变体SlIAA3[54]与马铃薯突变体StIAA2[55]的植株,其根系生长与向性运动均发生异常[56]。
Aux/IAAs在果实发育中有重要的调控作用。在果实形态发育方面,番茄SlIAA9表达抑制型突变体植株的复合叶片被简单叶片所替代,果实发育顺序发生颠倒从而产生单性结实的果实[57]。SlIAA9在转色期与红熟期时期高效表达,如沉默该因子的表达将会导致番茄单性结实[15, 51]。沉默SlIAA27可导致花粉与胚珠的生育能力明显降低,果实变小且胎座增大[58]。SlIAA17的组织定位结果揭示该基因在果肉中大量出现,沉默该基因后突变体果实较野生型果实明显增大,揭示该基因可能与果肉增厚有关[59]。SlIAA3随着番茄果实发育表达量逐步提升,在转色期对果实喷施乙烯信号阻遏剂1-MCP能显著抑制SlIAA3的表达,说明该基因还受到乙烯信号的调控[57]。在果实发育过程中,Aux/IAA的表达量变化差异明显。通过对草莓发育过程中FaAIAA1与FaIAA2的表达量进行检测,发现这两个基因在小绿期和白果期时期表达量较高,大绿期时期表达量最低,而FaIAA2在小绿期时期表达量最高,且随着发育进程的推进表达量逐步下降[60]。本实验室对桃Aux/IAA家族进行了鉴定与果实发育中的表达分析,结果显示PpIAA3和PpIAA17在桃果实成熟期中果皮的表达量显著升高,PpIAA26、PpIAA29Z种子中的表达量显著提高,表明以上4个因子在桃果实硬核期具有重要的调控作用[47]。由于PpIAA17与SlIAA17同源性较高,推测该因子也可能具有促进果实膨大的功能[59]。此外,史梦雅等认为PpIAA17还与桃内果皮在硬核期的木质化相关[29]。PpIAA3与SlIAA3在果实发育进程中的表达模式相似,推测两因子在果实发育也具有相似的调控功能[57]。Aux/IAA因子在植物体响应逆境胁迫的过程中也参与了重要的信号传导作用。经实验证实,拟南芥DREB/CBF家族可激活AtIAA5和AtIAA19表达以响应非生物胁迫,在IAAs基因表达抑制型突变体植株的抗逆性显著降低。该研究推测,Aux/IAA是协调植物响应胁迫以及生长素介导的调控网络的关键因子,为维持植物体稳定发育发挥重要的调控功能[61]。
3 ARF与Aux/IAA的互作调控目前普遍认为ARF与Aux/IAA因子间通过CTD结构域构成特定二聚体或直接形成二聚体调控下游基因表达,ARF因子自身也可直接调控下游基因表达[3-4]。生长素信号阻遏蛋白Aux/IAA结合并抑制ARF的转录调控活性是植物体响应生长素信号的主要机制[4, 32, 62]。近期在小立碗藓(Physcomitrella patens)的研究证实了生长素通过Aux/IAA影响ARF最终对下游基因进行调控这一机制,并发现了Aux/IAA对生长素信号的响应的专一性[63]。
近年来,基于互作模拟软件(Cytoscape,http://www.cytoscape.org)、酵母双杂交技术(Yeast two hybrid)与双分子荧光互补技术(Bimolecular fluorescence complementation,BiFC),在挖掘Aux/IAA-ARF互作关系上取得了一定的研究成果[27],目前以拟南芥、水稻及番茄等物种的研究居多。ARF与Aux/IAA互作网络非常复杂,有研究通过整合共表达图谱与蛋白-蛋白互作数据发现多达70%的ARF与Aux/IAA因子存在相互作用的可能性[64]。目前,拟南芥中共发现213对互作关系,水稻中8个ARF因子与15个Aux/IAA因子间可相互作用,推测番茄SlARF2A至少可与5个Aux/IAA因子互作,SlARF7A与SlARF16推测至少可与11个Aux/IAA因子互作[64-65]。在互作方式上,除了典型的ARF-Aux/IAA二聚体,还存在ARF-ARF和Aux/IAA-Aux/IAA的互作模式,但目前研究尚不深入[27]。不同ARF-Aux/IAA组合功能各异[66]。有实验表明拟南芥AtIAA3-AtARF7、AtIAA19-AtARF7、AtIAA17-AtARF1和AtIAA8-AtARFs(AtARF7、11、16及19)互作单元分别参与子叶下胚轴及根系发育调控[67-68]。AtARF6和8与一些Aux/IAA因子仅在发育中的花与花芽组织内互作,这表明ARF-Aux/IAA的互作具有组织特异性[64]。番茄SlARF5在成熟期表达量最高,该因子可与已证实的成熟相关因子SlIAA3发生互作[69]。此外SlARF7A-SlIAA8互作在番茄果实绿熟期的作用同样值得瞩目[69]。通过整合现有研究结果,刘振华等推测虽然ARF和Aux/IAA的互作网络非常复杂,但在特定的发育时期存在一对或几对优势组合起主要作用,其他组合起辅助作用[26]。迄今为止,ARF和Aux/IAA的互作研究在果实发育中依然存在较大空白。
在植物体的生长素信号转导过程中还存在ARF与Aux/IAA参与的其他信号转导途径。ETT因子(隶属于拟南芥ARF家族)缺失与Aux/IAA结合的关键结构域PBI,但依然在在雌蕊顶部具有生长素调节作用。经研究表明,ETT因子可与一类受生长素直接调控的具有典型碱性螺旋(bHLH)结构的转录因子IND(TF)发生互作,使之不需要通过泛素化途径就可响应生长素调控。该研究中,ETT-TE途径被认为有助于加快Aux/IAA因子的重新合成,重置生长素对下游基因的影响[70]。
4 展望生长素信号转导相关因子ARF和Aux/IAA在生长素介导的果实发育过程中具有非常重要的调控作用,包括果实与种子的形态建成、果肉膨大和糖分积累等。迄今为止,对ARF、Aux/IAA在果实发育中的调控方式与生物学功能方面的探究取得了一些进展,但是多集中于番茄与草莓等草本植物的转基因植株层面,而对具体生理指标相关基因的调控方面研究较为匮乏,因此具有广阔的研究前景。
此外,ARF与Aux/IAA的调控功能在核果类果树中的研究较少。其主要原因为核果类果树的转化体系尚未构建完全,进行ARF与Aux/IAA因子的基因功能验证比较困难。因此,目前在探究该类果实的ARF与Aux/IAA因子功能时,可根据果实生长类型,探究其在模式植物中的调控作用以及果实发育过程中转录水平的变化趋势,经过体外互作实验等,对这些因子的功能及作用进行相关研究。
在ARF-Aux/IAA互作方面,其复杂的互作关系赋予了生长素多样的调控功能和不同的调控方式。目前,有多种成熟的实验技术可以对蛋白互作关系进行研究,然而由于ARF与Aux/IAA本身互作组合数量比较庞大,且在大部分物种中的功能尚未研究透彻,以致ARF与Aux/IAA互作关系网络的构建与调控方式的阐明需要更为深入的研究。
| [1] |
Kazan K, Manners JM. Linking development to defense:auxin in plant-pathogen interactions[J]. Trends in Plant Science, 2009, 14(7): 373-382. DOI:10.1016/j.tplants.2009.04.005 |
| [2] |
Guilfoyle TJ. Aux/IAA proteins and auxin signal transduction[J]. Trends in Plant Science, 1998, 3(6): 205-207. DOI:10.1016/S1360-1385(98)01244-8 |
| [3] |
Guilfoyle TJ, Hagen G. Auxin response factors[J]. Journal of Plant Growth Regulation, 2001, 10(3): 453-460. |
| [4] |
Shiv B. Tiwari GHTG. The roles of auxin response factor domains in auxin-responsive transcription[J]. Plant Cell, 2003, 15(2): 533-543. DOI:10.1105/tpc.008417 |
| [5] |
Woodward AW, Bartel B. Auxin:regulation, action, and interaction[J]. Annals of Botany, 2005, 95(5): 707-735. DOI:10.1093/aob/mci083 |
| [6] |
Guilfoyle TJ. Chapter 19 -Auxin-regulated genes and promoters[J]. New Comprehensive Biochemistry, 1999, 33: 423-459. DOI:10.1016/S0167-7306(08)60499-8 |
| [7] |
Wang D, Pei K, Fu Y, et al. Genome-wide analysis of the auxin response factors(ARF)gene family in rice(Oryza sativa)[J]. Gene, 2007, 394(2): 13-24. |
| [8] |
Thakur JK, Jain M, Tyagi AK, et al. Exogenous auxin enhances the degradation of a light down-regulated and nuclear-localized OsIAA1, an AUX/IAA protein from rice, via proteasome[J]. Biochimica Et Biophysica Acta, 2005, 1730(3): 196-205. DOI:10.1016/j.bbaexp.2005.08.002 |
| [9] |
Van HaC, Le DT, Nishiyama R, et al. The auxin response factor transcription factor family in soybean:genome-wide identification and expression analyses during development and water stress[J]. DNA Research, 2013, 20(5): 511-524. DOI:10.1093/dnares/dst027 |
| [10] |
Singh VK, Jain M. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean[J]. Frontiers in Plant Science, 2015, 6: 918. |
| [11] |
Kalluri UC, Difazio SP, Brunner A M, et al. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa[J]. BMC Plant Biology, 2007, 7(1): 59. DOI:10.1186/1471-2229-7-59 |
| [12] |
Liu SQ, Hu LF. Genome-wide analysis of the auxin response factor gene family in cucumber[J]. Genetics & Molecular Research Gmr, 2013, 12(4): 4317-4331. |
| [13] |
王垒, 娄丽娜, 闫立英, 等. 黄瓜果实发育早期Aux/IAA家族部分基因的差异表达分析[J]. 南京农业大学学报, 2011, 34(4): 13-17. |
| [14] |
Kumar R, Tyagi AK, Sharma AK. Genome-wide analysis of auxin response factor(ARF)gene family from tomato and analysis of their role in flower and fruit development[J]. Molecular Genetics and Genomics, 2011, 285(3): 245-260. DOI:10.1007/s00438-011-0602-7 |
| [15] |
Audrandelalande C, Bassa C, Mila I, et al. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato[J]. Plant & Cell Physiology, 2012, 53(4): 659-672. |
| [16] |
Xie R, Pang S, Ma Y, et al. The ARF, AUX/IAA and GH3 gene families in citrus:genome-wide identification and expression analysis during fruitlet drop from abscission zone A[J]. Molecular Genetics and Genomics, 2015, 290(6): 2089-2105. DOI:10.1007/s00438-015-1063-1 |
| [17] |
Wan S, Li W, Zhu Y, et al. Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in Vitis vinifera[J]. Plant Cell Reports, 2014, 33(8): 1365-1375. DOI:10.1007/s00299-014-1622-7 |
| [18] |
Çakir B, Kiliçkaya O, Olcay A C. Genome-wide analysis of Aux/IAA genes in Vitis vinifera:cloning and expression profiling of a grape Aux/IAA gene in response to phytohormone and abiotic stresses[J]. Acta Physiologiae Plantarum, 2013, 35(2): 365-377. |
| [19] |
Li H, Ran K, Sun Q. Genome-wide identification and expression analysis of peach auxin response factor gene families[J]. Journal of Plant Biochemistry & Biotechnology, 2016, 25(4): 1-9. |
| [20] |
焦云, 马瑞娟, 沈志军, 等. 桃果实花色素苷积累与ABP1、ARF6表达分析[J]. 中国南方果树, 2015, 44(1): 12-16. |
| [21] |
Li SB, Ouyang WZ, Hou XJ, et al. Genome-wide identification, isolation and expression analysis of auxin response factor(ARF)gene family in sweet orange(Citrus sinensis)[J]. Frontiers in Plant Science, 2015, 6: 119. |
| [22] |
Liu K, Yuan C, Li H, et al. Genome-wide identification and characterization of auxin response factor(ARF)family genes related to flower and fruit development in papaya(Carica papaya L.)[J]. Bmc Genomics, 2015, 16(1): 1-12. DOI:10.1186/1471-2164-16-1 |
| [23] |
Hu W, Zuo J, Hou X, et al. The auxin response factor gene family in banana:genome-wide identification and expression analyses during development, ripening, and abiotic stress[J]. Frontiers in Plant Science, 2015, 6: 742. |
| [24] |
张敬虎, 潘一山, 王少峰, 等. 柚AUX/IAA基因族的基因注释分析[J]. 福建热作科技, 2015(4): 5-11. |
| [25] |
刘振华, 于延冲, 向凤宁. 生长素响应因子与植物的生长发育[J]. 遗传, 2011, 33(12): 1335-1346. |
| [26] |
Gray WM, del Pozo JC, Walker L, et al. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana[J]. Genes & Development, 1999, 13(13): 1678-1691. |
| [27] |
Vernoux T, Brunoud G, Farcot E, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex[J]. Molecular Systems Biology, 2011, 7(1): 508-508. |
| [28] |
Hardtke CS, Ckurshumova W, Vidaurre DP, et al. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4[J]. Development, 2004, 131(5): 1089-1100. DOI:10.1242/dev.00925 |
| [29] |
Schruff MC, Spielman M, Tiwari S, et al. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs[J]. Development, 2006, 133(133): 251-261. |
| [30] |
Schlereth A, Möller B, Liu W, et al. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor[J]. Nature, 2010, 464(7290): 913-916. DOI:10.1038/nature08836 |
| [31] |
Mallory AC, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes[J]. Plant Cell, 2005, 17(5): 1360-1375. DOI:10.1105/tpc.105.031716 |
| [32] |
Wang S, Tiwari SB, Hagen G, et al. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arab-idopsis leaf mesophyll protoplasts[J]. Plant Cell, 2005, 17(7): 1979-1993. DOI:10.1105/tpc.105.031096 |
| [33] |
杨俊. 拟南芥生长素响应因子ARF17调控花粉壁模式形成[D]. 上海: 上海师范大学, 2013.
|
| [34] |
Przemeck GKH, Mattsson J, Hardtke CS, et al. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization[J]. Planta, 1996, 200(2): 229-237. |
| [35] |
Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development[J]. Embo Journal, 1998, 17(5): 1405-1411. DOI:10.1093/emboj/17.5.1405 |
| [36] |
Zhao Z, Andersen S U, Ljung K, et al. Hormonal control of the shoot stem-cell niche[J]. Nature, 2010, 465(7301): 1089-1092. DOI:10.1038/nature09126 |
| [37] |
Donner TJ, Sherr I, Scarpella E. Regulation of preprocambial cell state acquisition by auxin signaling in Arabidopsis leaves[J]. Development, 2009, 136(19): 3235-46. DOI:10.1242/dev.037028 |
| [38] |
Wu J, Wang F, Cheng L, et al. Identification, isolation and expression analysis of auxin response factor(ARF)genes in Solanum lycopersicum[J]. Plant Cell Reports, 2011, 30(11): 2059-2073. DOI:10.1007/s00299-011-1113-z |
| [39] |
Goetz M, Hooper LC, Johnson SD, et al. Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato[J]. Plant Physiology, 2007, 145(2): 351-366. DOI:10.1104/pp.107.104174 |
| [40] |
Du L, Bao C, Hu T, et al. SmARF8, a transcription factor involved in parthenocarpy in eggplant[J]. Molecular Genetics and Genomics, 2016, 291(1): 1-13. DOI:10.1007/s00438-015-1110-y |
| [41] |
De J M, Wolters-Arts M, Feron R, et al. The Solanum lycopersicum auxin response factor7(Sl ARF7) regulates auxin signaling during tomato fruit set and development[J]. Plant Journal, 2009, 57(1): 160-170. DOI:10.1111/tpj.2008.57.issue-1 |
| [42] |
Sagar M, Chervin C, Roustant JP, et al. Under-expression of the Auxin Response Factor Sl-ARF4 improves postharvest behavior of tomato fruits[J]. Plant Signaling & Behavior, 2013, 8(10): 10-4161. |
| [43] |
Sagar M, Chervin C, Mila I, et al. Sl-ARF4, an Auxin Response Factor involved in the control of sugar metabolism during tomato fruit development[J]. Plant Physiology, 2013, 161(3): 1362-1374. DOI:10.1104/pp.113.213843 |
| [44] |
Nagpal P, Ellis CM, Weber H, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation[J]. Development, 2005, 132(18): 4107-4118. DOI:10.1242/dev.01955 |
| [45] |
李慧峰, 冉昆, 何平, 等. 苹果生长素响应因子(ARF)基因家族全基因组鉴定及表达分析[J]. 植物生理学报, 2015(7): 1045-1054. |
| [46] |
朱立新, 李光晨. 面向21世纪课程教材-园艺通论[M]. 第2版. 北京: 中国农业大学出版社, 2005.
|
| [47] |
史梦雅, 张巍, 余佳, 等. 桃生长素反应因子和生长素/吲哚乙酸蛋白家族基因的克隆及表达分析[J]. 园艺学报, 2014, 41(3): 536-544. |
| [48] |
Shen CJ, Wang SK, Bai YH, et al. Functional analysis of the structural domain of ARF proteins in rice(Oryza sativa L.)[J]. Journal of Experimental Botany, 2010, 61(14): 3971-3981. DOI:10.1093/jxb/erq208 |
| [49] |
Tiwari SB, Hagen G, Guilfoyle TJ. Aux/IAA proteins contain a potent transcriptional repression domain[J]. Plant Cell, 2004, 16(2): 533-543. DOI:10.1105/tpc.017384 |
| [50] |
Dreher KA, Brown J, Saw RE, et al. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness[J]. Plant Cell, 2006, 18(3): 699-714. DOI:10.1105/tpc.105.039172 |
| [51] |
Wang H, Bouzayen M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis[J]. Plant Cell, 2005, 17(10): 2676-2692. DOI:10.1105/tpc.105.033415 |
| [52] |
Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis[J]. Science, 2008, 319(5868): 1384-1386. DOI:10.1126/science.1151461 |
| [53] |
Rogg LE, Lasswell J, Bartel B. A Gain-of-function mutation in IAA28 suppresses lateral root development[J]. Plant Cell, 2001, 13(3): 465-480. DOI:10.1105/tpc.13.3.465 |
| [54] |
Zhang J, Chen R, Xiao J, et al. Isolation and characterization of SlIAA3, an Aux/IAA gene from tomato[J]. Mitochondrial DNA, 2007, 18(6): 407-414. |
| [55] |
Kloosterman B, Visser RG, Bachem CW. Isolation and characterization of a novel potato Auxin/Indole-3-Acetic Acid family member(StIAA2) that is involved in petiole hyponasty and shoot morphogenesis[J]. Plant Physiology & Biochemistry, 2006, 44(11-12): 766-775. |
| [56] |
Yang X, Lee S, So JH, et al. The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1)[J]. Plant Journal, 2004, 40(5): 772-782. DOI:10.1111/tpj.2004.40.issue-5 |
| [57] |
Chaabouni S, Jones B, Delalande C, et al. Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth[J]. Journal of Experimental Botany, 2009, 60(4): 1349-1362. DOI:10.1093/jxb/erp009 |
| [58] |
Bassa C, Mila I, Bouzayen M, et al. Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato[J]. Plant & Cell Physiology, 2012, 53(9): 1583-1595. |
| [59] |
Su L, Bassa C, Audran C, et al. The Auxin Sl-IAA17 transcriptional repressor controls fruit size via the regulation of endoreduplication-related cell expansion[J]. Plant & Cell Physiology, 2014, 55(11): 1969-1976. |
| [60] |
Liu DJ, Chen JY, Lu WJ. Expression and regulation of the early auxin-responsive Aux/IAA genes during strawberry fruit development[J]. Molecular Biology Reports, 2011, 38(2): 1187-1193. DOI:10.1007/s11033-010-0216-x |
| [61] |
Shani E, Salehin M, Zhang Y, et al. Plant stress tolerance requires auxin-sensitive Aux/IAA transcriptional repressors[J]. Current Biology Cb, 2017, 27(3): 437-444. DOI:10.1016/j.cub.2016.12.016 |
| [62] |
Guilfoyle TJ, Hagen G. Getting a grasp on domain Ⅲ/Ⅳ responsible for Auxin Response Factor-IAA protein interactions[J]. Plant Science, 2012, 190(3): 82-88. |
| [63] |
Lavy M, Prigge MJ, Tao S, et al. Constitutive auxin response in Physcomitrella reveals complex interactions between Aux/IAA and ARF proteins[J]. Elife, 2016, 5: e13325. |
| [64] |
Piya S, Shrestha SK, Binder B, et al. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis[J]. Frontiers in Plant Science, 2014, 5: 744. |
| [65] |
Llères D, Swift S, Lamond AI. Detecting protein-protein interactions in vivo with FRET using multiphoton fluorescence lifetime imaging microscopy(FLIM)[J]. Curr Protoc Cytom, 2007, Chapter 12:Unit12.
|
| [66] |
Weijers D, Benkova E, Jäger KE, et al. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators[J]. Embo Journal, 2005, 24(10): 1874-1885. DOI:10.1038/sj.emboj.7600659 |
| [67] |
Tatematsu K, Kumagai S, Muto H, et al. MASSUGU2 encodes aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana[J]. Plant Cell, 2004, 16(2): 379-393. DOI:10.1105/tpc.018630 |
| [68] |
Arase F, Nishitani H, Egusa M, et al. IAA8 involved in lateral root formation interacts with the TIR1 Auxin receptor and ARF transcription factors in Arabidopsis[J]. PLoS One, 2012, 7(8): e43414. DOI:10.1371/journal.pone.0043414 |
| [69] |
Shen CJ, Bai YH, Wang SK, et al. Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress[J]. Febs Journal, 2010, 277(14): 2954-2969. DOI:10.1111/j.1742-4658.2010.07706.x |
| [70] |
Simonini S, Deb J, Moubayidin L, et al. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis[J]. Genes & Development, 2016, 30(20): 2286-2296. |