2. 中国科学院植物研究所 北方资源植物重点实验室,北京 100093;
3. IWBT,Stellenbosch University,Cape Town,Republic of South Africa
2. Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093;
3. IWBT, Stellenbosch University, Cape Town, Republic of South Africa
水分是生物体的重要组成成分,除了维持细胞膨压外,还是生物体新陈代谢所必需的。水分的可利用率是影响植物产量的重要因素,同时也是决定物种分布的主要因素之一[1]。大多数陆生植物在其生活史的某一个阶段会遭遇缺水胁迫,为了生存,植物进化出各种保护机制及适应策略。例如,通过气孔调控和特化结构来增强保水能力,通过促进根系生长增强吸水能力,通过累积糖和脯氨酸等物质来提高渗透调节能力,通过抗氧化剂和活性氧清除酶来降低干旱造成的活性氧伤害等[2]。虽然这些机制对抵御轻度和中度干旱胁迫通常有效,但无法帮助植物有效应对严重持久的干旱胁迫。
自然界中仅有较少一部分植物被称为“复苏植物(resurrection plant)”,能够在极端缺水情况下生存。即使干旱到细胞已经丧失90%以上水分的程度,只要遇水,这类植物就能迅速恢复生活状态,其所具有的耐脱水性状被称为耐干性(desiccation tolerance,简称DT)[3]。近年来,国际上对复苏植物的研究日益广泛,不但因为复苏植物蕴含着强效抗旱基因资源,而且对其耐旱复苏机制的深入了解,将推动人们对植物抗逆机理的更广泛认识,并借此找到植物专门应对严重干旱的更有效的机制,最终找到使植物彻底摆脱干旱威胁的钥匙。本文就复苏植物的起源、进化、种类分布及耐脱水机制研究现状等进行综述,并对复苏植物未来的研究方向进行了展望。
1 复苏植物的起源、进化及种类分布国外对复苏植物的认识始于1914年Pickett[4]对一些蕨类植物的原叶体生态适应性的研究。随后Myrothamnus flabellifolia、Xerophyta humilis和Craterostigma plantagineum等一些复苏植物被陆续报道出来[5]。目前已发现的复苏植物约有1 300种,主要分布在苔藓和蕨类植物中,裸子植物中未见,被子植物中仅发现135种,分属于15个科[6]。这些复苏植物多为草本,木本植物只有M. flabellifolia一例,主要分布于非洲东部和南部、澳大利亚和南美地区,零散分布于东亚和巴尔干半岛。复苏植物原生境极其相似,都生长于岩石表层土壤(大约1 cm厚度),一年中会经历多次失水及复水过程,其中一些种类已趋于濒危[6]。
在中国,对于复苏现象的记录可以追溯到明朝李时珍《本草纲目》对巻柏(Seaslniealla tamarsicnia(P. Beauv.)spring)的记录:“卷柏,释名万岁、长生不死草,可以在晾干后,经浸水而生。”迄今,在我国已被报道过的复苏植物除卷柏[7]和小立碗藓[8]等蕨类和苔藓植物外,主要集中于苦苣苔科旋蒴苣苔属旋蒴苣苔(Boea hygrometrica,俗名牛耳草)[9-11]及其近缘种[12-18]。部分复苏植物的种类与分布情况见表 1。
苔藓、藻类、地衣、蕨类以及拟蕨类植物的生殖及营养器官都具有耐脱水性,干旱和复苏过程都很快,在几小时之内即发生明显的形态变化,被认为是一种“完全型”耐干性(Full DT)[20]。虽然几乎所有被子植物的种子都具有耐脱水性,但只有少数被子植物的营养器官耐脱水,这可能意味着维管植物本身结构和形态更为适应陆地环境,体内保水调控能力更强,营养器官耐脱水复苏能力逐渐退化丢失。因此,Oliver、Farrant和Moore等[20, 21]进一步提出,种子的耐脱水能力可能是从低等植物“完全型”耐干性进化而来,最终成为一种由发育程序调控的细胞保护系统;在干旱频发的生境中,一些植物通过对种子耐脱水性调控程序的“重新编程”,使之在营养组织中可响应干旱诱导,最终进化出复苏植物的“改良型”耐干性(Modified DT)。图 1展示了几种被子复苏植物脱水和复苏状态。

|
| 图 1 几种被子复苏植物脱水与复苏状态 A & B : Myrothamnus flabellifolia;C & D : Xerophyta humilis;E & F : Boea hygrometrica(牛耳草)。A,C,E :脱水状态;B,D,F :复苏状态 |
复苏被子植物大多为多年生、株型矮小的草本,生长于岩石表层土壤(大约1 cm厚度),一生中会经历多次失水及复水过程。与仙人掌等耐旱植物不同,复苏植物大多缺乏特殊的保水性结构,如特化为针形的叶片,或者较厚的蜡质层等。因此在干旱来临时,复苏植物叶片失水速度很快,迅速向枝干折叠(图 1-A),或干枯卷曲(图 1-C),或向叶柄方向卷曲(图 1-E)。单子叶复苏植物X. humilis脱水过程中叶片会沿叶中脉折叠成原来叶片的一半[22]。很多复苏植物的叶背有柔毛或者刚毛,在叶片脱水收缩后,这些毛密度和硬度增大,覆盖整个叶背,使植物的外观形态结构和硬度发生明显变化,其生化生理机制尚未被揭示。这些变化在雨后或者浇水后逆转,恢复正常状态,植物也恢复生长发育(图 1-B,D,F)。
3 复苏植物耐脱水的生理生化和分子机制复苏植物所要面对的干旱胁迫是一种极端干旱,叶片等营养组织可干旱至脱水状态,即含水量最低降至10%以下,相当于超干种子的含水量。Vicré等[23]总结脱水对植物的损伤主要表现为3个方面:(1)细胞失水达到一定阈值(细胞内含水量低于40%[19])后原生质体收缩引发机械损伤;(2)膜系统稳定性下降;(3)细胞失水后代谢物浓度变化、大分子结构和功能的破坏对代谢速率和方向的影响。
低等复苏植物应对脱水主要依赖于复水过程中的损伤修复机制,需消耗大量能量;而被子复苏植物在脱水时主要倾向于加强保护、减小损伤的机制,不需消耗过多能量[20]。这些植物中均发现干旱诱导大量基因表达,通过调节细胞壁成分和物理性质、渗透调节、抗氧化、光合保护以及蛋白质质量控制等过程参与耐旱保护。
3.1 细胞壁折叠植物细胞脱水时原生质体缩小会引发机械压力,其主要根源在于细胞壁的刚性结构。研究发现复苏植物如牛耳草和C. wilmsii,叶肉细胞的细胞壁可以在细胞脱水后发生折叠,不但可确保质壁分离的程度不至于引起质膜的撕裂,而且维持了胞间连丝的结构[24-27]。免疫细胞化学分析发现C. wilmsii脱水组织细胞壁可萃取性与水合组织有所不同:脱水组织细胞壁发生重组修饰,主要涉及一系列诸如钙-果胶交联物和木葡聚糖修饰等诱导因子,以促进细胞壁的交联和紧缩[24]。C. plantagineum脱水胁迫时α-expansin蛋白表达明显上调,被证明能够增加细胞壁的可延展性[25]。木本复苏植物M. flabellifolia脱水组织与水合组织中果胶、木葡聚糖以及阿拉伯糖抗原表位并没有差异,其特征主要是在脱水组织中聚集高浓度的阿拉伯糖,形成阿拉伯糖与阿拉伯半乳糖聚合物[26, 27]。
3.2 渗透调节与大分子保护渗透调节是水分胁迫下细胞避免机械伤害、维持结构完整和膜稳定的广泛机制。复苏植物脱水过程中也会积累蔗糖[28],一方面可以通过渗透调节和形成原生质体“玻璃化”溶胶状态来维持生物大分子结构的稳定从而保护生物膜系统的完整;另一方面作为信号分子来调控碳水化合物的水平、调节植物生长以及能量代谢。C. plantagineum新鲜叶片中含有高浓度、在其它植物中少见的C8-糖-2-辛酮糖,脱水过程中大量转化成蔗糖[29, 30]。很多被子复苏植物在脱水过程中也会积累棉子糖和海藻糖等低聚糖[31, 32]。
除渗透调节保护物质外,一些特定的亲水性蛋白如胚胎晚期丰富蛋白(LEA)、热激蛋白(HSPs)也对细胞结构以及包括膜脂和膜蛋白在内的大分子的稳定性起重要保护作用。当水分匮缺时,植物会同时激活不同种类的LEA蛋白,形成必要的互作关系网络在不同的组织或细胞结构中来保护和稳定生物大分子[33]。Xiao等[11]对牛耳草全基因组测序数据分析发现牛耳草基因组中存在大量的LEA基因,且其中2/3在脱水时表达明显增加,表明LEA蛋白对于牛耳草脱水响应起重要作用。另外,牛耳草中克隆到29个HSP蛋白基因。HSPs作为分子伴侣同其它蛋白质结合来促进蛋白质的正确折叠、组装并转运到特定的亚细胞区域,或者识别未折叠或错误折叠的蛋白防止其不可逆的聚集[34, 35]。
3.3 抗氧化系统复苏植物体内存在的抗氧化系统对防止膜脂过氧化、维持膜及细胞结构的稳定也起了重要作用。Kranner等[36]发现M. flabellifolia的复苏能力与其体内抗氧化防御系统有直接关系,脱水时间延长至8个月时,其体内抗氧化剂耗尽便不能复苏。
植物体内抗氧化防御系统由非酶组分和酶组分两部分组成。抗氧化剂和酶类的积累及其相关基因的表达在多种复苏植物中均已被检测到[37-41]。干旱及复水过程中,C. wilmsii和X. viscosa营养组织中APX,GR以及SOD等抗氧化酶基因的表达都会升高[37]。X. viscosa中还确定了一个新的干旱诱导的过氧化物酶基因XvPer1,其编码蛋白的功能是保护细胞核内的核酸免受氧化胁迫损伤[38]。牛耳草、H. rhodopensis、Ramonda serbica和R. nathaliae等苦苣苔科复苏植物离体叶片脱水过程中发现除谷胱甘肽-S-转移酶、抗坏血酸、GSH及维生素E(α生育酚)的含量明显增加外,多酚含量和多酚氧化酶活性也明显升高[39-41]。
3.4 光合保护植物遭受水分胁迫时,光合作用是最敏感的生理过程,而光合作用的降低有利于减少光合过程中产生的活性氧对细胞的损伤[42]。复苏植物失水后光合活性也会迅速下降,甚至在其叶片含水量尚未显著下降时就停止了,例如Tripogon loliiformis在叶片失水30%时,光合作用便完全停止[43]。尽管如此,复苏植物复水后光合作用可在几天之内重新恢复,而非复苏植物干旱造成的光合失活却不能逆转。这个现象意味着复苏植物叶绿体中存在某些特殊的保护机制。推测复苏植物叶片在干旱后发生的折叠或卷曲有助于减少叶片对光能的吸收,防止脱水过程中光诱导产生的叶绿体活性氧伤害。
蕨类植物、苔藓植物以及双子叶复苏植物叶绿体在脱水时结构保持完整,大部分叶绿素得以保持,叶绿体内部类囊体的垛叠结构清晰可见,膜结构只有轻微损伤,复水后可以迅速复苏并恢复光合作用,例如牛耳草[44]和C. wilmsii[45]等(表 1),这些植物被称为叶绿素保持型(Homoiochlorophyllous)。牛耳草脱水过程中类囊体膜色素-蛋白复合体虽然解聚但不降解[9],可能是其干旱后光合活性快速停止以及复水后迅速恢复的重要机制之一。LEA蛋白被发现在保护光合作用蛋白稳定方面具有明显效果[45]。复苏植物脱水过程中也大量积累ELIP(early light induced protein)蛋白,例如C. plantagineum类囊体膜上大量积累的一种22 kD的蛋白(dsp22)对于防止脱水过程导致的光抑制有重要作用[46]。牛耳草全基因组测序分析也发现大量ELIP基因,脱水过程中会大量转录,推测可能在PSII的保护中起作用[11]。
单子叶复苏植物在脱水时大部分叶绿素降解,膜结构破坏,类囊体解体,复水后需要重新合成叶绿素并修复膜结构,复苏时间较长,不能迅速恢复光合作用,被称为叶绿素变化型(Poikilochlorophyllous),如X. viscosa[22]等(表 1)。因此叶绿素保持型复苏植物比叶绿素变化型能更快地恢复光合作用,而且叶绿素保持型复苏植物离体叶片可以复苏但叶绿素变化型复苏植物离体叶片却不能复苏[19]。
4 复苏植物耐脱水相关的信号转导及分子调控网络脱落酸(ABA)是一种重要的植物激素,在调控植物的生长发育以及植物响应干旱、冷等逆境胁迫方面有重要作用[47]。研究认为ABA对于复苏植物耐脱水基因的激活发挥着重要的协调作用[48]。Wang等发现牛耳草体内肌醇半乳糖苷及棉子糖的合成是依赖ABA信号途径完成的:BhGoLS1和BhRFS的表达都受ABA诱导[32, 49],ABA诱导的转录因子WRKY能够结合BhGoLS1启动子区的W-box元件,从而调控BhGoLS1基因表达[32]。Bartels等[50]发现C. plantagineum干旱叶片中ABA大量积累,而且外源ABA可促进其愈伤组织获得耐脱水性。但是C. plantagineum脱水早期部分基因的表达与ABA并不相关,这表明可能有其它的信号途径参与植物耐脱水过程[51]。在另一种苦苣苔科复苏植物H. rhodopensis中茉莉酸比ABA更早响应干旱胁迫;而且水杨酸、细胞分裂素以及生长素等激素都参与了其脱水响应的调控[52]。
复苏植物体内的一系列转录因子都调控着脱水相关基因的表达。例如H. rhodopensis转录组数据分析显示NAC、NF-YA、MADS box、HSF、GRAS以及WRKY家族的转录因子在水分匮缺时被诱导表达[53]。C. plantagineum中也确定了几类脱水诱导的转录因子,包括3个MYB转录因子[54],7个亮氨酸拉链家族蛋白(HDZIP)[55-57]以及一类新的锌指因子[58]。CPHB-1和CPHB-2是从C. plantagineum中克隆到的两个HDZIP基因,都受脱水胁迫诱导,但只有CPHB-2受ABA诱导,这表明两个基因分别在ABA依赖途径与非ABA依赖途径中发挥作用;而这两种蛋白又可以形成异源二聚体,推测它们可能将ABA依赖途径与ABA非依赖途径联系起来共同发挥功能[55]。牛耳草中发现的热激因子BhHSF1可通过诱导抗逆基因的同时抑制细胞分裂相关基因的表达,实现对植物在干旱胁迫下能量和物质代谢方向的协同调控,促使其从生长需求转向抗逆性[59]。另外,牛耳草的一个含C2结构域的小蛋白BhC2DP1,可通过钙信号途径参与ABA对干旱的调控[60]。
除转录水平调控外,近年来研究发现转座子、小RNA与非编码RNA和表观遗传调控在复苏植物抗逆过程中也发挥了重要作用。C. plantagineum中发现的受脱水及ABA诱导的逆转座子CDT-1,能够指导一双链21 bp的siRNA合成,从而发挥其功能[50, 61, 62]。后续研究发现,C. plantagineum转录组数据中较大一部分无法匹配到已知序列的转录本可能是非编码RNAs,这些序列在基因组中存在大量的拷贝,并在脱水过程中被大量诱导表达[63];其中一个长非编码RNA,在脱水过程中大量表达,猜测可能通过表观遗传、转录水平或转录后水平来调控基因的表达从而在植物耐脱水中起重要作用[64]。与C. plantagineum相似,牛耳草中克隆到的逆转座子片段S21发挥功能的形式与CDT-1类似,可能是通过转录产生干扰RNA来发挥作用[65]。
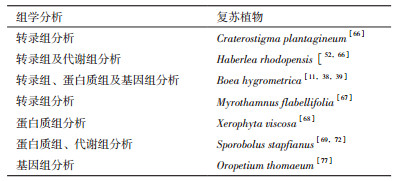
5 “组学”的方法来研究复苏植物耐脱水机制“组学”技术的发展实现了以高通量的方式来检测生物大分子在植物体内的丰度,所以研究者利用转录组、蛋白质组以及代谢组研究方法可以全面获取复苏植物在脱水及复水过程中转录本、蛋白质、代谢物的变化情况。转录组分析在多种复苏植物中鉴定了脱水复水诱导或抑制的转录本及其所富集的生物学过程和代谢过程,如逆境响应、氧化-还原反应、糖代谢和脂代谢、蛋白质降解与稳态维持和自噬等[38, 53, 66, 67]。蛋白质组学分析也发现,脱水过程中积累的胁迫保护蛋白涉及ROS清除、蔗糖积累、大分子保护、细胞壁折叠等不同生物学过程[39, 68, 69]。利用GC-MS、LC-MS、CE-MS以及NMR技术研究发现复苏植物脱水及复水过程中变化的代谢组分包括碳水化合物、氨基酸、核酸衍生物、脂类、多胺、抗氧化物以及防御化合物;而碳水化合物的代谢在复苏植物耐脱水过程中细胞保护方面起了关键作用[70, 71, 72]。复苏植物形态研究也从早期的显微镜技术观察推断结构及内部的变化[73, 74],发展至以高通量技术检测细胞壁、类囊体膜中糖类、蛋白的分布状态,并利用生物信息技术分析推测这些大分子在脱水状态下的排列[75, 76]。表 2中列出了已经利用“组学”技术研究过的复苏被子植物。另外,到目前为止,完成全基因组测序的复苏植物只有苦苣苔科的牛耳草[11]和禾本科的Oropetium thomaeum[77]。O. thomaeum基因组只有245 Mb,是已知具有最小基因组的禾本科草类;相比于其它禾本科植物,其基因组中含有较高的串联重复基因;这些串联重复基因主要涉及渗透胁迫响应,基因调控以及细胞代谢机制等并且对适应进化有重要作用[77]。而牛耳草基因组较大,约1 691 Mb,包含75.75%的重复序列(主要是转座子序列)和约占表达基因的10%的孤儿基因(Orphan gene),其中有128个孤儿基因与其复苏现象有关[11]。这表明复苏植物基因组在最新进化过程中可能通过基因组重排,基因组复制以及转座子或逆转座子的活性产生了独特的新基因。
复苏植物营养组织耐脱水机制与种子耐脱水机制有较大的相似性,例如高度依赖于抗氧化物、各种保护物质特别是蔗糖与棉子糖以及亲水性蛋白尤其是LEA大量积累的综合作用[20]。被子复苏植物营养组织耐脱水性可能来源于种子,但进化又使这种耐脱水性从发育程序调控转变为环境因子诱导调控,而且产生了CDT-1等新调控子。高通量技术数据的整合分析发现,脱水诱导基因表达、蛋白质及代谢物积累在复苏植物之间非常相似,再次证明了上述所阐述的复苏植物共有的耐脱水机制,同时也发现了很大一部分无法匹配到其它物种已知序列的转录本、或已鉴定的蛋白质和代谢物。这表明专门针对复苏植物进行的多物种基因组测序分析、进一步丰富和完善复苏植物特有蛋白质和代谢组等高通量数据库是现阶段深化复苏植物研究的前提和关键。因此利用“组学”技术及生物信息学方法,进一步分析复苏植物基因组序列所蕴含的遗传调控信息(包括基因序列和表观调控)、全面整合复苏植物转录组、蛋白质组、代谢组以及基因组数据并且与近缘的非复苏植物相关数据进行比较,确定其耐脱水性的关键成员/代谢组分和调控因子及其调控分子机制、比较复苏被子植物与种子耐脱水性之间以及复苏植物与非复苏植物的抗旱反应和调控机制之间的异同,将成为未来几年复苏植物研究的方向。这些研究将有助于理解植物抗旱调控机理和陆地植物的适应性进化,并为作物抗逆栽培和品种培育提供理论基础。
复苏植物中发现了大量与脱水复苏相关的基因,利用基因工程技术将其转化到模式植物拟南芥或烟草体内,大大提高了转基因植物自身的抗旱性。随着基因工程技术的发展,将复苏植物抗旱基因导入作物植株,使其稳定表达,可以培育出极度抗旱新品种,这对提高全球粮食产量具有重要意义。其中一些如BhLEA1、BhGOLS1、BhHsf1和BhDNAJC2等[31, 33, 45, 60]具有自主知识产权的复苏植物耐旱功能基因必将在我国植物抗逆分子育种上发挥极大作用。
| [1] | Delmer DP. Agriculture in the developing world: connecting innovations in plant research to downstream applications. Proceedings of the National Academy of Sciences of the United States of America , 2005, 102 (44) : 15739–15746. DOI:10.1073/pnas.0505895102 |
| [2] | Jaleel CA, Manivannan P, Wahid A, et al. Drought stress in plants: a review on morphological characteristics and pigments composition. International Journal of Agriculture & Biology , 2009, 11 (1) : 100–105. |
| [3] | Bewley JD. Physiological aspects of desiccation tolerance. Annual Review Plant Biology , 1979, 30 : 195–238. DOI:10.1146/annurev.pp.30.060179.001211 |
| [4] | Pickett FL. Some ecological adaptations of certain fern Prothallia-Camptosorus rhizophyllus Link., Asplenium platyneuron Oakes. American Journal of Botany , 1941, 1 (9) : 477–498. |
| [5] | Gaff DF. Desiccation-tolerant flowering plants in southern Africa. Science , 1971, 174 (4013) : 1033–1034. DOI:10.1126/science.174.4013.1033 |
| [6] | Gaff DF, Oliver M. The evolution of desiccation tolerance in angios-perm plants: a rare yet common phenomenon. Functional Plant Biology , 2013, 40 (4) : 315–28. DOI:10.1071/FP12321 |
| [7] | Wang X, Chen S, Zhang H, et al. Desiccation tolerance mechanism in resurrection fern-ally Selaginella tamariscina revealed by physiological and proteomic analysis. Journal of Proteome Research , 2010, 9 (12) : 6561–6577. DOI:10.1021/pr100767k |
| [8] | Wang XQ, Yang PF, Liu Z, et al. Exploring the mechanism of Physcomitrella patens desiccation tolerance through a proteomic strategy. Plant Physiology , 2009, 149 (4) : 1739–1750. DOI:10.1104/pp.108.131714 |
| [9] | Deng X, Hu Z, Wang H, et al. Effects of dehydration and rehydration on photosynthesis of detached leaves of the resurrective plant Boea hygrometrica. Acta Botanica Sinica , 1999, 42 (3) : 321–323. |
| [10] | Mitra J, Xu G, Wang B, et al. Understanding desiccation tolerance using the resurrection plant Boea hygrometrica as a model system. Frontiers in Plant Science , 2013, 4 : 446. |
| [11] | Xiao LH, Yang G, Zhang L C, et al. The resurrection genome of Boea hygrometrica: A blueprint for survival of dehydration. Proceedings of the National Academy of Sciences of the United States of America , 2015, 112 (18) : 5833–5837. DOI:10.1073/pnas.1505811112 |
| [12] | 张丹丹, 周守标, 周会, 等. 大花旋蒴苣苔对脱水与复水的生理响应. 生态学杂志 , 2016, 1 : 74–80. |
| [13] | Shen Y, Tang M J, Hu YL, et al. Isolation and characterization of a dehydrin-like gene from drought-tolerant Boea crassifolia. Plant Science , 2004, 166 (5) : 1167–1175. DOI:10.1016/j.plantsci.2003.12.025 |
| [14] | Chen BJ, Wang Y, Hu YL, et al. Cloning and characterization of a drought-inducible MYB gene from Boea crassifolia. Plant Science , 2005, 168 (2) : 493–500. DOI:10.1016/j.plantsci.2004.09.013 |
| [15] | Wu H, Shen Y, Hu Y, et al. A phytocyanin-related early nodulin-like gene, BcBCP1, cloned from Boea crassifolia enhances osmotic tolerance in transgenic tobacco. Journal of Plant Physiology , 2011, 168 (9) : 935–943. DOI:10.1016/j.jplph.2010.09.019 |
| [16] | Huang W, Yang SJ, Zhang SB, et al. Cyclic electron flow plays an important role in photoprotection for the resurrection plant Paraboea rufescens under drought stress. Planta , 2012, 235 (4) : 819–828. DOI:10.1007/s00425-011-1544-3 |
| [17] | Fu P, Zhang Y, Fan ZX, et al. Leaf gas exchange and xylem hydraulic traits of a resurrection plant(Paraboea rufescens, Gesneriaceae)and its responses to drought and re-watering. Oral presentation, 7th International workshop on desiccation sensitivity and tolerance across life forms. 2016. |
| [18] | Li A, Wang D, Yu B, et al. Maintenance or collapse: responses of extraplastidic membrane lipid composition to desiccation in the resurrection plant Paraisometrum mileense. PLoS ONE , 2014, 9 (7) : e103430. DOI:10.1371/journal.pone.0103430 |
| [19] | Dinakar C, Djilianov D, Bartel D. Photosynthesis in desiccation tolerant plants: Energy metabolism and antioxidative stress defense. Plant Science , 2012, 182 : 29–41. DOI:10.1016/j.plantsci.2011.01.018 |
| [20] | Oliver MJ, Tuba Z, Mishler BD. The evolution of vegetative desiccation tolerance in land plants. Plant Ecology , 2000, 151 (1) : 85–100. DOI:10.1023/A:1026550808557 |
| [21] | Farrant JM, Moore JP. Programming desiccation-tolerance: from plants to seeds to resurrection plants. Current Opinion in Plant Biology , 2011, 14 (3) : 340–345. DOI:10.1016/j.pbi.2011.03.018 |
| [22] | Sherwin HW, Farrant JM. Protection mechanisms against excess light in the resurrection plants Craterostigma wilmsii and Xerophyta viscosa. Plant Growth Regulation , 1998, 24 (3) : 203–210. DOI:10.1023/A:1005801610891 |
| [23] | Vicré M, Farrant J M, Driouich A. Insights into the cellular mechanisms of desiccation tolerance among angiosperm resurrection plant species. Plant, Cell & Environment , 2004, 27 (11) : 1329–1340. |
| [24] | Vicré M, Sherwin HW, Driouich A, et al. Cell wall characteristics and structure of hydrated and dry leaves of the resurrection plant Craterostigma wilmsii, a microscopical study. Journal of Plant Physiology , 1999, 155 (6) : 719–726. DOI:10.1016/S0176-1617(99)80088-1 |
| [25] | Jones L, McQueen-Mason S. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Letters , 2004, 559 (1-3) : 61–65. DOI:10.1016/S0014-5793(04)00023-7 |
| [26] | Moore JP, Nguema-Ona E, Chevalier L, et al. Response of the leaf cell wall to desiccation in the resurrection plant Myrothamnus flabellifolius. Plant Physiology , 2006, 141 (2) : 651–662. DOI:10.1104/pp.106.077701 |
| [27] | Moore JP, Farrant JM, Driouich A. A role for pectin-associated arabinans in maintaining the flexibility of the plant cell wall during water deficit stress. Plant Signaling & Behavior , 2008, 3 (2) : 102–104. |
| [28] | Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annual Review Plant Physiology and Plant Molecular Biology , 1996, 47 : 377–403. DOI:10.1146/annurev.arplant.47.1.377 |
| [29] | Bianchi G, Gamba A, Murelli C, et al. Novel carbohydrate metabo-lism in the resurrection plant Craterostigma plantagineum. The Plant Journal , 1991, 1 (3) : 355–359. DOI:10.1046/j.1365-313X.1991.t01-11-00999.x |
| [30] | Ingram J, Chandler JW, Gallagher L, et al. Analysis of cDNA clones encoding sucrose-phosphate synthase in relation to sugar interconversions associated with dehydration in the resurrection plant Craterostigma plantagineum Hochst. Plant Physiology , 1997, 115 (1) : 113–121. DOI:10.1104/pp.115.1.113 |
| [31] | Norwood M, Truesdale MR, Richter A, et al. Photosynthetic carbo-hydrate metabolism in the resurrection plant Craterostigma planta-gineum. Journal of Experimental Botany , 2000, 51 (343) : 159–165. DOI:10.1093/jexbot/51.343.159 |
| [32] | Wang Z, Zhu Y, Wang L, et al. A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase(BhGolS1)promoter. Planta , 2009, 230 (6) : 1155–1166. DOI:10.1007/s00425-009-1014-3 |
| [33] | Illing N, Denby KJ, Collett H, et al. The signature of seeds in resurrection plants: a molecular and physiological comparison of desiccation tolerance in seeds and vegetative tissues. Integrative & Comparative Biology , 2005, 45 (5) : 771–787. |
| [34] | 陈世璇, 张振南, 王波, 等. 复苏植物旋蒴苣苔J结构域蛋白编码基因BhDNAJC2的克隆, 表达与功能. 植物学报 , 2015, 50 (2) : 180–190. |
| [35] | Zhang Z, Wang B, Sun S, et al. Molecular cloning and differential expression of sHSP gene family members from the resurrection plant Boea hygrometrica in response to abiotic stresses. Biologia , 2013, 68 (4) : 651–661. |
| [36] | Kranner I, Beckett RP, Wornik S, et al. Revival of a resurrection plant correlates with its antioxidant status. The Plant Journal , 2002, 31 (1) : 13–24. DOI:10.1046/j.1365-313X.2002.01329.x |
| [37] | Mowla SB, Thomson JA, Farran JM, et al. A novel stress-inducible antioxidant enzyme identified from the resurrection plant Xerophyta viscosa Baker. Planta , 2002, 215 (5) : 716–726. DOI:10.1007/s00425-002-0819-0 |
| [38] | Zhu Y, Wang B, Phillips J, et al. Global transcriptome analysis reveals acclimation-primed processes in the acquisition of desiccation tolerance in Boea hygrometrica. Plant Cell Physiology , 2015, 56 (7) : 1429–1441. DOI:10.1093/pcp/pcv059 |
| [39] | Jiang G, Wang Z, Shang H, et al. Proteome analysis of leaves from the resurrection plant Boea hygrometrica in response to dehydration and rehydration. Planta , 2007, 225 (6) : 1405–1420. DOI:10.1007/s00425-006-0449-z |
| [40] | Jovanovic Z, Rakic T, Stevanovic B, et al. Characterization of oxidative and antioxidative events during dehydration and rehydration of resurrection plants Ramonda nathaliae. Plant Growth Regulation , 2011, 64 (3) : 231–240. DOI:10.1007/s10725-011-9563-4 |
| [41] | Farrant JM, Vander Willigen C, Loffell DA, et al. An investigation into the role of light during desiccation of three angiosperm resurrection plants. Plant, Cell & Environment , 2003, 26 (8) : 1275–1286. |
| [42] | Karbaschi MR, Williams B, Taji A, et al. Tripogon loliiformis elicits a rapid physiological and structural response to dehydration for desiccation tolerance. Functional Plant Biology , 2016, 43 (7) : 643–655. DOI:10.1071/FP15213 |
| [43] | Wang L, Shang H, Liu Y, et al. A role for a cell wall localized glycine-rich protein in dehydration and rehydration of the resurrection plant Boea hygrometrica. Plant Biology , 2009, 11 (6) : 837–848. DOI:10.1111/plb.2009.11.issue-6 |
| [44] | Liu X, Wang Z, Wang L, et al. LEA 4 group genes from the resurrection plant Boea hygrometrica confer dehydration tolerance in transgenic tobacco. Plant Science , 2009, 176 (1) : 90–98. DOI:10.1016/j.plantsci.2008.09.012 |
| [45] | Alamillo JM, Bartels D. Effects of desiccation on photosynthesis pigments and the ELIP-like dsp 22 protein complex in the resurrection plant Craterostigma plantagineum. Plant Science , 2001, 160 (6) : 1161–1170. DOI:10.1016/S0168-9452(01)00356-9 |
| [46] | Sherwin HW, Farrant JM. Differences in rehydration of three desiccation-tolerant angiosperm species. Annals of Botany , 1996, 78 (6) : 703–710. DOI:10.1006/anbo.1996.0180 |
| [47] | Bartels D, Phillips J, Chandler J. Desiccation tolerance: Gene expression, pathways, and regulation of gene expression. [M]// Jenks MA, Wood AJ. Plant desiccation tolerance. Ames, Iowa: Blackwell Publishing, 2007: 115-148. |
| [48] | Toldi O, Tuba Z, Scott P. Vegetative desiccation tolerance: is it a goldmine for bioengineering crops?. Plant Science , 2009, 176 (2) : 187–199. DOI:10.1016/j.plantsci.2008.10.002 |
| [49] | Wang Z, Liu Y, Wei J, et al. Cloning and expression of a gene encoding a raffinose synthase in the resurrection plant Boea hygrometrica. Chinese Bulletin Botany , 2012, 47 (1) : 44–54. DOI:10.3724/SP.J.1259.2012.00044 |
| [50] | Furini A, Koncz C, Salamini F, et al. High level transcription of a member of a repeated gene family confers dehydration tolerance to callus tissue of Craterostigma plantagineum. The EMBO Journal , 1997, 16 (12) : 3599–3608. DOI:10.1093/emboj/16.12.3599 |
| [51] | Frank W, Munnik T, Kerkmann K, et al. Water deficit triggers phospholipase D activity in the resurrection plant Craterostigma plantagineum. The Plant Cell , 2000, 12 (1) : 111–124. DOI:10.1105/tpc.12.1.111 |
| [52] | Djilianov DL, Dobrev PI, Moyankova DP, et al. Dynamics of endogenous phytohormones during desiccation and recovery of the resurrection plant species Haberlea rhodopensis. Journal of Plant Growth Regulation , 2013, 32 (3) : 564–574. DOI:10.1007/s00344-013-9323-y |
| [53] | Gechev TS, Benina M, Obata T, et al. Molecular mechanisms of desiccation tolerance in the resurrection glacial relic Haberlea rhodopensis. Cellular and Molecular Life Sciences , 2013, 70 (4) : 689–709. DOI:10.1007/s00018-012-1155-6 |
| [54] | Iturriaga G, Leyns L, Villegas A, et al. A family of novel myb-related genes from the resurrection plant Craterostigma plantagineum are specifically expressed in callus and roots in response to ABA or desiccation. Plant Molecular Biology , 1996, 32 (4) : 707–716. DOI:10.1007/BF00020211 |
| [55] | Frank W, Phillips J, Salamini F, et al. Two dehydration-inducible transcripts from the resurrection plant Craterostigma plantagineum encode interacting homeodomain-leucine zipper proteins. The Plant Journal , 1998, 15 (3) : 413–421. DOI:10.1046/j.1365-313X.1998.00222.x |
| [56] | Deng X, Phillips J, Meijer A H, et al. Characterization of five novel dehydration-responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum. Plant Molecular Biology , 2002, 49 (6) : 601–610. DOI:10.1023/A:1015501205303 |
| [57] | Deng X, Phillips J, Brautigam A, et al. A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses. Plant Molecular Biology , 2006, 61 (3) : 469–489. DOI:10.1007/s11103-006-0023-x |
| [58] | Hilbricht T, Salamini F, Bartels D. CpR18, a novel SAP-domain plant transcription factor, binds to a promoter region necessary for ABA mediated expression of the CDeT27-45 gene from the resurrection plant Craterostigma plantagineum Hochst. The Plant Journal , 2002, 31 (3) : 293–303. DOI:10.1046/j.1365-313X.2002.01357.x |
| [59] | Zhu Y, Wang Z, Jing YJ, et al. Ectopic over-expression of BhHsf1, a heat shock factor from the resurrection plant Boea hygrometrica, leads to increased thermotolerance and retarded growth in transgenic Arabidopsis and tobacco. Plant Molecular Biology , 2009, 71 : 451–467. DOI:10.1007/s11103-009-9538-2 |
| [60] | Zhang L, Ji F, Wang L, et al. A small C2-domain protein from the resurrection plant Boea hygrometrica promotes plant responses to abscisic acid. Chinese Bulletin of Botany , 2012, 47 (1) : 11–27. DOI:10.3724/SP.J.1259.2012.00011 |
| [61] | Smith-Espinoza CJ, Phillips JR, Salamini F, et al. Identification of further Craterostigma plantagineum cdt mutants affected in abscisic acid mediated desiccation tolerance. Molecular Genetics and Genomics , 2005, 274 (4) : 364–372. DOI:10.1007/s00438-005-0027-2 |
| [62] | Hilbricht T, Varotto S, Sgaramella V, et al. Retrotransposons and siRNA have a role in the evolution of desiccation tolerance leading to resurrection of the plant Craterostigma plantagineum. New Phytologist , 2008, 179 (3) : 877–887. DOI:10.1111/nph.2008.179.issue-3 |
| [63] | Giarola V, Bartels D. What can we learn from the transcriptome of the resurrection plant Craterostigma plantagineum?. Planta , 2015, 242 (2) : 427–434. DOI:10.1007/s00425-015-2327-z |
| [64] | Giarola V, Krey S, Frerichs A, et al. Taxonomically restricted genes of Craterostigma plantagineum are modulated in their expression during dehydration and rehydration. Planta , 2015, 241 (1) : 193–208. DOI:10.1007/s00425-014-2175-2 |
| [65] | Zhao Y, Xu T, Shen CY, et al. Identification of a retroelement from the resurrection plant Boea hygrometrica that confers osmotic and alkaline tolerance in Arabidopsis thaliana. PLoS ONE , 2014, 9 (5) : e98098. DOI:10.1371/journal.pone.0098098 |
| [66] | Rodriguez MCS, Edsg?rd D, Hussain SS, et al. Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. The Plant Journal , 2010, 63 (2) : 212–228. DOI:10.1111/j.1365-313X.2010.04243.x |
| [67] | Ma C, Wang H, Macnish AJ, et al. Transcriptomic analysis reveals numerous diverse protein kinases and transcription factors involved in desiccation tolerance in the resurrection plant Myrothamnus flabellifolia. Horticulture Research , 2015, 2 : 15034. |
| [68] | Ingle R, Schmidt U, Farrant J, et al. Proteomic analysis of leaf proteins during dehydration of the resurrection plant Xerophyta viscosa. Plant, Cell & Environment , 2007, 30 (4) : 435–446. |
| [69] | Oliver MJ, Jain R, Balbuena TS, et al. Proteome analysis of leaves of the desiccation-tolerant grass, Sporobolus stapfianus, in response to dehydration. Phytochemistry , 2011, 72 (10) : 1273–1284. DOI:10.1016/j.phytochem.2010.10.020 |
| [70] | Moyankova D, Mladenov P, Berkov S, et al. Metabolic profiling of the resurrection plant Haberlea rhodopensis during desiccation and recovery. Physiologia Plantarum , 2014, 152 (4) : 675–687. DOI:10.1111/ppl.2014.152.issue-4 |
| [71] | Yobi A, Wone BWM, Xu W, et al. Comparative metabolic profiling between desiccation-sensitive and desiccation tolerant species of Selaginella reveals insights into the resurrection trait. The Plant Journal , 2012, 72 (6) : 983–999. DOI:10.1111/tpj.2012.72.issue-6 |
| [72] | Oliver MJ, Guo L, Alexander DC, et al. A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. The Plant Cell , 2011, 23 (4) : 1231–1248. DOI:10.1105/tpc.110.082800 |
| [73] | Liu YB, Wang G, Liu J, et al. Anatomical, morphological and metabolic acclimation in the resurrection plant Reaumuria soongorica during dehydration and rehydration. Journal of Arid Environments , 2007, 70 : 183–194. DOI:10.1016/j.jaridenv.2006.12.020 |
| [74] | Moore JP, Hearshaw M, Ravenscroft N, et al. Desiccation-induced ultrastructural and biochemical changes in the leaves of the resurrection plant Myrothamnus flabellifolia. Australian Journal of Botany , 2007, 55 : 482–491. DOI:10.1071/BT06172 |
| [75] | Moore JP, Nguema-Ona EE, Vicré-Gibouin M, et al. Arabinose-rich polymers as an evolutionary strategy to plasticize resurrection plant cell walls against desiccation. Planta , 2013, 273 (3) : 739–54. |
| [76] | Zia A, Walker B J, Oung H M, et al. Protection of the photosynthetic apparatus against dehydration stress in the resurrection plant Craterostigma pumilum. The Plant Journal , 2016 . DOI:10.1111/tpj.13227 |
| [77] | VanBuren R, Bryant D, Edger PP, et al. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature , 2015, 527 (7579) : 508–511. DOI:10.1038/nature15714 |