b. Puer University, Puer 665000, Yunnan, China;
c. Institute of Comparative Study of Traditional Materia Medica, Institutes of Integrative Medicine, Fudan University, Shanghai, 200040, China;
d. Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan, China;
e. Central Herbarium of Uzbekistan, Institute of Botany, Academy Sciences of Uzbekistan, Tashkent 100125, Uzbekistan
Rosaceae is a globally distributed family found mostly in the northern hemisphere that comprises approximately 2950 species in 91 genera (Christenhusz and Byng, 2016). Up to 55 genera and 950 species of Rosaceae, of which 55% are endemics, are listed in the Flora of China (Lingdi et al., 2003). Rosaceae has been divided into four subfamilies: Amygdaloideae, Rosaceae, Maloideae and Prunoideae (Potter et al., 2007). The genus Prunus L. belongs to subfamily Prunoideae.
The genus Prunus L. senso lato (s.l) consists of 430 species (Chen et al., 2013). Species within the genus Prunus L. are mainly trees or shrubs. The genus is defined based on a combination of characters, including the presence of leaf glands, having a superior ovary, a solitary carpel with two pendulous anatropous ovules per carpel, and drupes for fruits, which have succulent mesocarp that is fleshy, or dry, and does not split, or more rarely, splits when ripe (Rehder, 1940; Li and Bartholomew, 2003; Chin et al., 2013; Zhao et al., 2016). In Shi et al. (2013) used molecular analysis to divide the genus Prunus senso lato into three subgenera: subg. Padus, subg. Cerasus, and subg. Prunus. Cerasus is the most important subgenus of the genus Prunus. It consists of 45 species in China (Yu and Li, 1986; Li and Bartholomew, 2003; Chen et al., 2013). These species have been grouped into 11 sections by Li and Bartholomew (2003). The subg. Cerasus is characterized by having inflorescences with usually conspicuous bracts, solitary flowers or several flowers in short racemes or corymbs, and by having smooth drupes that are glabrous but not glaucous, with endocarp that is not compressed or is slightly compressed (Rehder, 1940; Yu and Li, 1986; Li and Bartholomew, 2003).
During field studies in Hunan province (Yongshun County, Xiaoxi National Nature Reserve and Suining County, Huangsang National Nature Reserve) and Hubei Province (Wufeng County) conducted by Tao Deng, Xiao-Shuang Zhang, Meng-Hua Zhang, DaiGui Zhang between 2013 and 2015, several specimens of an unknown species of Prunus were found. The new taxon was determined to be most morphologically similar to Prunus cerasoides Buch.-Ham. ex D.Don of Prunus subg. Cerasus sect. Serrula based on the presence of characteristic features such as having 1-4 flowered, umbellate or subumbellate inflorescences, petals apex divided. The results of morphological and phylogenetical analyses support the status of the taxon as a new species, which is described below.
2. Material and methods 2.1. Morphological analysesMorphological observations and measurement of leaves, calyx, flowers and fruits were randomly made on flowering and fruiting plants. A total of 9 diagnostic characters were compared (Table 1). Specimens were investigated in herbaria from KUN and JIU.
| Characters | Prunus sunhangii | Prunus cerasoides |
| Leaf margin | apex acuminate | apex acuminate to long acuminate |
| Petals | white | white or pink |
| Sepals margin | laxly dentate | entire |
| Petals apex | longitudinally 2-lobed | emarginate |
| Stamens | 17-25 | 32-34 |
| Hypanthium color | brown | red to dark red |
| Drupe | black | purplish black |
| Flowering | March-April | October-December |
| Fruiting | April-May | February-March |
The voucher information of the studied taxa is listed in Appendix A1. In total, 12 species were sampled to represent Prunus subg. Cerasus, and one outgroup Prunus henryi (Prunus subg. Laurocerassus Koehne, Chin et al., 2014).
2.3. DNA extraction, amplification, and sequencingGenomic DNA was isolated using TaKaRa Universal Genomic DNA Extraction Kit Ver. 3.0 (DV811A) (TaKaRa, Osaka, Japan) according to the manufacturer's protocol. The selected DNA regions were amplified, following standard polymerase chain reaction (PCR) protocols (Kusukawa et al., 1990), and were sequenced using primers for the ITS protocols as described by White et al. (1990). Thermocycling conditions were 4 min at 94 ℃, followed by 35 cycles at 94 ℃ for 1 min, 54 ℃ for 1 min, and 72 ℃ for 1.5 min, with a final extension at 72 ℃ for 7 min. The cpDNA regions were amplified using primers trnG-S (Hamilton and Baulcombe, 1999), rps16 (Shaw and Small, 2004), trnLetrnF (Taberlet et al., 1991) and psbAetrnH (Sang et al., 1997). The thermocycling conditions for all genes were 4 min at 94 ℃, followed by 35 cycles at 94 ℃ for 1 min, 53 ℃ for 1 min, and 72 ℃ for 1.5 min, with a final extension at 72 ℃ for 7 min.
2.4. Phylogenetic analysesThe sequences were edited in SequencherTM 4.12 (Gene Codes Corporation, Ann Arbor, MI, USA) and aligned using the program Mega 7.0 (Hall, 1999). A homogeneity test was conducted in PAUP v4.0 (Swofford, 2013) to check for a congruence of ITS and Chloroplast trees (P = 0.87; P > 0.5). The homogeneity test revealed a positive correlation for combined datasets. The best-fitting substitution models for Bayesian inference were selected using MrModeltest 2.3 (Nylander et al., 2004). Bayesian inference (BI) employed MrBayes v.3.2.6 and Maximum likelihood (ML) employed RAxML v.8.2.10 at the Cipres Portal (https://www.phylo.org/portal2).
Bayesian analysis was performed with four Markov chains each initiated with a random tree and two independent runs each for 10, 000, 000 generations, sampled every 100th generation. When the log-likelihood scores stabilized, the first 20% of trees were discarded as burn-in, and the retained trees were imported into PAUP to produce a 50% majority-rule consensus tree.
3. Results 3.1. Taxonomic treatmentPrunus sunhangii D. G. Zhang & T. Deng, sp. nov (Figs. 1 and 2).
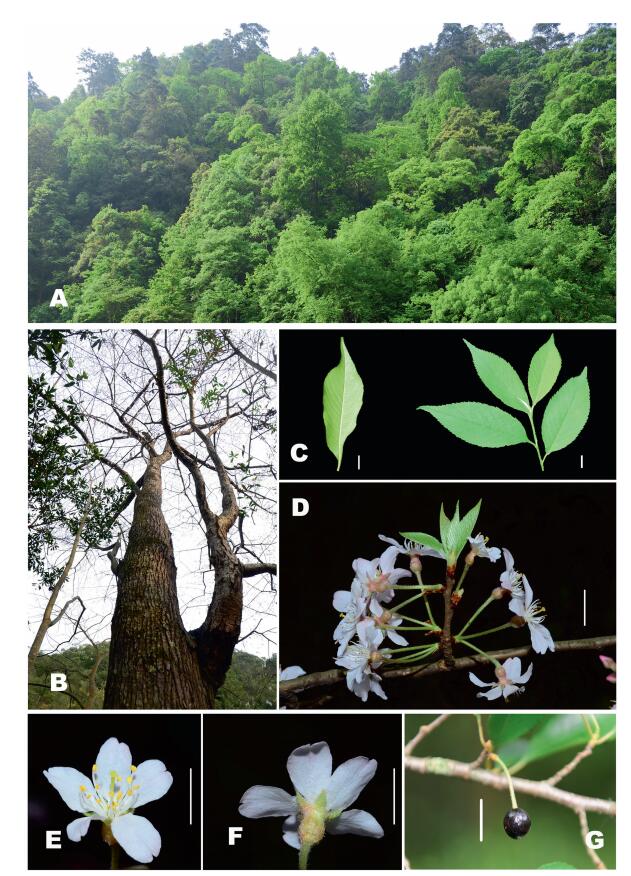
|
| Fig. 1 Living images of Prunus sunhangii D. G. Zhang & T. Deng sp. nov. (A) Habitat; (B) Stem; (C) Leaf, showing abaxial indumentums; (D) Inflorescence; (E-F) Opening white flower: front and back side; (G) Ripe fruit. Scale bars: 1 cm in C, D, E, F, G. |

|
| Fig. 2 Illustration of Prunus sunhangii D. G. Zhang & T. Deng sp. nov. (A) Fruiting shoot; (B) Leaf, showing abaxial indumentums; (C) Flower, showing with pistil and stamens; (D-E) Fruit, showing with pedicel. |
Type: CHINA, Hunan, Yong Shun, XiaoXi National Nature Reserve, along the roadside, 28°46'27.10"N, 110°15'21.18"E, 319 m, 17 March 2016, D.G. Zhang, T. Deng 107 (Type KUN!, Holotype KUN!)
3.2. DiagnosisP. sunhangii is easily differentiated from P. cerasoides by its apically longitudinally 2-lobed petals and other features: white flowers, black drupe, stamens number (17-25), brown hypanthium and with phenology.
3.3. DescriptionTrees 20-25 m tall, stems 40 cm in diam. Bark gray, longitudinally fissured. Young branchlets green, gray pilose. Stipules brown, linear, shorter than petiole, margin glandular dentate. Petiole, 0.9-1.2 cm long, densely white pubescent leaf blades ovatelanceolate, 7-12 × 3-4 cm, abaxially pale green and appressed white pilose or more densely so along midvein, adaxially dark green and glabrous or appressed pilose on veins, margin sharply biserrate, apex acuminate. Secondary veins 12-19 on each side, straight and parallel. Inflorescences umbellate, 3- or 5-flowered rare 2, involucre bracts obovate, outside pilose, soon deciduous after flowering. Flowers opening at same time as leaves. Peduncle short 0.19-0.35 cm. Pedicels 0.68-1.90 cm, densely pilose. Hypanthium brown, urceolate, 3.2-5.5 × 2.0-3.9 mm, outside densely pubescent. Sepals ovate or oblong-triangle, 1.9-3.0 × 1.0-2.0 mm, nearly as long as hypanthium or slightly shorter, open and flat, margin laxly dentate, apex acute. Petals white, apically longitudinally 2-lobed (rare emarginate), 0.75-1.21 × 0.47-0.80 cm. Stamens 17-25. Drupe black, ovoid.
Phenology-Flowering from March to April, fruiting from April to May.
Distribution and habitat- P. sunhangii is restricted to the Wuling Mountains in South China, which is a center of the Metasequoia Flora (Chen et al., 2017). Plants were growing on limestone soil, scattered along a slope adjoining the valley at 300-600 m in Hunan province and at 1000-1200 m in Hubei province (Fig. 4).
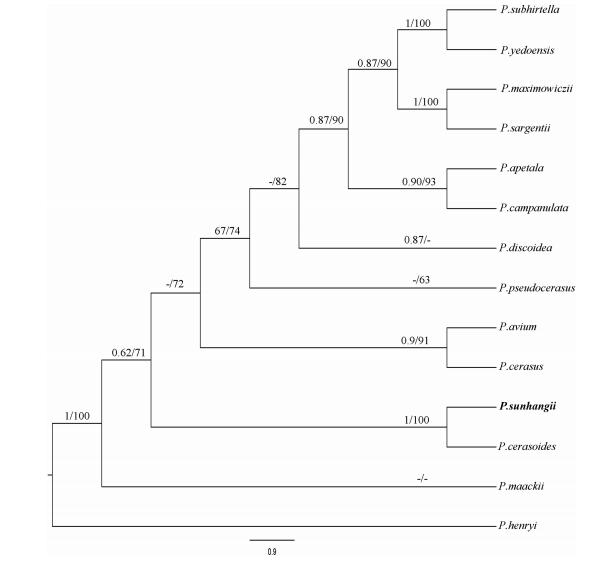
|
| Fig. 3 Bayesian 50% majority-rule consensus tree of Prunus subg. Cerasus and closely related taxa, inferred from combined sequences data from plastid (trnG-S, psbA-trnH, rps16, trnL-F) and nuclear (ITS) markers. Bayesian posterior probabilities/maximum likelihood values are given above the branches. The new species is shown in bold. |
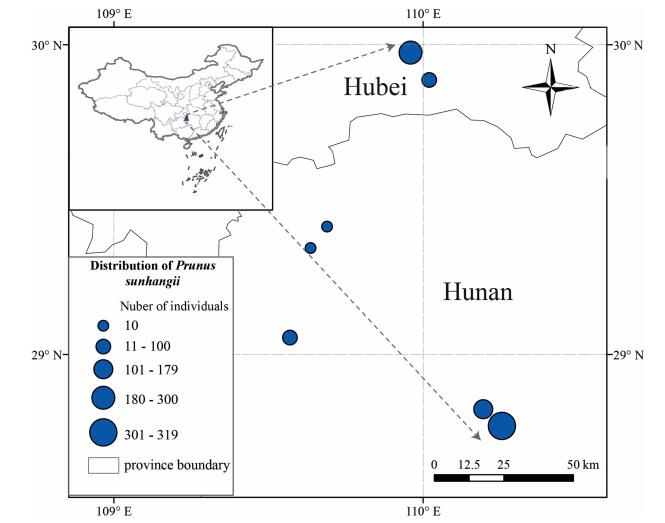
|
| Fig. 4 Distribution of Prunus sunhangii. |
| Taxa | ITS | psbA-trnH | trnG-trnS | rps16 | trnL-trnF |
| Prunus apetala (Siebold & Zucc.) Franch & Sav. | AF411509 | AB254601 | - | AB254554 | AF429902 |
| Prunus avium (L.) L. | FJ899097 | FN675831 | HQ244223 | JQ776739 | KT875773 |
| Prunus campanulata Maxim. | AF318658 | AB254632 | - | AB254585 | AF429903 |
| Prunus cerasoides Buch.-Ham. ex D.Don | KF186459 | AB254636 | HQ244236 | AB254589 | JQ034182 |
| Prunus cerasus L. | FJ899099 | FN675832 | HQ244241 | - | FJ899125 |
| Prunus discoidea (T.T.Yu & C.L.Li) Z.Wei & Y.B.Chang | KT887495 | HQ427051 | - | JQ776741 | JQ034176 |
| Prunus henryi (C.K.Schneid.) Koehne | JQ926620 | HQ188785 | HQ244268 | - | HQ244039 |
| Prunus maackii Rupr. | AY864831 | HQ188801 | AY871251 | JQ776783 | JQ034175 |
| Prunus maximowiczii Rupr. | KT887510 | AB254637 | - | AB254595 | AF429915 |
| Prunus pseudocerasus Lindl. | KF241133 | AB254631 | HQ244332 | AB254584 | AF318684 |
| Prunus sargentii Rehder | AY052507 | AB254623 | - | AB254577 | AF429918 |
| Prunus subhirtella Miq | AF179520 | AB254614 | - | AB254567 | AF429917 |
| Prunus × yedoensis Matsum. | KT887511 | EF590735 | - | JQ776749 | AF429936 |
| Prunus sunhangii D. G. Zhang et T. Deng | MK411814 | MK411810 | MK411813 | MK411812 | MK 411811 |
Etymology- The epithet of the new species refers to the chinese botanist Prof. Hang Sun, who made a significant contribution to our knowledge of the flora of China.
Vernacular name - Chinese mandarin: Sūn Háng Yīng (孙航樱).
Morphological evidence - As shown in Figs. 1 and 2, the new species is morphologically similar to P. cerasoides and appears to belong to the same section, Serrula (Koehne) Yu et Li (Yu and Li, 1986), but differs from the latter by petal shape and color, hypanthium color, number of stamens, phenology and other traits (Table 1).
Molecular phylogenetic evidence- The aligned combined data matrix included 4209 bp (ITS-799; cpDNA-3410). The best fit model of nucleotide substitutions for the combined ITS and cpDNA datasets was found to be the generalized time reversible model GTR þ G (the hierarchical likelihood ratio test, MrModeltest). The phylogenetic tree generated by these data placed P. sunhangii in the same clade with P. cerasoides (PP = 1, ML = 100) (Fig. 3).
Distribution evidence- P. cerasoides is found in China (South Xizang, North-West Yunnan), Bhutan, North India, Kashmir, North Laos, Myanmar, Nepal, Sikkim, North Thailand, North Vietnam. P. cerasoides is not found in the provinces of Hunan and Hubei (Wuling Mountains), where P. sunhangii was found.
Conservation Significance: P. sunhangii is an endemic species known from 4 localities (Fig. 4). The area of occupancy (AOO) of the new species is approximately 75 km2. Therefore, P. sunhangii should be assigned a risk of extinction of "Endandgered" [criterion B2 (a)] following the IUCN Red List (International Union for Conservation of Nature, 2016). We used species distribution modeling to predict the geographic distribution of suitable habitat for P. sunhangii under current climatic conditions. The MAXENT (Phillips et al., 2006; Phillips and Dudik, 2008) was used to generate an estimate of probability of presence of the species that varies from 0 to 1, where 0 is the lowest and 1 the highest probability. In our analyses, we set number of iterations to 500 and used ten replicates under the 'crossvalidate' option. The summary map was generated by averaging Maxent outputs (Fig. 5).
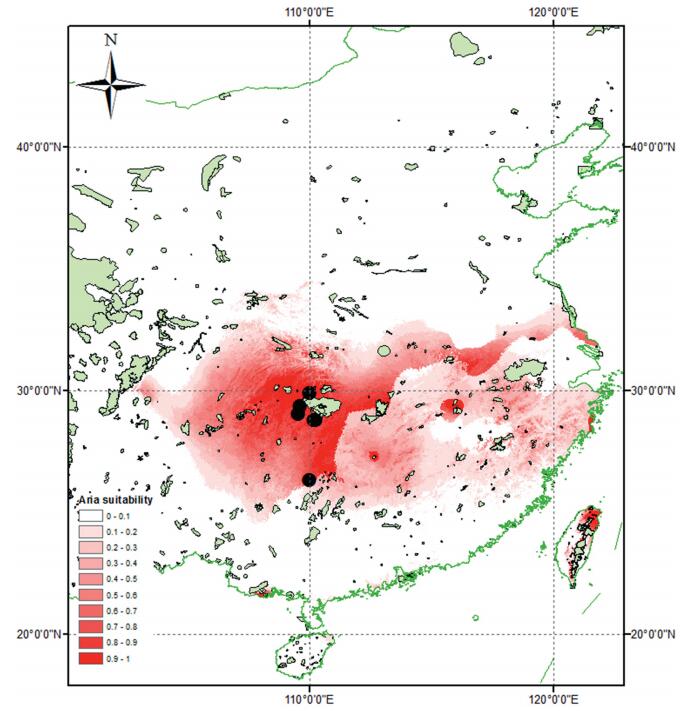
|
| Fig. 5 Predicted range of Prunus sunhangii based on bioclimatic modeling. Green areas denote protected areas and dots - species occurrences. |
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Peter Fritsch and two anonymous reviewers for constructive comments. This study was supported by the Major Program of the National Natural Science Foundation of China (31590823), the National Key R & D Program of China (2017YFC0505200), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA20050203), the National Natural Science Foundation of China (31700165), the Fund for Reserve Talents of Young and Middle-aged Academic and Technical Leaders of Yunnan Province (2014HB027), the CAS "Light of West China" Program, the Comprehensive Scientific Investigation of Biodiversity from the Wuling Mountains (2014FY110100), and the survey on baseline resources of Wufeng Houhe National Nature Reserve in Hubei Province.
Chen Z.L., Chen W.J., Chen H., Zhou Y.Y., Tang M.Q., Fu M.Q., Jin X.F., 2013. Prunus pananensis (Rosaceae), a new species from Pan'an of central Zhejiang, China.. PLoS One, 8: e54030. DOI:10.1371/journal.pone.0054030 |
Chen Y.S., Deng T., Zhou Z., Sun H., 2018. Is the East Asian flora ancient or not? Nat. Sci. Rev, 5: 920-932. |
Chin S.W., Shaw J., Haberle R., Wen J., Potter D., 2014. Diversification of almonds, peaches, plums and cherriesemolecular systematics and biogeographic history of Prunus (Rosaceae). Mol. Phylogenet. Evol, 76: 34-48. DOI:10.1016/j.ympev.2014.02.024 |
Chin S.W., Lutz S., Wen J., Potter D., 2013. The bitter and the sweet:inference of homology and evolution of leaf glands in Prunus (Rosaceae) through anatomy, micromorphology, and ancestralecharacter state reconstruction. Int. J. Plant Sci, 174: 27-46. DOI:10.1086/668219 |
Christenhusz M.J., Byng J.W., 2016. The number of known plants species in the world and its annual increase.. Phytotaxa, 261(3): 201-217. DOI:10.11646/phytotaxa.261.3 |
Hall, A.T., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic Acids Symposium, 41.Oxford University Press, pp. 95-98.
|
Hamilton A., Baulcombe D., 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants.. Science, 286(5441): 950-952. DOI:10.1126/science.286.5441.950 |
International Union for Conservation of Nature, 2016. Guidelines for Using the IUCN Red List Categories and Criteria.
|
Kusukawa N., Uemori T., Asada K., Kato I., 1990. Rapid and reliable protocol for direct sequencing of material amplified by the polymerase chain reaction.. Biotechniques, 9: 66-68. |
Li, C., Bartholomew, B., 2003. Cerasus mill. In: Wu, C.Y., Raven, P.H. (Eds.), Flora of China, vol. 9. Science Press, Beijing & Missouri Botanical Garden Press, St. Louis, pp. 404-420.
|
Lingdi, L., Cuizhi, G., Chaoluan, L., Alexander, C., Bartholomew, B., Brach, A.R., Spongberg, S.A., 2003. Rosaceae. Flora of China. Science Press, Beijing, and Missouri Botanical Garden Press, St. Louis, pp. 46-434.
|
Nylander J.A., Ronquist F., Huelsenbeck J.P., Nieves-Aldrey J.L., 2004. Bayesian phylogenetic analysis of combined data. Syst. Biol, 53(1): 47-67. |
Phillips S.J., Anderson R.P., Schapire R.E., 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model, 190: 231-259. DOI:10.1016/j.ecolmodel.2005.03.026 |
Phillips S.J., Dudik M., 2008. Modeling of species distributions with Maxent:new extensions and a comprehensive evaluation.. Ecography, 31: 161-175. DOI:10.1111/j.0906-7590.2008.5203.x |
Potter D., Eriksson T., Evans R.C., Oh S.H., Smedmark J.E.E., Morgan D.R., et al, 2007. Phylogeny and classification of Rosaceae. Plant Systemat. Evol, 266: 5-43. DOI:10.1007/s00606-007-0539-9 |
Rehder, A., 1940. Manual of Cultivated Trees and Shrubs Hardy in North America Exclusive of the Subtropical and Warmer Temperate Regions, second ed. MacMillan, New York.
|
Sang T., Crawford D.J., Stuessy T.F., 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot, 84(9): 1120-1136. |
Shaw J., Small R.L., 2004. Addressing the "hardest puzzle in American pomology:"Phylogeny of Prunus sect. Prunocerasus (Rosaceae) based on seven noncoding chloroplast DNA regions. Am. J. Bot, 91(6): 985-996. DOI:10.3732/ajb.91.6.985 |
Shi S., Li J., Sun J., Yu J., Zhou S., 2013. Phylogeny and classification of Prunus sensu lato (Rosaceae). J. Integr. Plant Biol, 55(11): 1069-1079. DOI:10.1111/jipb.12095 |
Swofford, D.L., 2013. PAUP. Phylogenetic Analysis UsingParsimony (And OtherMethods), Version 4, vol. 325. INSECTA MUNDI, Sinauer, Sunderland, Massachusetts.
|
Taberlet P., Gielly L., Pautou G., Bouvet J., 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol, 17: 1105-1109. DOI:10.1007/BF00037152 |
White, T.J., Bruns, T., Lee, S., Taylor, J., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J. (Eds.), PCR Protocols-A Guide to Methods and Applications. Academic Press, San Diego, California, pp. 315-322.
|
Yu, T.T., Li, C.L., 1986. Cerasus mill. In: Yu, T.T. (Ed.), Flora Reipublicae Popularis Sinicae, vol. 38. Science Press, Beijing, pp. 41-89.
|
Zhao L., Jiang X.W., Zuo Y.J., Liu X.L., Chin S.W., Haberle R. R., Potter D., Chang Wen J., 2016. Multiple events of allopolyploidy in the evolution of the racemose lineages in Prunus (Rosaceae) based on integrated evidence from nuclear and plastid data.. PLoS One, 11(6): e0157123. DOI:10.1371/journal.pone.0157123 |



