b. College of Life Sciences, Nanchang University, Nanchang, 330031, China
Stemonaceae Engl., a monocotyledonous flowering plant family, belongs to Pandanales and consists of four genera: Croomia Torr., Stemona Lour., Stichoneuron Hook. f. and Pentastemona Steenis, with about 37 known species (Chase et al., 2016; Christenhusz et al., 2016). The family is native to seasonal climate areas across Southeast Asia and tropical Australia, with one species [Croomia pauciflora (Nutt.) Torr.] in North America (Li et al., 2008). Because the roots and rhizomes of some Stemona and Croomia species are found to contain many bioactive and structurally unique alkaloids, such as protostemonine, stichoneurine, and croomine groups (Greger, 2006; Lin et al., 2006, 2008; Kongkiatpaiboon et al., 2011; Chen et al., 2017), they are widely used in Southeast Asian folk medicine to treat cough, traumatic injury, and enteric helminth worms (Lee et al., 2008; Chen et al., 2017). Thus, the family Stemonaceae has been a focus of phytochemical and pharmacological research (Kongkiatpaiboon et al., 2011; Chen et al., 2017). Among the four genera, Croomia exhibits a typical EA-ENA disjunct distribution across the Pacific Ocean, comprised of three herbaceous perennial species: Croomia heterosepala (Bak.) Oku. and Croomia japonica from East Asia; C. pauciflora from Southeastern North America (Li et al., 2008; Fang et al., 2013). Due to their small range size and small number of populations, all three species of Croomia are treated as rare and endangered in China, Japan, and the United States (Estill and Cruzan, 2001; Wang and Xie, 2004). However, except for a few traditional molecular markers (e.g. ISSR, nSSR) developed and used in former studies (Li et al., 2008; Fang et al., 2013), no genomic resources have been reported for these endangered Croomia species, which hinders the studies on population genetics, conservation management, and adaptive divergence of these species.
In addition, our previous molecular phylogenetic analyses using chloroplast DNA sequence variation of the trnL-F region have supported the monophyly of Croomia and a sister relationship of C. pauciflora to the two Asian species (Li et al., 2008). Molecular dating suggests the two Asian species diverged in the Mid-to-Late Pleistocene (0.84e0.13 mya), and C. pauciflora diverged at the Plio-Pleistocene boundary (< 2.6 mya). Although chloroplast markers are often used to infer phylogenetic relationships, very low resolution of the dataset hampered any further statistical inference. Moreover, since the possible presence of multiple haplotypes and/or independent chloroplast lineages does not correspond with the actual species divergence, our previous molecular dating may only represent the divergence of a particular genetic locus but not the divergence of species. Thus, application of coalescent-based models or related simulation-based approaches to multi-locus sequence data should allow us to explore in more detail the prevalent time scales, geographic modes, and demographic history of population and species divergences (Li et al., 2012; Dolman and Joseph, 2016). However, the genus lacks sequence data at a genomic scale to facilitate such investigations.
The two Asian species form a parapatric species pair with abutting ranges in South Japan, but C. japonica also occurs disjunctively on the adjacent Asiatic mainland in East China. There is considerable difference in floral characters of the two species (Rogers and Harpending, 1992). For example, all four tepals of C. japonica are homomorphic with a re-curved edge, whereas those of C. heterosepala have a straight edge, and one tepal is much larger than the other three (Okuyama, 1944; Ohwi, 1965; Li et al., 2008). Thus, like other species of Stemonaceae, the flowers of C. japonica are radially symmetrical (i.e., actinomorphic). By contrast, C. heterosepala represents the sole species with bilaterally symmetrical (i.e., zygomorphic) flowers within this family. Croomia and its related genera, therefore, provide an ideal model to investigate the origin and maintenance of flower symmetry; the first step to addressing this problem is to identify the genes responsible for flower symmetry. More recently, functional studies in model [e.g. Antirrhinum majus (Preston and Hileman, 2009); Oryza sativa (Yuan et al., 2009)] and non-model species [e.g. Caprifoliaceae and Lonicera (Howarth et al., 2011); Commelinaceae (Preston and Hileman, 2012); Ranunculaceae (Jabbour et al., 2014)] have demonstrated that CYC/TB1-like genes (sometimes referred to as TB1-like or TBL in monocots), which belong to the class Ⅱ TCP (TB1, CYC and PCF) family, are involved in the establishment and maintenance of zygomorphic flowers. However, to date, few studies are available for monocots (Bartlett and Specht, 2011; Preston and Hileman, 2012; Hoshino et al., 2014), and no TCP or CYC/TB1 genes have been identified from Stemonaceae.
Transcriptome sequencing or RNA-Seq is one of the most efficient and cost-effective methods currently available for gene discovery and developing massive genome-wide markers in non-model organisms (Wen et al., 2015). Recent studies have demonstrated the utility of these data for resolving the relationships of diverse lineages of organisms (i.e. RNA-Seq phylogenetics) (Zhou et al., 2017), inferring demographic histories (Zhu et al., 2016), estimating genomic variation (Kawakami et al., 2014), and identifying genetic bases of adaptive divergence (Wen et al., 2015; Mao et al., 2016). Furthermore, transcriptome analysis provides valuable insights into genes and gene activities responsible for differences in organ morphology during the developmental processes (Liu et al., 2013; Ma et al., 2014). In this study, using the Illumina Hiseq 2000 platform, we obtained the RNA sequence data for two flower samples (C. heterosepala and C. japonica) and one leaf sample (C. japonica). De novo assembly of these transcripts was conducted to characterize the transcriptomes of the two flowers and one leaf, respectively. Based on transcriptome data of Croomia, a set of putative single-copy genes were screened in a genus framework by filtering strict orthologs. By pairwise comparison of the orthologous sequences from each species pair, candidate genes under adaptive selection in speciation or population differentiation were identified. In addition, we developed a large set of EST-SSR loci and validated a subset of them, yielding massive potential molecular markers. Finally, we isolated and sequenced CYC/TB1 (TCP) genes across four genera (7 species) of Stemonaceae to reconstruct the CYC/TB1 gene tree and estimate its molecular evolution in this family.
2. Materials and Methods 2.1. Plant samples, cDNA library preparation, and illumina sequencingDuring April 2010, living plants of C. heterosepala from Nara, Japan (34°31'N, 135°41'E) and C. japonica from Mt. Tianmu, China (30°20'N, 119°26'E) were transplanted to the Botanical Garden of Zhejiang University, Hangzhou, China. Fresh flowers of C. heterosepala (ChFlower) and C. japonica (CjFlower) at the anthesis stage, as well as juvenile leaf samples of C. japonica (CjLeaf), were harvested from these living plants and immediately frozen in liquid nitrogen for storage at -80 ℃ until total RNA extraction. For each tissue, total RNA was extracted from a mixture of three individuals using TRIzol Reagent (Invitrogen Life Technologies, USA) according to the manufacturer's recommendations. RNA quality and quantity were assessed using gel electrophoresis and a NanoDrop spectrophotometer 2000 (Thermo Scientific, Wilmington, DE, USA). Sequencing libraries were generated from 3 μg total RNA using NEBNext®UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer's protocol, and index codes were added to attribute sequences to each sample. After validating the library quality with an Agilent 2100 Bioanalyzer system, the constructed libraries were sequenced on an Illumina Hiseq 2000 platform and paired-end reads were generated. All of the sequencing data were deposited in the Sequence Read Archive (SRA) of the NCBI database under accession numbers: SRX3328777 (ChFlower), SRX3328635 (CjFlower), and SRX3328351 (CjLeaf).
2.2. Raw data processing and de novo assemblyRaw sequence reads were firstly filtered through in-house Perl scripts. In this step, clean reads were obtained by removing reads containing adaptors, reads with more than 5% unknown bases (N bases), and reads with more than 20% of low-quality bases (quality value ≤ 20). At the same time, Q20, Q30, GC content and the sequence duplication level of the clean data were calculated. All the downstream analyses were based on these high-quality clean reads. The high-quality clean reads from ChFlower, CjFlower, and CjLeaf were then assembled de novo separately using TRINTY (Haas et al., 2013) with "min_kmer_cov" set to 2 and all other parameters set to default. These assembled unigene sets were further processed by sequence splicing and redundancy removal using TGICL software (Pertea et al., 2003) to retrieve non-redundant unigenes. Furthermore, the individual assembled unigene sets from CjFlower and CjLeaf were pooled together and assembled using TGICL software (Pertea et al., 2003) into non-redundant unigenes that represented the C. japonica transcriptome (CjTranscriptome).
2.3. Functional annotation and prediction of protein-coding sequence (CDS) regionsTo predict the putative functions of the assembled unigenes in ChFlower, CjFlower, CjLeaf and CjTranscriptome, the unigenes were utilized for homology searches against the National Center for Biotechnology Information (NCBI) non-redundant (Nr) protein database, Swiss-Prot protein database (http://www.expasy.ch/sprot), the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database (Kanehisa et al., 2008) and the Cluster of Orthologous Groups (COG) database (http://www.ncbi.nlm.nih.gov/COG/) using BLASTX (Altschul et al., 1990) with an E-value cut-off of 1e-5. Based on the best BLASTX hits from the NCBI Nr database, Gene Ontology (GO) terms (Ashburner et al., 2000) of the unigenes were performed using BLAST2GO version 2.6.0 (Conesa et al., 2005) with an E-value cut-off of 1e-5. To obtain information on general functional categories, the distributions of level-2 GO terms for all unigenes were plotted with the Web Gene Ontology Annotation Plot (WEGO) (Ye et al., 2006) for three main categories: biological process, molecular function and cellular component (http://www.geneontology.org). In addition, unigenes were further queried against the Nt (NCBI nonredundant nucleotide sequences) database using BLASTN with an Evalue cut-off of 1e-5.
The protein-coding region sequences (CDS) of unigenes were predicated according to the Blast results against the Nr, Swiss-Prot, KEGG and COG databases (E-value < 1e-5) and translated into amino acid sequences using the standard codon table. If the results between different databases were in conflict with each other, a priority order of Nr, Swiss-Prot, KEGG, and COG was followed. For those unigenes that could not align to any of the above databases, ESTScan (Iseli et al., 1999) was used to predict CDS regions and determine the amino acid sequences.
2.4. Ka/Ks ratios of orthologous pairs between ChFlower and CjFlower transcriptomes and mining of single copy nuclear genes (SCNGs)Based on the predicted CDS regions of ChFlower and CjFlower transcriptomes, putative orthologous groups between these two flower transcriptomes were obtained using OrthoMCL v2.0.9 (Li et al., 2003) with default parameters by identifying clusters with only one sequence from each flower transcriptome. Those orthologs were removed following three criteria: (A) the alignment lengths were < 150 bp, (B) Ka or Ks values were not applicable, (C) Ks values were more than 0.1, which is a benchmark of potential paralogs (Bustamante et al., 2005; Ai et al., 2015). The YN algorithm (Yang and Nielsen, 2000) implemented in KAKS_CALCULATOR v1.2 (Zhang et al., 2006) was employed to calculate non-synonymous rates (Ka), synonymous rates (Ks) and Ka/Ks (ω) ratios of each putative pair of orthologs.
For the mining of putative single copy nuclear genes (SCNGs), 959 single copy nuclear genes shared by Arabidopsis thaliana, Populus trichocarpa, Vitis vinifera, and O. sativa (usually referred as APVO genes, Duarte et al., 2010) were used for our analysis. We retrieved the protein sequences encoded by APVO genes from the TAIR10 database (Duarte et al., 2010) and queried these sequences against the putative orthologous genes between ChFlower and CjFlower using TBLASTN with a threshold E-value of 1e-10. All the queries with BLAST hits were considered to be putative single copy nuclear genes (SCNGs) in the Croomia species.
2.5. Detection and validation of EST-SSRsEST-SSRs were identified from unigenes of CjTranscriptome using the program MISA (http://pgrc.ipk-gatersleben.de/misa) (Dieringer and Schlotterer, 2003) with thresholds of 12, 6, 5, 5, 4, and 4 € repeat units for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide motifs, respectively. Primer pairs for the SSRs were designed using Primer Premier v5.0 (Premier Biosoft International, Palo Alto, California, USA). To assess the reliability and cross-species transferability of the identified EST-SSRs, 100 primer pairs were randomly selected to test for amplification across 15 populations (two samples per population) of C. heterosepala, C. japonica and C. pauciflora (Table S1). The PCR amplification reactions were carried out in 30 μL reaction volume containing 17 μL 2x master mix (TSINGKE, Beijing, China), 9 μL of ddH2O, 1 μL of each primer, and 2 μL of genomic DNA. PCR amplifications were performed using a thermal cycler GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) following the procedures in Zhu et al. (2016). The PCR products, along with a 100 bp marker (TaKaRa, Dalian, Liaoning, China), were electrophoresed on 12% non-denaturing polyacrylamide gels stained with silver staining to assess amplification success and polymorphism. The polymorphic primer pairs were synthesized again with fluorescent dyes (TAMRA, HEX, or 6-FAM) at the 5' end of the forward primer and were then used to amplify DNA from all 15 populations (240 individuals in total) of C. heterosepala, C. japonica and C. pauciflora for genetic diversity analysis (Table S1). PCR amplification followed the conditions described above. Fragments of PCR product were separated on a 3730xl DNA Analyzer (Applied Biosystems) and then the alleles were manually scored and determined using GENEMARKER v2.2.0 (SoftGenetics, PA, USA) with GeneScan 500 LIZ as an internal size standard. Finally, the number of observed alleles (NA), as well as observed (HO) and expected (HE) heterozygosities, and the polymorphism information content (PIC) values were calculated using CERVUS v3.0.3 (Kalinowski et al., 2007). The significance of departures from HardyeWeinberg equilibrium (HWE) and linkage disequilibrium (LD) between all pairs of polymorphic loci were analyzed using GENEPOP v4.0.7 (Rousset, 2008). Frequencies of null alleles were estimated in the FREENA package (Chapuis and Estoup, 2007) following the expectation maximization (EM) method (Dempster et al., 1977).
2.6. Analysis of the CYC/TB1-like genes of TCP transcription factors in StemonaceaeIn order to identify CYC/TB1-like genes in C. heterosepala and C. japonica, three strategies were adopted in this study. First, TCP domain sequences of O. sativa (23 accessions) and A. thaliana (24 accessions) downloaded from PlantTFDB v4.0 (planttfdb.cbi.pku.edu.cn) were used as query entries to perform BLASTP searches against the protein sequences from both ChFlower and CjFlower transcriptomes (E-value < 1e-5). In addition, all the output protein sequences were scanned using InterProScan (https://www.ebi.ac.uk/interpro/search/sequence-search) to remove those sequences without a whole TCP domain. Second, high-quality reads that mapped to Commelinaceae TB1-like genes (Preston and Hileman, 2009) were directly assembled into CYC/TB1-like genes. Third, CYC/TB1-like genes were amplified from genomic DNA of C. heterosepala, C. japonica, and five other Stemonaceae species (C. pauciflora, Stemona tuberosa, S. japonica, Stichoneuron caudatum, and Pentastemona egregia) representing all four genera. The primers and the methodology for amplification used in this study were described in Howarth and Donoghue (2005). To distinguish and classify the TCP genes of ChFlower and CjFlower transcriptomes, TCP genes obtained from the first strategy were divided into class Ⅰ PCF-like, class Ⅱ CIN-like, and class Ⅱ CYC/TB1-like groups. The amino acid sequences of TCP domains from O. sativa, A. thaliana, ChFlower and CjFlower were aligned using ClustalW (Thompson et al., 1994) and manually edited if necessary. Then, the Neighbor-Joining (NJ) phylogenetic tree was constructed using MEGA v7 (Kumar et al., 2016) with 1000 bootstrap replicates. To further determine ortholog or copy number of newly-determined Stemonaceae CYC/TB1-like sequences, a total of 72 amino acid sequences, which have been confirmed to be related to the formation of flower symmetry (Yuan et al., 2009; Bartlett and Specht, 2011; Preston and Hileman, 2012; Hoshino et al., 2014; Citerne et al., 2017) plus 7 newly-determined Stemonaceae sequences were used to generate Bayesian inference (BI) and maximum likelihood (ML) trees. The best-fitting model (JTT + G + Ⅰ) was selected by ProtTest v2.4 (Abascal et al., 2005). Bayesian inteference analyses were conducted in MrBayes v3.2 (Ronquist and Huelsenbeck, 2003). The Markov chain Monte Carlo (MCMC) algorithm was run for two million generations with trees sampled every 500 generations. The first 25% of generations were discarded as burn-in. A 50% majority-rule consensus tree was constructed from the remaining trees to estimate posterior probabilities (PPs). Maximum likelihood analysis was conducted using PhyML v3.0 (Guindon et al., 2010) with 1000 bootstrap replicates.
Both the CYC/TB1-like phylogeny and codon alignments were used to estimate the synonymous (dS) and nonsynonymous (dN) substitution rates (ω) in CODEML as part of the PAML package (Yang, 2007). Analyses were conducted under three models: the 'one-ratio' model (M0) assumes the equal ω (dN/dS) value for the entire tree, the 'free-ratios' model (M1) assumes an independent ω value for each branch and the 'two-ratio' model (M2) assumes that one or more of branches (the 'foreground' branch) have a distinct ω value different from the remaining branches (the 'background' branches). Moreover, 12 hypotheses of the 'two-ratio' model (M2) were tested by selecting each clade (clades labeled in Fig. 3) as foreground: Hypothesis 1 (H1): ZinTBL2, H2: ComTB1a, H3: PoaTBL2, H4: ZinTBL1a, H5: ZinTBL1, H6: AlsTCP1, H7: SteTBL1, H8: PoaTBL1, and H9: AcoTBL were selected as foreground, respectively; H10: clade (ZinTBL1a and ZinTBL1b) was selected as foreground; H11: clade (ZinTBL2, ComTB1a and PoaTBL2) was selected as foreground; H12: specific ω values are estimated for each of clades leading to ZinTBL2, ComTB1a, PoaTBL2, ZinTBL1a, ZinTBL1, AlsTCP1, SteTBL1, PoaTBL1, and AcoTBL. To evaluate the goodness of fit of the data, a likelihood ratio test (LRT) was performed by using the latter models with respect to the M0 model.
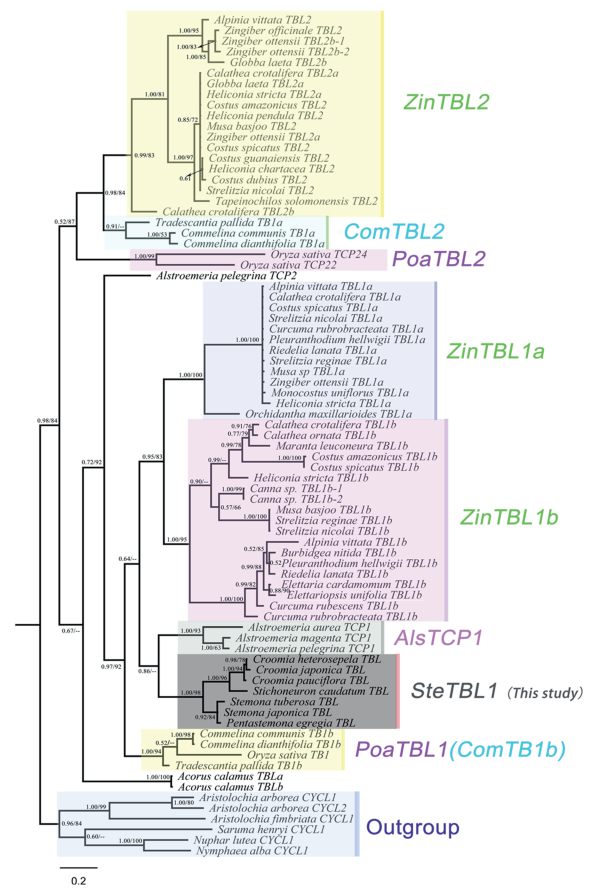
|
| Fig. 3 Bayesian inference (BI) phylogeny of monocot CYC/TB1 genes rooted with basal eudicot CYC/TB1 genes. Posterior probabilities (pp, first) and bootstrap values (BS, second) > 50% are indicated (– < 0.5). Where a particular relationship was not shown in ML analysis, only the Posterior probability is presented. |
The Illumina sequencing of cDNA libraries of ChFlower, CjFlower, and CjLeaf yielded 49, 881, 224, 50, 481, 150 and 60, 582, 220 raw reads, respectively. After filtering and trimming raw reads, 43, 683, 202, 44, 355, 666 and 54, 416, 848 clean reads of ChFlower, CjFlower and CjLeaf were generated, with the Q20 percentage over 97% (Table 1). Through de novo assembly, 76, 976, 77, 344, 79, 538 contigs with an N50 length of 1, 452, 1, 461, 1641 bp and mean length of 916, 951, 1027 bp were obtained for ChFlower, CjFlower and CjLeaf, respectively. These contig sequences were further assembled into 57, 193 unigenes (average length = 1042 bp, N50 = 1603 bp) for ChFlower, 62, 131 unigenes (average length = 1044 bp, N50 = 1575 bp) for CjFlower and 64, 448 unigenes (average length = 1106 bp, N50 = 1737 bp) for CjLeaf (Table 1).
| Characteristics | C. heterosepala | C. japonica | ||
| ChFlower | CjFlower | CjLeaf | CjTranscriptome | |
| Raw reads | 49 881 224 | 50 481 150 | 60 582 220 | 111 063 370 |
| Clean reads | 43 683 202 | 44 355 666 | 54 416 848 | e |
| GC percentage (%) | 50.46 | 49.68 | 50.24 | e |
| Q20 percentage | 97.60 | 97.60 | 98.55 | e |
| Total number of contigs | 76 976 | 77 344 | 79 538 | e |
| N50 length of contigs (bp) | 1452 | 1461 | 1641 | e |
| Mean length of contigs (bp) | 916 | 951 | 1027 | e |
| Total number of unigenes (bp) | 57 193 | 62 131 | 64 448 | 86 457 |
| 200-500 bp (% of total unigenes) | 21 357 (37.34%) | 22 099 (35.57%) | 23 801 (36.93%) | 30 335 (35.09%) |
| 500-1000 bp (% of total unigenes) | 13 622 (23.82%) | 14 847 (23.90%) | 14 830 (23.01%) | 19 700 (22.79%) |
| 1000-1500 bp (% of total unigenes) | 8655 (15.13%) | 10 000 (16.10%) | 9412 (14.60%) | 13 244 (15.32%) |
| 1500-2000 bp (% of total unigenes) | 6076 (10.62%) | 7053 (11.35%) | 6469 (10.04%) | 9521 (11.01%) |
| 2000-2500 bp (% of total unigenes) | 3301 (5.77%) | 3936 (6.34%) | 3907 (6.06%) | 5786 (6.69%) |
| 2500-3000 bp (% of total unigenes) | 1871 (3.27%) | 2076 (3.34%) | 2303 (3.57%) | 3313 (3.83%) |
| ≥3000 bp (% of total unigenes) | 2311 (4.04%) | 2120 (3.41%) | 3726 (5.78%) | 4558 (5.27%) |
| N50 length of unigenes (bp) | 1603 | 1575 | 1737 | 1732 |
| Mean length of unigens (bp) | 1042 | 1044 | 1106 | 1121 |
By further combining and reassembling the CjFlower and CjLeaf unigene sets, we obtained 86, 457 unigenes with a mean length of 1121 bp and an N50 of 1732 bp, which represented the C. japonica transcriptome (CjTranscriptome). Additionally, not only the number of unigenes (86, 457) but also the percentage of longer unigenes (> 1000 bp, 42.12%) in CjTranscriptome were greater than those in CjFlower and CjLeaf transcriptomes (Table 1). The distribution of unigene length also indicated a common pattern that smaller size groups possessed more unigenes, and more than half ranged from 200 to 1000 bp (Table 1).
3.2. Functional annotation and CDS predicationBased on a BLAST search, 39, 919 (69.80%) unigenes from ChFlower, 44, 209 (71.15%) unigenes from CjFlower, 43, 375 (67.30%) unigenes from CjLeaf and 58, 895 (68.12%) unigenes from CjTranscriptome had at least one Blast match against Nr, Swiss-Prot, KEGG, COG, GO or Nt databases (Table 2). Based on GO function classifications of the two flower transcriptomes, 18, 436 (32.23%) unigenes from ChFlower and 20, 376 (32.80%) unigenes from CjFlower were assigned into three main categories and 54 subcategories, and the distribution of GO terms was consistent with each other (Fig. 1). Among these subcategories, 'metabolic process' (ChFlower/CjFlower: 11, 178, 60.63%/12, 352, 60.62%) and 'cellular process' (9, 971, 54.08%/ 11, 106, 54.51%) were the two mostly dominant GO terms for the biological process category, while 'cell' (9, 143, 49.59%/10, 131, 49.72%) and 'cell part' (9, 143, 49.59%/10, 131, 49.72%) were most abundant in the cellular component category, and 'catalytic activity' (9, 640, 52.29%/10, 575, 51.90%) and 'binding' (8, 199, 44.47%/8, 993, 44.14%) were highly representative for the molecular function category (Fig. 1). Beyond that, the distributions of CjLeaf and CjTranscriptome unigenes in three categories are displayed in Fig. S1.
| Characteristics | C. heterosepala | C. japonica | ||||
| ChFlower | CjFlower | CjLeaf | CjTranscriptome | |||
| Functional annotations | Nr | 37 970 (66.39%) | 42 032 (67.65%) | 40 760 (63.24%) | 55 405 (64.08%) | |
| Swiss-Prot | 26 629 (46.56%) | 29 573 (47.60%) | 29 008 (45.01%) | 42 708 (49.40%) | ||
| KEGG | 25 382 (44.38%) | 27 938 (44.97%) | 27 652 (42.91%) | 37 106 (42.92%) | ||
| COG | 17 633 (30.83%) | 19 404 (31.23%) | 19 412 (30.12%) | 26 073 (30.16%) | ||
| GO | 18 436 (32.23%) | 20 376 (32.80%) | 26 640 (41.34%) | 34 595 (40.01%) | ||
| Nt | 32 432 (56.71%) | 35 556 (57.22%) | 35 794 (55.54%) | 48 039 (55.56%) | ||
| ALL | 39 919 (69.80%) | 44 209 (71.15%) | 43 375 (67.30%) | 58 895 (68.12%) | ||
| CDS annotations | Homolog | 37 111 (64.89%) | 41 059 (66.08%) | 40 029 (62.11%) | 54 155 (62.64%) | |
| ESTscan | 1248 (2.18%) | 1390 (2.24%) | 1187 (1.84%) | 1607 (1.86%) | ||
| ALL | 38 359 (67.07%) | 42 449 (68.32%) | 41 216 (63.95%) | 55 762 (64.50%) | ||

|
| Fig. 1 Gene ontology categories of unigenes in flowers of Croomia heterosepala (ChFlower) and C. japonica (CjFlower). |
Based on the BLAST searches against the four protein databases, the coding regions of 37, 111, 41, 059, 40, 029 and 54, 155 unigenes were separately extracted from ChFlower, CjFlower, CjLeaf and CjTranscriptome. Moreover, 1248 unigenes in ChFlower, 1390 unigenes in CjFlower, 1187 unigenes in CjLeaf and 1607 unigenes in CjTranscriptome with CDS were gained according to the ESTScan methods (Table 2), and the most abundant size class was 200-500 bp for each transcriptome (Table S2).
3.3. Orthologous pairs detection, Ka/Ks ratios, and mining of SCNGsAfter filtering the preliminary orthologous pairs based on the three criteria (see before), we detected 11, 566 potential orthologous groups between ChFlower and CjFlower, and proceeded to estimate the Ka, Ks and Ka/Ks values of these groups (Table S3). The mean values of the Ka, Ks and Ka/Ks ratios were 0.007, 0.024 and 0.670, respectively. Although there are 827 orthologous pairs with Ka/Ks ratios > 1, only six pairs showed Ka/Ks ratios significantly > 1 (P < 0.05), suggesting that these pairs are likely under positive selection. Taking a more appropriate threshold of 0.5 for the Ka/Ks ratio as an indicator of positive selection (Swanson et al., 2004; Zhang et al., 2013), another 13 orthologous pairs with Ka/Ks values between 0.5 and 1 and P values < 0.05 were identified (Table 3). The functions of these positively selected genes are described based on the best hits against the Nr database (Table 3).
| Gene ID of orthologous genes | Ka/Ks value | P-value (Fisher) | Description | |
| ChFlower | CjFlower | |||
| CL3372.Contig2 | CL7786.Contig2 | 1.904 | 0.0027 | uncharacterized protein |
| CL7139.Contig1 | CL4557.Contig4 | 667.825 | 0.0142 | RING-H2 ωnger protein ATL58-like |
| Unigene30940 | Unigene7233 | 5.860 | 0.0142 | hypothetical protein Csa_7G372310 |
| Unigene132 | CL5006.Contig1 | 2.262 | 0.0263 | extensin-2-like, partial |
| Unigene9837 | Unigene3498 | 4.382 | 0.0230 | protein RALF-like 19 |
| CL3076.Contig2 | CL1809.Contig7 | 1.647 | 0.0301 | probable disease resistance protein At1g12280 |
| CL428.Contig4 | CL1228.Contig3 | 0.503 | 0.0082 | uncharacterized protein LOC105055788 |
| CL1969.Contig4 | CL159.Contig3 | 0.556 | 0.0130 | uncharacterized ATP-dependent helicase C17A2.12 isoform X1 |
| CL3529.Contig5 | CL1483.Contig4 | 0.527 | 0.0171 | hypothetical protein VITISV_015618 |
| Unigene31095 | CL3236.Contig1 | 0.511 | 0.0204 | uncharacterized protein LOC105040955 |
| Unigene766 | CL1582.Contig5 | 0.522 | 0.0275 | polypyrimidine tract-binding protein homolog 2 |
| CL6003.Contig1 | CL4260.Contig1 | 0.635 | 0.0328 | pentatricopeptide repeat-containing protein At5g62370 |
| CL3248.Contig4 | CL7151.Contig1 | 0.571 | 0.0329 | protein STICHEL-like 2 |
| Unigene22720 | CL1751.Contig4 | 0.501 | 0.0350 | la-related protein 6 A |
| CL2431.Contig1 | CL6743.Contig2 | 0.682 | 0.0362 | protein LONGIFOLIA 1 isoform X1 |
| CL6630.Contig10 | CL6598.Contig2 | 0.529 | 0.0366 | uncharacterized protein LOC103701275 |
| CL4997.Contig1 | Unigene3839 | 0.657 | 0.0397 | phytochrome A isoform X1 |
| Unigene24430 | Unigene8493 | 0.507 | 0.0407 | ethylene-responsive transcription factor RAP2-13-like |
| Unigene11165 | Unigene22312 | 0.513 | 0.0495 | RNA-binding protein 47-like |
Of the APVO SCNG dataset (Duarte et al., 2010) used to perform TBLASTN queries against orthologous pairs between ChFlower and CjFlower (see above), a total of 429 genes were found to have hits against our orthologous genes (Table S4), which are most likely the single copy nuclear genes in Croomia species.
3.4. Identification, validation and cross-species transferability of EST-SSRsIn this study, we identified a total of 16, 384 sequences containing 21, 460 EST-SSRs from the C. japonica (CjTranscriptome) unigenes, with 3681 sequences containing more than one SSR. Among the 21, 460 EST-SSRs, the most frequent repeat types were dinucleotide (10, 169, 47.4%), followed by trinucleotide (6, 789, 30.2%), and mononucleotide (2, 871, 13.4%) (Fig. 2, Table S5). The dominant dinucleotide repeat types were AG/CT, accounting for 77.6% of repeats, followed by AT/AT (14.6%), and AC/GT (7%), while the CG/CG motifs (0.8%) were least abundant. Among the trinucleotide motifs, the most common repeats were CGG/CGG (32.0%), followed by AGG/CCT (21.1%), and AAG/CTT (14.0%) (Fig. 2, Table S5).
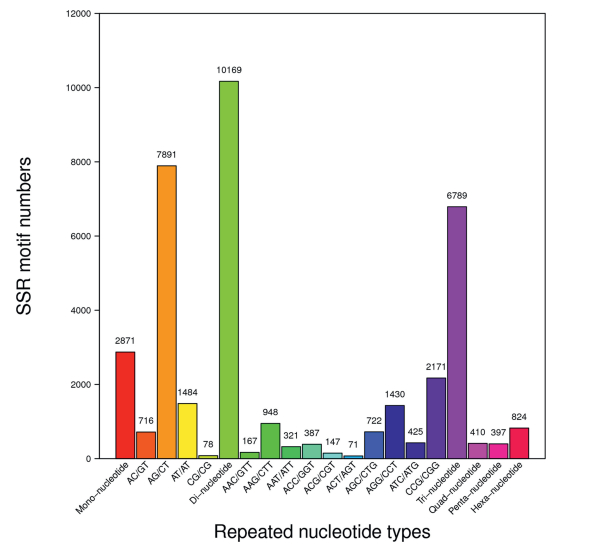
|
| Fig. 2 Total numbers of different EST-SSR motifs in the transcriptomes of C. japonica flowers + leaves (CjTranscriptome). |
Of the 100 primer pairs (Table S6) selected for testing polymorphism, 46 primer pairs (46%) successfully produced PCR fragments with expected size, and 10 primer pairs (10%) exhibited polymorphism in all three Croomia species (Fig. S2). Therefore, these 10 polymorphic loci were further used to evaluate the diversity of all 240 Croomia individuals (Table 4). Across the 10 analyzed loci, the observed number of alleles (NA) varied from 5 to 16, with a mean of 9.5 alleles per locus. The observed (HO) and expected (HE) heterozygosities ranged from 0.271 to 0.758 and from 0.303 to 0.897, respectively. The PIC values varied between 0.289 and 0.866, with an average of 0.678. In addition, 6 out of 10 loci showed significant deviation from HWE (P < 0.001) (Table 4), although no null alleles were found at these EST-SSR loci.
| Locus | Primer pair | Repeat motif | Size range (bp) | NA | HO | HE | PIC | HWE P value |
| Cro_10 | F: CATCAATATCCGAAACTCTCCAG R: CGCCATAAATACTCTCACGAGTC |
AG (2*12) | 118-146 | 16 | 0.733 | 0.879 | 0.866 | 0.0001 |
| Cro_21 | F: GGGACATTATTGGATCACAACTT R: GTGTATGGGTGGATAGTTTGTGG |
GAC (3*5) | 125-143 | 5 | 0.454 | 0.695 | 0.637 | 0.0232 |
| Cro_22 | F: GGGTGGAAGGAGATAGATGAGAT R: CCACCTCCACTCTTAACACACTC |
GTG (3*6) | 104-116 | 8 | 0.271 | 0.303 | 0.289 | 0.1184 |
| Cro_30 | F: CTCTTCCCCATCTTGTTCACTC R: GGATAATAATAACGCGAGAAGCC |
GCT (3*6) | 89-110 | 8 | 0.433 | 0.81 | 0.784 | 0.0001 |
| Cro_38 | F: GTCACAGAGCACCCATCTCC R: GACAGAGACTCCGATCATCTCAT |
GGC (3*8) | 104-146 | 10 | 0.421 | 0.669 | 0.638 | 0.0000 |
| Cro_50 | F: AACGAAAAACAGAAAGCCAAAAG R: GATCCCCAATTCTCGATCTATTC |
GCT (3*5) | 95-155 | 12 | 0.758 | 0.725 | 0.694 | 0.0000 |
| Cro_56 | F: CGACCCTCCTCTTCTCCTATTTA R: GTGAGTAAAAGAACGACGCCAT |
CCG (3*5) | 86-116 | 8 | 0.275 | 0.632 | 0.581 | 0.0003 |
| Cro_61 | F: CAGCAGCAGCAGCAGCAGCAG R: GCTGAGGTTGAGAGAGTTGGATA |
CAG (3*7) | 89-122 | 12 | 0.646 | 0.834 | 0.813 | 0.0000 |
| Cro_77 | F: ATCGCTCCACCAACAACAG R: CTGCTAGGGTTTGTCACCATC |
CAG (3*8) | 98-119 | 7 | 0.254 | 0.749 | 0.71 | 0.2675 |
| Cro_78 | F: ATCCAAAACCGACTACCAAGATT R: GGACTTCCGAAATGAAAAACTCT |
ACC (3*5) | 95-119 | 9 | 0.392 | 0.802 | 0.772 | 0.0068 |
| average | 9.5 | 0.464 | 0.710 | 0.678 | 0.0416 | |||
| Note: NA number of alleles per locus, HO observed heterozygosity, HE expected heterozygosity, PIC polymorphism information content, HWE Hardy-Weinberg equilibrium. | ||||||||
In total, 10 and 11 TCP proteins with an open reading frame (ORF) exceeding the TCP domain were retrieved from ChFlower and CjFlower, respectively, through the first strategy (Supplementary data). The Neighbor-Joining (NJ) tree constructed from multiple amino acid alignment of the TCP domain sequences classified the TCP proteins into two distinct clades (Class Ⅰ and Class Ⅱ), however, all the Class Ⅱ transcripts presented in subclade CIN-like group with none of the transcripts belonging to the Class Ⅱ CYC/TB1-like group (Fig. S3). As expected, we also failed to obtain any CYC/TB1-like transcript by direct assembly of high-quality reads that were aligned to Commelinaceae references (the second strategy); to be exact, only one read (150 bp) of CjFlower was successfully mapped to the references. Potential reasons for the failure of the first and second strategies will be discussed below. Through the third strategy, the single amplified product with a length of 417 bp for C. heterosepala (named C. heterosepala TBL gene) and 404 bp for C. japonica (C. japonica TBL) were acquired, and both NeighborJoining phylogenetic analysis (Fig. S3) and online BLAST search confirmed their homology with CYC/TB1-like genes. Furthermore, no cases of multiple amplification fragments of the CYC/TB1-like gene were found for the remaining five Stemonaceae species (four genera), indicating that the CYC/TB1-like gene is most likely a single copy gene in this family. Newly-generated CYC/TB1-like sequences were submitted to GenBank under accession numbers MG322167—MG322173.
Bayesian inference and maximum likelihood analyses based on 79 monocot CYC/TB1 protein sequences yielded a highly similar tree topology. All the newly-generated Stemonaceae CYC/TB1 genes formed a strongly-supported SteTBL1 clade (PP = 1.0, BS = 98%) that was sister to Alstroemeria TCP1 (AlsTCP1) genes. Three Croomia TBL genes were resolved as a monophyletic clade, in which TBL genes of C. heterosepala and C. japonica were identified as sister groups with strong support (PP = 0.98, BS = 78%), despite different flower symmetry types for these two species (Fig. 3). Moreover, phylogenetic analyses resolved TBL1 and TBL2 clades of TB1 genes from our full CYC/TB1 data set. The TBL1 clade mainly includes six subclades: ZinTBL1a, ZinTBL1b, AlsTCP1, SteTBL1 (this study), PoaTBL1, and AcoTBL, while the TBL2 clade comprised three subclades containing genes from Zingiberales (ZinTBL2 subclade), Commelinaceae (ComTB1a subclade), and two PoaTBL2 subclade genes.
The branch models of Yang (2007) explored the variation of selective pressure on CYC/TBL genes from monocot groups (Table 5, branches labeled in Fig. 3). Three hypotheses were significantly more likely (P < 0.01) than the M0 model and used in subsequent analyses: hypothesis 4 of M2 model (M2-H4), in which the ZinTBL1a branch has a distinct ω value; hypothesis 8 of M2 model (M2-H8), in which the PoaTBL1 branch has a distinct ω value; and hypothesis 12 of M2 model (M2-H12), in which all the main branches have distinct ω values. Under hypotheses H4 and H8 of the M2 model, estimates of ω values on the ZinTBL1a branch were far greater than the remaining branches, which were considered to be under positive selection. Furthermore, SteTBL1 genes supported by all these three best models (H4, H8, and H12 of the M2 model) were under strong purifying selection with ω values ranging from 0.1402 to 0.1523, as expected for a gene that codes for a functionallyimportant protein.
| ωZ2 | ωC1a | ωp2 | ωZ1a | ωZ1b | ωA1 | ωSteTBL | ωP1 | ωAcoTBL | LnL | df | P | |
| M0 | 0.1463 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | -2207.81 | ||
| M1 | 0.0013 | 0.0001 | 0.0011 | 999 | 0.0001 | 0.1473 | 999 | 0.9159 | 0.0748 | -2137.08 | 131 | 0.2511 |
| M2 H1: Z2 | 2.0201 | 0.1463 | =ωC1a | =ωC1a | =ωC1a | =ωC1a | =ωC1a | =ωC1a | =ωC1a | -2207.81 | 1 | 0.9936 |
| M2 H2: C1a | 0.1460 | 0.0657 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | -2207.81 | 1 | 0.9335 |
| M2 H3: P2 | 0.1486 | =ωZ2 | 0.0690 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | -2207.66 | 1 | 0.5907 |
| M2 H4: Z1a | 0.1402 | =ωZ2 | =ωZ2 | 999 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | -2203.06 | 1 | 0.0021 |
| M2 H5: Z1b | 0.14630 | =ωZ2 | =ωZ2 | =ωZ2 | 2.3813 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | -2207.81 | 1 | 0.9935 |
| M2 H6: A1 | 0.1474 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | 0.0514 | =ωZ2 | =ωZ2 | =ωZ2 | -2207.51 | 1 | 0.4394 |
| M2 H7: SteTBL | 0.1460 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | 0.1613 | =ωZ2 | =ωZ2 | -2207.81 | 1 | 0.9111 |
| M2 H8: P1 | 0.1464 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | 0.0016 | =ωZ2 | -2204.26 | 1 | 0.0068 |
| M2 H9: AcoTBL | 0.1447 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | 0.2714 | -2207.68 | 1 | 0.6093 |
| M2 H10: Z1a & Z1b | 0.1463 | =ωZ2 | =ωZ2 | 1.8274 | =ωZ1a | =ωZ2 | =ωZ2 | =ωZ2 | =ωZ2 | -2207.81 | 1 | 0.9906 |
| M2 H11: Z2 & C1a & P2 | 999 | =ωZ2 | =ωZ2 | 0.1444 | 0.1444 | 0.1444 | 0.1444 | 0.1444 | 0.1444 | -2207.63 | 1 | 0.4455 |
| M2 H12: all independent ω | 0.0933 | 0.1523 | 0.0611 | 999 | 0.0001 | 0.067 | 0.1523 | 0.1523 | 0.1523 | -2196.80 | 8 | 0.0048 |
| Note: Z2 = ZinTBL2, C1a =ComTB1a, P2 = PoaTBL2, Z1a = ZinTBL1a, Z1b = ZinTBL1b, A1 = AlsTCP1 and P1=PoaTBL1. | ||||||||||||
Since next-generation sequencing (NGS) technologies appeared on the market ten years ago, dramatic increases have been made in terms of speed, read length and throughput, along with a sharp reduction in per-base cost, which paved the way for the wide application of NGS technologies in basic science (Dijk et al., 2014). Illumina-based transcriptome sequencing technology has proved to be a powerful tool for obtaining large amounts of transcriptome data, and is widely used in gene discovery (Clark et al., 2010; Mao et al., 2016), molecular marker development (Huang et al., 2014; Ai et al., 2015), phylogenomic analysis (Teasdale et al., 2016), differential gene expression (Akashi et al., 2016) and so on. In this study, we presented the first reference transcriptome (CjTranscriptome) in Croomia and the first comparative analysis of flower transcriptomes in Stemonaceae, for which, no genomic resources have been reported to date.
Through Illumina transcriptome sequencing analysis of ChFlower, CjFlower, and CjLeaf, we obtained 57, 193e64, 448 unigenes with an average length of 1042e1106 bp and an N50 of 1575e1737 bp for these three samples (Table 1). Our results closely resembled the transcriptome assembly outcome of other monocots using similar technologies (Huang et al., 2015, 2016). By pooling the CjFlower and CjLeaf unigenes into a CjTranscriptome, we obtained the longer unigenes of CjTranscriptome than those of CjFlower and CjLeaf, possibly due to a relative increase of read coverage. For each transcriptome, more than 67% of the unigenes had significant Blast hits against the protein databases, however, a fraction of unigenes from our study had no BLAST matches to these databases. This phenomenon is common in many other plants and these "nonBLASTable" genes were thought to be Croomia-specific genes, rapidly evolving genes, or untranslated regions (UTRs) (Parchman et al., 2010; Logacheva et al., 2011; Zhang et al., 2013). Overall, our study is the first to obtain high-quality transcriptomes of Croomia, offering a platform to perform comparative analysis, evolutionary biology, and phylogenomic studies.
4.2. Orthologous genes and SCNGs between ChFlower and CjFlowerTo evaluate the effects of selection on these two Croomia species with different flower symmetry types, the Ka/Ks ratio, an indicator of selective pressure on protein-coding genes, was calculated between ChFlower and CjFlower orthologs. Of 11, 566 potential orthologous pairs between ChFlower and CjFlower, only six orthologs had a Ka/Ks ratio significantly > 1, and an additional 13 pairs were above the relaxed threshold of 0.5 (Table 3), suggesting that these genes are likely under the effect of positive selection and may be related to the adaptive divergence of these two species. According to the Nr annotation of these positively selected genes, more than half were involved in several biological functions (e.g., disease resistance, RNA-binding), while the remaining seven were recognized as uncharacterized or hypothetical proteins. Thus, further studies on expression analysis, cellular localization, molecular interactions and three-dimensional structures are needed on these uncharacterized proteins (Schluepen et al., 2013).
The identification of orthologous genes is a crucial prerequisite for reliable phylogenetic investigation (Teasdale et al., 2016), and the advantages of single or low copy nuclear genes for resolving deep evolutionary relationships have been proven at the species level to the order level (Duarte et al., 2010; Ai et al., 2015; Xiang et al., 2016; Zeng et al., 2017). Previous phylogenetic analyses using sequence data from a few chloroplast genes and nrDNA ITS, as well as morphological characters, revealed that Stemonaceae was monophyletic; Pentastemona firstly diverged from the remaining genera with Stemona sister to (Croomia + Stichoneuron) (Caddick et al., 2002; Rudall et al., 2005). However, the detailed picture of species relationships within each genus has remained unclear due to too few genetic markers employed in previous studies. In this study, the 429 putative SCNGs identified in the transcriptomes of Croomia species may represent more desirable choices for specieslevel phylogenetic reconstruction for Croomia and its related genera. Moreover, since multiple unlinked genetic loci provide independent realizations of divergence history, accounting for mutational and coalescent stochasticity (McCormack et al., 2011; Leavitt et al., 2012), these putative SCNGs are suitable candidates for studies aimed at understanding molecular phylogeography and population genetics in Croomia.
4.3. EST-SSR markersUnlike SSRs, EST-SSR markers are easier to obtain, more widely transferable among species, and related to phenotypic variation (Andersen and Lübberstedt, 2003; Li et al., 2004; Duran et al., 2009). Therefore, in recent years, EST-SSR markers have been developed in many species for genetic analysis (Poncet et al., 2006; Qiu et al., 2010; Chen et al., 2015). However, prior to this study, neither ESTs nor EST-SSR markers were available on public databases for medicinally important, endangered species of Croomia. This lack of genomic resources has impeded our understanding of Croomia conservation, phylogeography, and population genetics. Of the 21, 460 EST-SSRs identified in our work, dinucleotide (47.4%) repeats were the most frequent SSR repeat type, followed by trinucleotide (30.2%), and mononucleotide (13.4%) repeats (Fig. 2, Table S5). This result is in contrast to most results observed in previous studies, in which tri-nucleotide repeats were the most abundant EST-SSR repeat motif (Varshney et al., 2005; Koilkonda et al., 2012; Guo et al., 2014; Mao et al., 2016). Morgante et al. (2002) investigated the distribution of SSRs across different genomic fractions and found that, excluding tri-nucleotides, SSRs rarely occurred in coding regions compared with non-coding regions. Therefore, the dominance of di-nucleotide repeats detected here may be caused by an over-representation of untranslated regions (UTRs) compared with open reading frames (Kumpatla and Mukhopadhyay, 2005; Chen et al., 2015). Recently, 126 genomic SSR primer pairs were designed and only 11 (8.7%) primer pairs showed inter-species transferability among the three Croomia species (Fang et al., 2013). In this study, of the 100 primer pairs, 46 (46%) primer pairs successfully produced PCR fragments of expected size, suggesting EST-SSRs are more transferable than genomic SSRs in Stemonaceae. Moreover, another important advantage of EST-SSRs is the possibility to track adaptive divergence processes for their functional involvement (Ai et al., 2015). Thus, these newly identified EST-SSR markers will be powerful tools for future population genetics across a wide range of taxa in Stemonaceae.
4.4. Analysis of CYC/TB1-like genes in Stemonaceae speciesZygomorphy has evolved repeatedly from actinomorphy throughout angiosperms (Zhang et al., 2010). During the evolution of angiosperms, floral zygomorphy has evolved independently at least 130 times, while reversal to actinomorphy may have evolved at least 69 times. Among the origins of floral zygomorphy, 29 cases have been in monocots (Reyes et al., 2016). CYC/TB1-like genes, which are associated with the evolution and maintenance of flower monosymmetry have not been reported in Stemonaceae. Previous phylogenetic analyses of the reduced CYC/TB1 dataset resolve two clades of TBL genes from monocots, named TBL1 and TBL2 (Bartlett and Specht, 2011). This ancient duplication in the TBL gene lineage is found to predate the divergence of the commelinid monocots (Bartlett and Specht, 2011). The first two strategies of our study (see Materials and Methods) failed to obtain CYC/TB1-like genes in both flower and leaf transcriptomes, suggesting that these genes are not expressed, or are expressed at an extremely low level at the stages of anthesis and leaf. The expression patterns of CYC/TB1-like genes are found to vary across different plant species. For example, asymmetric expression of TB1a in Commelina communis and C. dianthifolia (zygomorphic flowers) is initiated in early flower development (Preston and Hileman, 2012), while asymmetric expression of CYC/TB1-like genes in the only zygomorphic magnoliid, Aristolochia, is only detected after the establishment of perianth zygomorphy at later stages of floral development (Horn et al., 2015). Future studies should detect and compare the expression patterns at different stages (primordium, 0.5 cm buds, 1.0 cm buds etc.), and using dissected tissues (lateral petals, carpels and stamens etc.). Even though we have not detected CYC/TB1-like genes from the transcriptomes of Croomia, based on PCR-based methods using different sets of TBL primers (Howarth and Donoghue, 2005), we have isolated a single copy of the CYC/TB1-like gene from each of the Stemonaceae taxa surveyed. The resulting tree topology (Fig. 3) strongly supports the monophyly of the 7 SteTBL sequences (PP = 1.00), which clearly belong to the TBL1 clade (Bartlett and Specht, 2011). This phylogenetic pattern provides additional evidence for the homology of these sequences by representing orthologous genes in Stemonaceae that descended from a single ancestral copy. The association between CYC-like copy number and floral symmetry has been discovered in the Dipsacales (Howarth and Donoghue, 2005) and Plantaginaceae (Reardon et al., 2009). In fact, the TBL1 gene is also found to be single copy in a large number of grasses, e.g., Orzya, Sorghum, and Brachypodium (Lukens and Doebley, 2001). The single TBL gene in Stemonaceae and Poaceae differs from the inferred homologs of TB1 in the Zingiberales, ZinTBL1a and ZinTBL1b, which have both been maintained in the Zingiberales genome following a duplication event in the TBL1 gene lineage (Bartlett and Specht, 2011). Taken together, the shifts of floral symmetry in Stemonaceae do not appear to be correlated with TBL copy number. Within the TBL1 clade, multiple sequences are clustered in different subclades, and generally grouped according to taxonomy (e.g., Stemonaceae, Poaceae, Alstroemeriaceae). This phylogenetic pattern suggests that a common ancestral copy of TB1 has undergone significant diversification following the diversification of these families. On the other hand, these results imply that functional diversification of the TBL genes may play an important role in the diversification of these families, resulting in a new protein function that was maintained through extreme purifying selection acting on the TBL gene (Bartlett and Specht, 2011).
AcknowledgementsThis research was supported by the National Natural Science Foundation of China (Grant Nos. 31370241, 31570214), the International Cooperation and Exchange of the National Natural Science Foundation of China (Grant Nos. 31511140095, 31561143015), and National Science Foundation of China (grant no. 30960027). We are very grateful to the editor and reviewers for critically evaluating the manuscript and providing constructive comments for its improvement.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2018.11.006.
Abascal F., Zardoya R., Posada D., 2005. ProtTest:selection of best-fit models of protein evolution. Bioinformatics, 21, 2104-2105.
DOI:10.1093/bioinformatics/bti263 |
||
Ai B., Gao Y., Zhang X., Tao J., Kang M., Huang H., 2015. Comparative transcriptome resources of eleven Primulina species, a group of 'stone plants' from a biodiversity hot spot. Mol. Ecol. Resour, 15, 619-632.
DOI:10.1111/1755-0998.12333 |
||
Akashi H.D., Cádiz D.A., Shigenobu S., Makino T., Kawata M., 2016. Differentially expressed genes associated with adaptation to different thermal environments in three sympatric Cuban Anolis lizards. Mol. Ecol, 25, 2273-2285.
DOI:10.1111/mec.2016.25.issue-10 |
||
Altschul S.F., Altschul S.F., Gish W., Gish W., Miller W., Miller W., et al., 1990. Basic local alignment search tool. J. Mol. Biol, 215, 403-410.
DOI:10.1016/S0022-2836(05)80360-2 |
||
Andersen J.R., Lübberstedt T., 2003. Functional markers in plants. Trends Plant Sci, 8, 554-560.
|
||
Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., et al., 2000. Gene ontology:tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet, 25, 25.
|
||
Bartlett M.E., Specht C.D., 2011. Changes in expression pattern of the TEOSINTE BRANCHED1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. Am. J. Bot, 98, 227-243.
DOI:10.3732/ajb.1000246 |
||
Bustamante C.D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M.T., Glanowski S., et al., 2005. Natural selection on protein-coding genes in the human genome. Nature, 437, 1153-1157.
DOI:10.1038/nature04240 |
||
Caddick L.R., Rudall P.J., Wilkin P., Hedderson T.A.J., Chase M.W., 2002. Phylogenetics of Dioscoreales based on combined analyses of morphological and molecular data. Bot. J. Linn. Soc, 138, 123-144.
DOI:10.1046/j.1095-8339.2002.138002123.x |
||
Chapuis M., Estoup A., 2007. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol, 24, 621-631.
DOI:10.1093/molbev/msl191 |
||
Chase M.W., Christenhusz M.J.M., Fay M.F., Byng J.W., Judd W.S., Soltis D.E., et al., 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants:APG IV. Bot. J. Linn. Soc, 181, 1-20.
DOI:10.1111/boj.2016.181.issue-1 |
||
Chen G., Brecker L., Felsinger S., Cai X., Kongkiatpaiboon S., Schinnerl J., et al., 2017. Morphological and chemical variation of Stemona tuberosa from southern China - evidence for heterogeneity of this medicinal plant species. Plant Biol, 19, 835-842.
DOI:10.1111/plb.2017.19.issue-5 |
||
Chen L., Cao Y., Yuan N., Nakamura K., Wang G., Qiu Y., 2015. Characterization of transcriptome and development of novel EST-SSR makers based on nextgeneration sequencing technology in Neolitsea sericea (Lauraceae) endemic to East Asian land-bridge islands. Mol. Breed, 35, 1-15.
DOI:10.1007/s11032-015-0202-z |
||
Christenhusz M., Byng J., 2016. The number of known plants species in the world and its annual increase. Phytotaxa, 261, 201-217.
DOI:10.11646/phytotaxa.261.3 |
||
Citerne H., Reyes E., Le G.M., Delannoy E., Simonnet F., Sauquet H., et al., 2017. Characterization of CYCLOIDEA-like genes in Proteaceae, a basal eudicot family with multiple shifts in floral symmetry. Ann. Bot, 119, 367-378.
DOI:10.1093/aob/mcw219 |
||
Clark M.S., Thorne M.A.S., Vieira F.A., Cardoso J.C.R., Power D.M., Peck L.S., 2010. Insights into shell deposition in the Antarctic bivalve Laternula elliptica:gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics, 11, 362.
DOI:10.1186/1471-2164-11-362 |
||
Conesa A., Götz S., García J.M., Teról J., Talón M., Robles M., 2005. Blast2GO:a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674-3676.
DOI:10.1093/bioinformatics/bti610 |
||
Dempster A.P., Laird N.M., Rubin D.B., 1977. Maximum likelihood from incomplete data via the EM algorithm. J. Roy. Stat. Soc.B, 39, 1-38.
|
||
Dieringer D., Schlötterer C., 2003. MICROSATELLITE ANALYSER (MSA):a platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes, 3, 167-169.
DOI:10.1046/j.1471-8286.2003.00351.x |
||
Dijk E.L.V., Auger H., Yan J., Thermes C., 2014. Ten years of next-generation sequencing technology. Trends Genet, 30, 418-426.
DOI:10.1016/j.tig.2014.07.001 |
||
Dolman G., Joseph L., 2016. Multi-locus sequence data illuminate demographic drivers of Pleistocene speciation in semi-arid southern Australian birds (Cinclosoma spp.). BMC Evol. Biol, 16, 226.
DOI:10.1186/s12862-016-0798-6 |
||
Duarte J.M., Wall P.K., Edger P.P., Landherr L.L., Ma H., Pires J.C., et al., 2010. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol. Biol, 10, 61.
DOI:10.1186/1471-2148-10-61 |
||
Duran C., Appleby N., Edwards D., Batley J., 2009. Molecular genetic markers:discovery, applications, data storage and visualization. Curr. Bioinf, 4, 16-27.
DOI:10.2174/157489309787158198 |
||
Estill J.C., Cruzan M.B., 2001. Phytogeography of rare plant species endemic to the Southeastern United States. Castanea, 66, 3-23.
|
||
Fang M., Fu C., Fu C., Zhu Y., Naiki A., Li E., 2013. Development of microsatellite markers for Croomia japonica and cross-amplification in its congener. Sci. Hortic, 161, 228-232.
DOI:10.1016/j.scienta.2013.07.014 |
||
Greger H., 2006. Structural relationships, distribution and biological activities of Stemona alkaloids. Planta Med, 72, 99-113.
DOI:10.1055/s-2005-916258 |
||
Guindon S., Dufayard J., Lefort V., Anisimova M., Hordijk W., Gascuel O., 2010. New algorithms and methods to estimate maximum-likelihood phylogenies:assessing the performance of PhyML 3.0. Syst. Biol, 59, 307-321.
DOI:10.1093/sysbio/syq010 |
||
Guo R., Mao Y., Cai J., Wang J., Wu J., Qiu Y., 2014. Characterization and crossspecies transferability of EST-SSR markers developed from the transcriptome of Dysosma versipellis (Berberidaceae) and their application to population genetic studies. Mol. Breed, 34, 1733-1746.
DOI:10.1007/s11032-014-0134-z |
||
Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., et al., 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc, 8, 1494-1512.
DOI:10.1038/nprot.2013.084 |
||
Horn S., Pabón-Mora N., Theuß V.S., Busch A., Zachgo S., 2015. Analysis of the CYC/TB1 class of TCP transcription factors in basal angiosperms and magnoliids. Plant J, 81, 559-571.
DOI:10.1111/tpj.2015.81.issue-4 |
||
Hoshino Y., Igarashi T., Ohshima M., Shinoda K., Murata N., Kanno A., et al., 2014. Characterization of CYCLOIDEA-like genes in controlling floral zygomorphy in the monocotyledon Alstroemeria. Sci. Hortic, 169, 6-13.
DOI:10.1016/j.scienta.2014.01.046 |
||
Howarth D.G., Donoghue M.J., 2005. Duplications in CYC-like genes from dipsacales correlate with floral form. Int. J. Plant Sci, 166, 357-370.
DOI:10.1086/428634 |
||
Howarth D.G., Martins T., Chimney E., Donoghue M.J., 2011. Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann. Bot, 107, 1521-1532.
DOI:10.1093/aob/mcr049 |
||
Huang D., Zhang Y., Jin M., Li H., Song Z., Wang Y., et al., 2014. Characterization and high cross-species transferability of microsatellite markers from the floral transcriptome of Aspidistra saxicola (Asparagaceae). Mol. Ecol. Resour, 14, 569-577.
DOI:10.1111/men.2014.14.issue-3 |
||
Huang J., Gao Y., Jia H., Zhang Z., 2016. Characterization of the teosinte transcriptome reveals adaptive sequence divergence during maize domestication. Mol. Ecol. Resour, 16, 1465-1477.
DOI:10.1111/men.2016.16.issue-6 |
||
Huang L.K., Yan H.D., Zhao X.X., Zhang X.Q., Wang J., Frazier T., et al., 2015. Identifying differentially expressed genes under heat stress and developing molecular markers in orchardgrass (Dactylis glomerata L.) through transcriptome analysis. Mol. Ecol. Resour, 15, 1497-1509.
DOI:10.1111/1755-0998.12418 |
||
Iseli C., Jongeneel C.V., Bucher P., 1999. ESTScan:a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol, 138-148.
|
||
Jabbour F., Cossard G., Le G.M., Sannier J., Nadot S., Damerval C., 2014. Specific duplication and dorsoventrally asymmetric expression patterns of cycloidealike genes in zygomorphic species of Ranunculaceae. PLoS One, 9, e95727.
DOI:10.1371/journal.pone.0095727 |
||
Kalinowski S.T., Taper M.L., Marshall T.C., 2007. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol, 16, 1099-1106.
DOI:10.1111/j.1365-294X.2007.03089.x |
||
Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., et al., 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res, 36, 480-484.
|
||
Kawakami T., Backström N., Burri R., Husby A., Olason P., Rice A.M., et al., 2014. Estimation of linkage disequilibrium and interspecific gene flow in Ficedula flycatchers by a newly developed 50k single-nucleotide polymorphism array. Mol. Ecol. Resour, 14, 1248-1260.
DOI:10.1111/men.2014.14.issue-6 |
||
Koilkonda P., Sato S., Tabata S., Shirasawa K., Hirakawa H., Sakai H., et al., 2012. Large-scale development of expressed sequence tag-derived simple sequence repeat markers and diversity analysis in Arachis spp. Mol. Breed, 30, 125-138.
DOI:10.1007/s11032-011-9604-8 |
||
Kongkiatpaiboon S., Schinnerl J., Felsinger S., Keeratinijakal V., Vajrodaya S., Gritsanapan W., et al., 2011. Structural relationships of stemona alkaloids:assessment of species-specific accumulation trends for exploiting their biological activities. J. Nat. Prod, 74, 1931-1938.
DOI:10.1021/np2004374 |
||
Kumar S., Stecher G., Tamura K., 2016. MEGA7:molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol, 33, 1870-1874.
DOI:10.1093/molbev/msw054 |
||
Kumpatla S.P., Mukhopadhyay S., 2005. Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome, 48, 985-998.
DOI:10.1139/g05-060 |
||
Leavitt S.D., Esslinger T.L., Divakar P.K., Lumbsch H.T., 2012. Miocene and Pliocene dominated diversification of the lichen-forming fungal genus Melanohalea(Parmeliaceae, Ascomycota) and Pleistocene population expansions. BMC Evol. Biol, 12, 176.
DOI:10.1186/1471-2148-12-176 |
||
Lee S., Xiao C., Pei S., 2008. Ethnobotanical survey of medicinal plants at periodic markets of Honghe Prefecture in Yunnan Province, SW China. J. Ethnopharmacol, 117, 362-377.
DOI:10.1016/j.jep.2008.02.001 |
||
Li E., Yi S., Qiu Y., Guo J., Comes H.P., Fu C., 2008. Phylogeography of two East Asian species in Croomia (Stemonaceae) inferred from chloroplast DNA and ISSR fingerprinting variation. Mol. Phylogenet. Evol, 49, 702-714.
DOI:10.1016/j.ympev.2008.09.012 |
||
Li L., Stoeckert C.J., Roos D.S., 2003. OrthoMCL:identification of ortholog groups for eukaryotic genomes. Genome Res, 13, 2178-2189.
DOI:10.1101/gr.1224503 |
||
Li Y., Korol A.B., Fahima T., Nevo E., 2004. Microsatellites within genes:structure, function, and evolution. Mol. Biol. Evol, 21, 991-1007.
DOI:10.1093/molbev/msh073 |
||
Li Z., Zou J., Mao K., Lin K., Li H., Liu J., et al., 2012. Population genetic evidence for complex evolutionary histories of four high altitude juniper species in the Qinghai-Tibetan plateau. Evolution, 66, 831-845.
DOI:10.1111/evo.2012.66.issue-3 |
||
Lin G., Lin L., Pak-Ho L.H., Zhu J., Tang C., Ke C., et al., 2008. Croomine-and tuberostemonine-type alkaloids from roots of Stemona tuberosa and their antitussive activity. Tetrahedron, 64, 10155-10161.
DOI:10.1016/j.tet.2008.08.046 |
||
Lin L., Zhong Q., Cheng T., Tang C., Ke C., Lin G., et al., 2006. Stemoninines from the roots of Stemona tuberosa. J. Nat. Prod, 69, 1051-1054.
DOI:10.1021/np0505317 |
||
Liu Z., Ma L., Nan Z., Wang Y., 2013. Comparative transcriptional profiling provides insights into the evolution and development of the zygomorphic flower of Vicia sativa (Papilionoideae). PLoS One, 8, e57338.
DOI:10.1371/journal.pone.0057338 |
||
Logacheva M.D., Kasianov A.S., Vinogradov D.V., Samigullin T.H., Gelfand M.S., Makeev V.J., et al., 2011. De novo sequencing and characterization of floral transcriptome in two species of buckwheat (Fagopyrum). BMC Genomics, 12, 30.
DOI:10.1186/1471-2164-12-30 |
||
Lukens L., Doebley J., 2001. Molecular evolution of the teosinte branched gene among maize and related grasses. Mol. Biol. Evol, 18, 627-638.
DOI:10.1093/oxfordjournals.molbev.a003843 |
||
Ma C., Xin M., Feldmann K.A., Wang X., 2014. Machine learning-based differential network analysis:a study of stress-responsive transcriptomes in Arabidopsis. Plant Cell, 26, 520-537.
DOI:10.1105/tpc.113.121913 |
||
Mao Y., Zhang Y., Xu C., Qiu Y., 2016. Comparative transcriptome resources of two Dysosma species (Berberidaceae) and molecular evolution of the CYP719A gene in Podophylloideae. Mol. Ecol. Resour, 16, 228-241.
DOI:10.1111/1755-0998.12415 |
||
McCormack J.E., Heled J., Delaney K.S., Peterson A.T., Knowles L.L., 2011. Calibrating divergence times on species trees versus gene trees:implication for speciation history of Aphelocoma Jays. Evolution, 65, 184-202.
DOI:10.1111/j.1558-5646.2010.01097.x |
||
Morgante M., Hanafey M., Powell W., 2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet, 30, 194-200.
DOI:10.1038/ng822 |
||
Okuyama S., 1944. On the Japanese species of Croomia. J. Jpn. Bot, 20, 31-32.
|
||
Ohwi, J., 1965. Croomia. Flora of Japan. Smithsonian Institution, Washington, p. 279.
|
||
Parchman T.L., Geist K.S., Grahnen J.A., Benkman C.W., Buerkle C.A., 2010. Transcriptome sequencing in an ecologically important tree species:assembly, annotation, and marker discovery. BMC Genomics, 11, 180.
DOI:10.1186/1471-2164-11-180 |
||
Pertea G., Huang X., Liang F., Antonescu V., Sultana R., Karamycheva S., et al., 2003. TIGR gene indices clustering tools (TGICL):a software system for fast clustering of large EST datasets. Bioinformatics, 19, 651-652.
DOI:10.1093/bioinformatics/btg034 |
||
Poncet V., Rondeau M., Tranchant C., Cayrel A., Hamon S., Kochko A., et al., 2006. SSR mining in coffee tree EST databases:potential use of EST-SSRs as markers for the Coffea genus. Mol. Genet. Genom, 276, 436-449.
DOI:10.1007/s00438-006-0153-5 |
||
Preston J.C., Hileman L.C., 2009. Developmental genetics of floral symmetry evolution. Trends Plant Sci, 14, 147-154.
DOI:10.1016/j.tplants.2008.12.005 |
||
Preston J.C., Hileman L.C., 2012. Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. EvoDevo, 3, 6.
DOI:10.1186/2041-9139-3-6 |
||
Qiu L., Yang C., Tian B., Yang J., Liu A., 2010. Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol, 10, 278.
DOI:10.1186/1471-2229-10-278 |
||
Reardon W., Fitzpatrick D.A., Fares M.A., Nugent J.M., 2009. Evolution of flower shape in Plantago lanceolata. Plant Mol. Biol, 71, 241-250.
|
||
Reyes E., Sauquet H., Nadot S., 2016. Perianth symmetry changed at least 199 times in angiosperm evolution. Taxon, 65, 945-964.
DOI:10.12705/655.1 |
||
Rogers A.R., Harpending H., 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol, 9, 552-569.
|
||
Ronquist F., Huelsenbeck J.P., 2003. MrBayes 3:Bayesian phylogenetic inference under mixed models. Bioinformatics, 19, 1572-1574.
DOI:10.1093/bioinformatics/btg180 |
||
Rousset F., 2008. Genepop'007:a complete re-implementation of the genepop software for windows and linux. Mol. Ecol. Resour, 8, 103-106.
DOI:10.1111/j.1471-8286.2007.01931.x |
||
Rudall P.J., Cunniff J., Wilkin P., Caddick L.R., 2005. Evolution of dimery, pentamery and the monocarpellary condition in the monocot family Stemonaceae (Pandanales). Taxon, 54, 701-711.
DOI:10.2307/25065427 |
||
Schluepen C., Malito E., Marongiu A., Schirle M., McWhinnie E., Lo Surdo P., et al., 2013. Mining the bacterial unknown proteome:identification and characterization of a novel family of highly conserved protective antigens in Staphylococcus aureus. Biochem. J, 455, 273-284.
|
||
Swanson W.J., Wong A., Wolfner M.F., Aquadro C.F., 2004. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics, 168, 1457-1465.
DOI:10.1534/genetics.104.030478 |
||
Teasdale L.C., Köhler F., Murray K.D., O'Hara T., Moussalli A., 2016. Identi fication and qualification of 500 nuclear, single-copy, orthologous genes for the Eupulmonata (Gastropoda) using transcriptome sequencing and exon capture. Mol. Ecol. Resour, 16, 1107-1123.
DOI:10.1111/1755-0998.12552 |
||
Thompson J.D., Higgins D.G., Gibson T.J., 1994. ClustalW:improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res, 22, 4673-4680.
DOI:10.1093/nar/22.22.4673 |
||
Varshney R.K., Graner A., Sorrells M.E., 2005. Genic microsatellite markers in plants:features and applications. Trends Biotechnol, 23, 48-55.
DOI:10.1016/j.tibtech.2004.11.005 |
||
Wang, S., Xie, Y., 2004. Red List. In: China Species Red List, vol. 1. Higher Education Press, Beijing.
|
||
Wen, J., Egan, A.N., Dikow, R.B., Zimmer, E.A., 2015. Utility of transcriptome sequencing for phylogenetic inference and character evolution. In: Elvira, H., Marc, S.A. (Eds.), Next-generation Sequencing in Plant Systematics, pp. 1-42.
|
||
Xiang Y., Huang C., Hu Y., Wen J., Li S., Yi T., et al., 2016. Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol. Biol. Evol, 34, 262-281.
|
||
Yang Z., 2007. PAML 4:phylogenetic analysis by maximum likelihood. Mol. Biol. Evol, 24, 1586-1591.
DOI:10.1093/molbev/msm088 |
||
Yang Z., Nielsen R., 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol, 17, 32-43.
DOI:10.1093/oxfordjournals.molbev.a026236 |
||
Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., et al., 2006. WEGO:a web tool for plotting GO annotations. Nucleic Acids Res, 34, W293-W297.
DOI:10.1093/nar/gkl031 |
||
Yuan Z., Gao S., Xue D., Luo D., Li L., Ding S., et al., 2009. RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol, 149, 235.
|
||
Zeng L., Zhang N., Zhang Q., Endress P.K., Huang J., Ma H., 2017. Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic and genomic datasets. New Phytol, 214, 1338-1354.
DOI:10.1111/nph.14503 |
||
Zhang J., Franks R., Liu X., Kang M., Keebler J., Schaff J., et al., 2013a. De novo sequencing, characterization, and comparison of inflorescence transcriptomes of Cornus canadensis and C. florida (Cornaceae). PLoS One, 8, e82674.
DOI:10.1371/journal.pone.0082674 |
||
Zhang L., Yan H., Wu W., Yu H., Ge X., 2013b. Comparative transcriptome analysis and marker development of two closely related Primrose species (Primula poissonii and Primula wilsonii). BMC Genomics, 14, 329.
DOI:10.1186/1471-2164-14-329 |
||
Zhang W., Kramer E.M., Davis C.C., Donoghue M.J., 2010. Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. P. Natl. Acad. Sci. USA, 107, 6388-6393.
DOI:10.1073/pnas.0910155107 |
||
Zhang Z., Li J., Zhao X., Wang J., Wong G.K.S., Yu J., 2006. KaKs_Calculator:calculating Ka and Ks through model selection and model averaging. Dev. Reprod. Biol, 4, 259-263.
|
||
Zhou S., Yan B., Li F., Zhang J., Ma H., Liu W., et al., 2017. RNA-seq analysis provides the first insights into the phylogenetic relationship and interspecific variation between Agropyron cristatum and Wheat. Front. Plant Sci, 8, 1644.
|
||
Zhu S., Ding Y., Yap Z., Qiu Y., 2016. De novo assembly and characterization of the floral transcriptome of an economically important tree species, Lindera glauca(Lauraceae), including the development of EST-SSR markers for population genetics. Mol. Biol. Rep, 43, 1243-1250.
DOI:10.1007/s11033-016-4056-1 |



