The Cycadales (cycads), whose origin can be dated to the Late Paleozoic (~265-290 Ma), are the most primitive living seed plants. The earliest Cycad fossil in the world was found in China and dates to Lower Permian, whereas peak abundance and diversity of cycads date to the Mesozoic (Martínez et al., 2012; Gao and Barry, 1989). Extant cycads are distributed in tropical and subtropical regions of Africa, Asia, Oceania, and America. Ten genera and ~300 species are currently accepted (Hill et al., 2004; Norstog and Nicholls, 1997). Cycads, which originated from seed ferns, have many of same characteristics as ferns. Such characteristics include pinnately compound leaves, circinate vernation, unique girdling leaf traces, no axillary buds, dichotomous branching (versus axillary branching in higher plants). However, cycads have recognizable intermediate morphological traits between angiosperms and gymnosperms, and have therefore been classified as gymnosperms. Of the four order that comprise the gymnosperms-Ginkgoales, Gnetales, Coniferales, and Cycadales-Cycadales are considered the most ancestral (Nixon et al., 1994; Soltis et al., 2002). Their pollen tubes possess multiciliate sperm, and their ovules are borne on the margins of leaflike megasporophylls (Loconte and Stevenson, 1990). Clearly, cycads represent a key node in the phylogeny of seed plants.
Previous research has also indicated the importance of understanding cycad biological nitrogen fixation (BNF). Cycads have a particular root type, referred to as coralloid roots due to their 'corallike' appearance. Nitrogen-fixing cyanobacteria invade coralloid roots, where BNF processes occur (Vessey et al., 2005; Lindblad and Costa, 2002). To date, no symbiosis-related genes in cycads have been identified (or searched) and the molecular mechanisms of symbiotic nitrogen fixation in these plants are still largely unclear (Rai et al., 2000).
In non-model species with a large genome size, EST (expressed sequence tag) sequencing and annotation is a means for gene discovery and a way to understand the transcription and expression patterns of specific genes. Normalization techniques reduce the frequency of highly expressed genes and increase the rate of rare gene discovery (Soares et al., 1994). Cycads have a large genome size (~20-30 Gbp) and are evolutionarily and horticulturally important, long-lived plants (Zonneveld, 2012). Cycads are also the only early seed plants that have evolved a specialized coralloid root to host endophytic bacteria that fix nitrogen for the plant.
Microsatellites, also called SSRs (simple sequence repeats) or STRs (short tandem repeats), are highly polymorphic genetic markers. They have been extensively used for plant population genetic studies because of their co-dominant inheritance, relative abundance, multi-allelic nature, high reproducibility, and ease of detection (Powell et al., 1996). The development of SSR markers has improved the characterization and use of genetic variation in cycads (Ju et al., 2011; Yang et al., 2008). SSRs have been adopted to evaluate genetic diversity and to reconstruct the population structure, allowing researchers to design reasonable conservation and management protocols.
Cycas is at the basal node of the Cycadales (Treutlein and Wink, 2002). Cycas debaoensis, endemic to southern China, is a rare and endangered plant species (Chen and Zhong, 1997; Ma et al., 2003). In this study, we constructed a normalized full-length cDNA library using RNA derived from the coralloid roots of C. debaoensis. We assembled ESTs into contigs and singletons, and subsequently performed comparative protein annotations. Using this database, we identified a total of 22 ESTs that encode putative receptor-like protein kinase (RLK) genes, which play a variety of important defensive and symbiotic roles in plant-microbe interactions (Shiu and Bleecker, 2003; Shiu et al., 2004). Finally we used EST data to detect SSRs.
2. Materials and methods 2.1. Plants and total RNA extractionCoralloid roots of C. debaoensis were collected from National Cycad Germplasm Conservation Center, Shenzhen Fairylake Botanical Garden. Tissue was obtained from the topsoil, washed with sterile water, and then 70% ethanol, wrapped with aluminum foil bags, frozen in liquid nitrogen for more than 2 h, and stored at -80 ℃. Total RNA was extracted from pulverized, frozen tissue using trizol reagent (Invitrogen). The integrity of the total RNA was confirmed by examining the ratio of 28S and 18S ribosomal RNA with 1% agarose gel electrophoresis. Quality and quantity of the isolated RNA were determined using a spectrophotometer. The extracted RNA was found to be of high quality (OD260/ OD280 = 2.02).
2.2. Construction of full-length normalized cDNA libraryThe cDNA library was constructed using Creator SMART cDNA library construction kit (Clontech, USA). Double stranded cDNA was synthesized according to the manufacturer's protocol. DSN normalization was applied according to the instructions of Trimmer-Director kit (Evrogen, Cat. No. NK002). The cDNA inserts were directionally cloned in pDNR-LIB vector and transformed using DH10B electrocompetent cells of Escherichia coli. The recombinants were selected in LB agar plates supplemented with ampicillin. The quality and quantity of the isolated plasmid DNA was confirmed on 0.8% agarose gels before sequencing.
2.3. EST sequencing and assemblyPlasmid DNA was sequenced by 5'end single-pass sequencing on automated DNA capillary sequencer ABI 3730XL (Applied Biosystems) according to the manufacturer's instructions using T7 primer (50TAATACGACTCACTATAGG3'). All EST sequences were deposited in the GenBank database under the accession numbers dbEST JZ917721eJZ922731. Sequence analysis was performed at Beijing Luhe Huada Gene Sci-Tech Company. ESTs were scanned and trimmed for vector sequences using NCBI's VecScreen tool. Low quality and short (< 100 bp) sequences were also removed. The processed sequences were assembled into contigs and singletons using Contig Assembly Program CAP3. Then EST and contig redundancy was calculated.
2.4. Annotation and classification of singletsAll contigs and singletons were compared against the NCBI nonredundant protein (Nr) and Swiss-Prot database (Boeckmann et al., 2003). Based on the Nr annotations, GO annotations of the unigenes were obtained using the Blast2GO program. GO terms were assigned to each unigene and classified into three functional annotation categories: biological process, cellular component, and molecular function. All unigenes were also compared to the proteins in the Cluster of Orthologous Groups (COG) databases (Tatusov et al., 2000) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2008).
2.5. EST-SSR search and primer designSSR loci of unigenes were identified and analyzed by MISA software (http://pgrc.ipk-gatersleben.de/misa/). Premier 5.0 software (PREMIER Biosoft International, Palo Alto, CA) was employed to design PCR primers for the conserved flanking regions of the SSRs. The minimum repeats of sequences with di-, tri-, tetra-, penta- and hexanucleotide motifs was set as 6, 5, 5, 5 and 5, respectively. The range of PCR product size was set between 100 and 300 bp.
2.6. Analysis of sequencesDatabases used to perform BLASTN analyses included Cycas rumphii PUT (10, 901), Zamia vazquezii PUT (7, 657), Gnetum gnemon PUT (6, 193), and Ginkgo biloba PUT (10, 210). All databases as described above were downloaded from plantGDB (http://www.plantgdb.org). All BLAST searches were subject to an expect value < 1e-5.
3. Results and discussion 3.1. EST sequence quality, contigs and singletsA total of 5393 ESTs were generated from this cDNA library; removing vector and low quality sequences resulted in 5011 highquality ESTs. The EST length was distributed from 100 to 500 bp (19.04%), 501 to 800 bp (80.42%) and 801 to 840 bp (0.54%). The cluster analyses of ESTs generated 2366 singlets and 618 contigs. The average lengths of singlets and contigs were respectively 538.87 bp and 720.66 bp. The redundancy of the library was calculated as 40.45% ((1 - Number of Unigenes/Number of ESTs) × 100%) (Tatusov et al., 2000). This is lower than that reported in other gymnosperms studies like C. rumphii (Brenner et al., 2003), Ginkgo biloba (Brenner et al., 2005), and Picea glauca (Birol et al., 2013). The number of ESTs in a contig ranged from 2 to 150. About 96.28% of the contigs had less than 10 ESTs with 338 having only 2 sequences. EST sequencing and assembly results are shown in Table 1 and the distribution of ESTs in contigs is shown (Fig. 1).
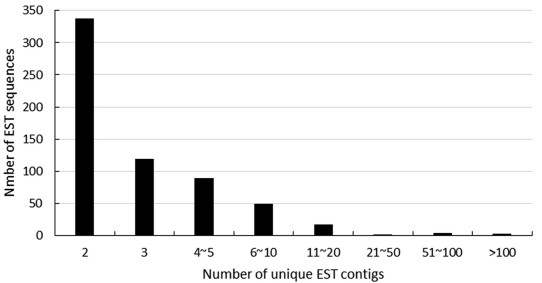
|
| Fig. 1 Distribution of individual EST sequences among the clustered contigs |
| EST sequences and contigs | Number |
| Number of EST sequences | 5011 |
| Number of Contigs | 618 |
| Number of singletons | 2366 |
| Average assembled EST length | 600.59 |
| Average number of sequences per contig | 4.23 |
| Number of contigs containing: | |
| 2 ESTs | 338 |
| 3 ESTs | 119 |
| 4~5 ESTs | 89 |
| 6~10 ESTs | 49 |
| 11~20 ESTs | 17 |
| 21~50 ESTs | 1 |
| 51~100 ESTs | 3 |
| > 100 ESTs | 2 |
All unique sequences (including 2366 singlets and 618 contigs) were subjected to BLASTX searches against the NCBI Nr (nonredundant) protein database. A total of 2333 unigenes had significant hits. Among the 2333 unique sequences, 577 matched proteins of known function and 1756 matched predicted proteins of unknown function. These unique sequences were also compared to proteins in Swiss-port, KEGG, and COG databases, which revealed there were 1, 901, 2255 and 951 unigenes that showed high similarity, respectively. Of these, 938 sequences had annotations in all four databases, 2343 sequences in at least one of the four database, and 641 sequences had no annotation in any database (Fig. 2).
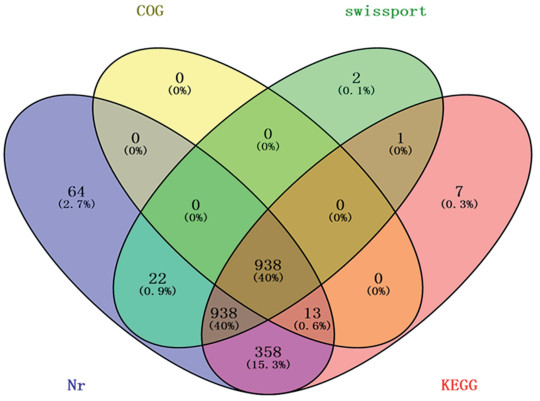
|
| Fig. 2 Venn diagram of annotation results against Nr, Swiss-Prot, COG, and KEGG databases. The numbering each color block indicates the number of unigenes that is annotated by single or multiple databases. |
Gene Ontology (GO) is a standard system defining gene classes (Ashburner et al., 2000). Fifty percent of 2984 unique sequences were successfully annotated with 4082 GO terms (Fig. 3). The unigenes were thus functionally classified with one or more ontologies: 954 sequences (63.8%) were assigned GO terms associated with biological processes, 1329 sequences (88.9%) were involved in molecular functions, and 415 sequences (27.8%) were cellular components. Under the category "biological process", 51.2% sequences were associated with "physiological process", 39.4% with "cellular process", 5.7% with "regulation of biological process", and 3.5% with "response to stimulus". Under the category "molecular functions", sequences associated with "binding" (805), "catalytic activity" (739), and "transporter activity" (111) were respectively 35.8%, 32.8%, and 4.9%. Under the category "cellular component", 57.3% sequences were associated with "cell", 23.7% with "organelle", and 17.8% with "protein complex".

|
| Fig. 3 GO analysis and functional classification of the C. debaoensis unigenes |
The COG protein database is an attempt to classify orthologous gene products (Tatusov et al., 2000). All unigenes were compared to proteins in the COG database. The results showed that a total of 847 sequences were assigned to 22 COG categories (Fig. 4). The cluster for "general function prediction only" (154, 18.18%) was the largest group, followed by "posttranslational modification, protein turnover, chaperones" (132, 15.58%), "translation, ribosomal structure and biogenesis" (90, 10.63%), "carbohydrate transport and metabolism" (73, 8.62%), "energy production and conversion" (61, 7.2%), and "amino acid transport and metabolism" (56, 6.61%). In addition, the clusters for "signal transduction" (29, 3.42%) and "defense mechanisms" (5, 0.59%) were found.
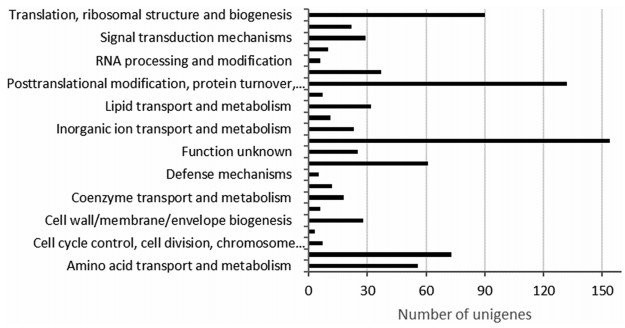
|
| Fig. 4 COG functional classification of the C. debaoensis unigenes |
The expression levels of corresponding genes can be estimated preliminarily according to the distribution frequency of transcribed ESTs in the cDNA library (Mann et al., 2013). Among the annotated sequences, 22 contained at least 10 supporting ESTs (Table 2). A family of metallothionein-like proteins was the most abundant transcript, with 470 ESTs detected. These are small metal ion binding proteins. By binding heavy metals, metallothionein-like proteins can effectively reduce the toxicity of heavy metals to the body (Zhou et al., 2005; Liu et al., 2002). Other highly abundant transcripts were genes associated with DJ-1 protein, antimicrobial proteins, germin-like proteins, aquaporin proteins, ERD (early responsive to dehydration) proteins, WAT1-related proteins, and membrane steroid-binding proteins. High abundance of genes involved in stress and defence responses is quite expected because the cDNA library was constructed from coralloid roots of C. debaoensis under natural conditions. The genes identified in our study provide a valuable transcriptomic resource for structural and functional genomics studies in C. debaoensis.
| Putative protein identification | Number of ESTs | Number of unique ESTs |
| Metallothionein-like protein EMB30 | 470 | 90 |
| Protein DJ-1 homolog D | 101 | 26 |
| Antifungal protein ginkbilobin-2 | 55 | 17 |
| Germin-like protein 9-3 | 50 | 19 |
| Ubiquitin-conjugating enzyme E2 28 | 42 | 10 |
| Glycine cleavage system H protein, mito | 38 | 6 |
| WAT1-related protein At5g07050 | 23 | 4 |
| Glucoamylase | 23 | 5 |
| Protein early responsive to dehydration | 17 | 1 |
| Chitotriosidase-1 | 15 | 7 |
| High mobility group B protein 1 | 14 | 2 |
| Clavaminate synthase-like protein At3g2 | 14 | 6 |
| Subtilisin-like protease SDD1 | 13 | 4 |
| Retrovirus-related Pol polyprotein from | 13 | 13 |
| Probable aquaporin PIP2-8 | 12 | 7 |
| Flavanone 3-dioxygenase | 11 | 3 |
| Small nuclear ribonucleoprotein E | 10 | 1 |
| Probable aquaporin PIP1-5 | 10 | 1 |
| Non-functional NADPH-dependent codeinon | 10 | 4 |
| Membrane steroid-binding protein 2 | 10 | 1 |
| EndochitinaseA2 | 10 | 2 |
| EC protein homolog 1 | 10 | 3 |
The BLASTX comparisons of unique C. debaoensis sequences were conducted against the published PUT (putative unique transcripts) of four major gymnosperms (C. rumphii, Z. vazquezii, Ginkgo biloba, and Gnetum gnemon). Phylogenetially, C. debaoensis is closest to C. rumphii, next to Z. vazquezii, Ginkgo biloba, and last to Gnetum gnemon.
Accordingly, C. rumphii and C. debaoensis had the highest matches in 1354 sequences (45.4%). The similarity results indicate that 45.4% of C. debaoensis genes discovered are homologues (orthologs) of C. rumphii genes and may have originated from a common ancestor. Ginkgo biloba followed, with 436 (14.6%) unique sequence similarities. The number of unique C. debaoensis sequences that were similar to Z. vazquezii PUTs was 429 (14.4%). Ginkgo biloba had higher similarity than Z. vazquezii since the EST resource generated for Ginkgo biloba (10, 210 PUTs) is much larger than that of Z. vazquezii (7, 657). Only 50 (1.7%) sequences had statistically significant similarity between C. rumphii and Gnetum gnemon (Fig. 5).
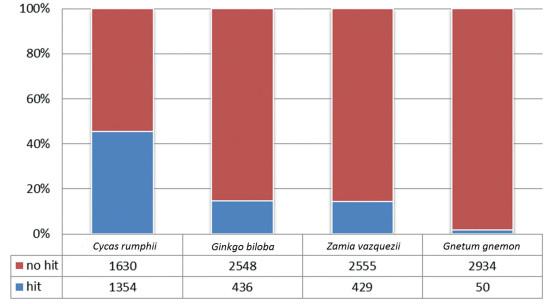
|
| Fig. 5 Conservation between PUT sequences of C. debaoensis and other gymnosperms |
A total of 94 different 2-6 nucleotide repeats were developed from unigenes obtained in this experiment (Table 3). Trinucleotide repeats were the most frequent (46, 49%), followed by dinucleotide repeats (38, 40%), tetranucleotide repeats (8, 8.5%), pentanucleotide repeats (1, 1.1%), and hexanucleotide repeats (1, 1.1%). Among the various SSRs, AG/CT repeats were the most abundant (23, 24.5%), followed by AAG/CTT (10, 10.6%), and ATC/ATG (9, 9.6%). Our results are in agreement with previous studies that have shown that angiosperms and gymnosperms have a higher abundance of AG/CT and AAG/CTT motifs. These sequences are also known methylation targets in plants (Ranade et al., 2014). In gymnosperms, some studies report the AG/CT motif as the most abundant in Cycas (von Stackelberg et al., 2006) and Gnetum (Victoria et al., 2011). In other studies, the AAG/CTT motif has been shown to be the most abundant trimer in Picea (Rungis et al., 2004) and Cycas (von Stackelberg et al., 2006). Finally, 60 pairs of primers were designed for PCR amplification. Of these primer pairs, 20 successfully amplified the target regions using genomic DNA of C. debaoensis (Table 4). As newly developed molecular markers, they provide valuable resources for the population and conservation genetics of C. debaoensis and other cycads.
| Repeats motif | Number of repeats | total | ||||||
| 5 | 6 | 7 | 8 | 9 | 10 | > 10 | ||
| AC/GT | — | 2 | 3 | 1 | 1 | — | 7 | |
| AG/CT | — | 14 | 4 | — | 1 | 2 | 2 | 23 |
| AT/AT | — | 5 | 2 | — | 1 | — | — | 8 |
| AAC/GTT | 3 | 1 | — | — | — | — | 4 | |
| AAG/CTT | 6 | 2 | 1 | — | 1 | — | — | 10 |
| AAT/ATT | 4 | 3 | — | — | — | — | — | 7 |
| ACG/CGT | 2 | — | — | — | — | — | — | 2 |
| AGC/CTG | 4 | 2 | 1 | — | — | — | — | 7 |
| AGG/CCT | 3 | 3 | 1 | — | — | — | — | 7 |
| ATC/ATG | 5 | 1 | 1 | 1 | 1 | — | — | 9 |
| AAAT/ATTT | 6 | — | — | — | — | — | — | 6 |
| AATT/AATT | 1 | — | — | — | — | — | — | 1 |
| ACAT/ATGT | 1 | — | — | — | — | — | — | 1 |
| AAAAT/ATTTT | — | 1 | — | — | — | — | — | 1 |
| ACAGCC/CTGTGG | 1 | — | — | — | — | — | — | 1 |
| Total | 36 | 34 | 13 | 2 | 4 | 3 | 1 | 94 |
The receptor-like protein kinase (RLKs) is the main receptor for plant extracellular signals (Shiu et al., 2004; Shiu and Bleecker, 2003). Plant RLKs function in diverse signaling pathways, including the responses to microbial signals in symbiosis and defense (Antolínllovera et al., 2012). Because the cDNA library was constructed from coralloid root tissues, genes involved in plantemicrobe interactions were expected to be expressed. In this study, we focus on RLKs involved in plantemicrobe interactions. Three types of the RLKs highlighted here include the leucine-rich repeat (LRR) type, lysin-motif (LysM) type, and lectin (Lec) domain type. The LRR-RLKs contain a tandemly repeated (9-26) Leu-rich motif, which plays an important role in plant development, defense symbiosis, and other biological processes (Liu et al., 2017). RLKs with lysin-motif (LysM) ectodomains confer recognitional specificity toward N-acetylglucosamine-containing signaling molecules, such as chitin, peptidoglycan (PGN) and rhizobial nodulation factor (NF), which induce immune or symbiotic responses (Antolínllovera et al., 2012). The lectin receptor-like kinases (LecRLKs) possess a characteristic extracellular carbohydratebinding lectin domain. LecRLKs play important roles in plant development and innate immunity (Vaid et al., 2013; Prashant and Laurent, 2013). Using this database, we identified a total of 22 ESTs that encoded putative RLK genes (LRR-RLKs, LysM-RLKs, and LecRLKs), which play a variety of important defensive and symbiotic roles in plant-microbe interaction (Table 5). Further study of these genes will help us to understand the signaling pathways leading to symbiosis and defense.
| Locus | Primer sequence (5'—3') | Repeat m | product | Ta (℃) | GenBank Accession No. |
| Cdb01 | F:CGCCCCATnTAGATCTCTC R:AAACGATGTGAGCCAAAACC |
(TC)6 | 155 | 55 | JZ918061 |
| Cdb02 | F:CAATGCCAACGCTGTGTCTA R:CCCTCAACCTGCAATTTCTC |
(CAT)9 | 222 | 57 | JZ918389 |
| Cdb04 | F:TTGCACCTGCCATTAGTCAA R:TGATCGGTCTCAACAGGTAATG |
(AATA)5 | 196 | 55 | JZ918792 |
| Cdb05 | F:TTGCACCTGCCATTAGTCAA R:TGATCGGTCTCAACAGGTAATG |
(AATA)5 | 196 | 55 | JZ919036 |
| Cdb07 | F:ATCCAAGCTAAAGGGTTCGG R:TGAACTGCTGCTGCTATAAAAA |
(TGA)5 | 141 | 55 | JZ919105 |
| Cdb08 | F:CGACTGATCTCGTCCCAAAT R:AGACATAATCCGCCACGAAG |
(GA)6 | 221 | 57 | JZ920586 |
| Cdb09 | F:AAATCCAAGCCAAAGGGTTC R:CCCCCAACAACAACTGAACT |
(TGA)5 | 157 | 55 | JZ921520 |
| Cdb11 | F:TTGCACCTGCCATTAGTCAA R:TGATCGGTCTCAACAGGTAATG |
(AATA)5 | 196 | 55 | JZ921918 |
| Cdb12 | F:CCTGTACCAGGGACGAAGAA R:CCCTCAACCTGCAATTTCTC |
(CAT)8 | 273 | 57 | JZ922529 |
| Cdb13 | F:CGGACCCTCAATGTGTOTT R:CAGCAGCCAAATGAGCACTA |
(CT)6 | 163 | 57 | JZ920486 |
| Cdb18 | F:ATTGTATATGCAGCAGCCCC R:CAAGACCACGCGTTGAGATA |
(GCA)6 | 265 | 57 | JZ920178 |
| Cdb19 | F:ATTGTATATGCAGCAGCCCC R:CAAGACCACGCGTTGAGATA |
(CCT)7 | 265 | 57 | JZ920178 |
| Cdb33 | F:AAGTTCCGTGCCAACCATAA R:GATCTGCTGCCnCACCTTC |
(ATA)5 | 164 | 55 | JZ918520 |
| Cdb45 | F:TGGATTCATGAGCATTGGAA R:TAATGCAAACAGGGCAATGA |
(CAT)5 | 148 | 53 | JZ920472 |
| Cdb48 | F:AAGCCAAAAAGGGCAAGATT R:CTTCTACTTCGCCCCTCOT |
(CAA)5 | 186 | 53 | JZ921258 |
| Cdb50 | F:TACTTACAGCAGGGGGAAGG R:CACATGACAGAGGTCTAGTGGG |
(TATG)5 | 263 | 59 | JZ921416 |
| Cdb53 | F:TCTGTAGCGAGTTTGGGGTT R:CCGCTAAGATTGCCACATTT |
(TAT)6 | 255 | 55 | JZ921726 |
| Cdb54 | F:TACATCAGGCAATGGCAAAA R:TGCAAACTCCAATAATTCAAGAGA |
(AT)7 | 259 | 53 | JZ922036 |
| Cdb55 | F:CCTCCGAGGAACACAAACAT R:ATATCGCCCTCGCTCCTAAT |
(AAG)7 | 241 | 57 | JZ922127 |
| Cdb56 | F:ATCGGTCTCAACTTGGATGC R:CGTCGTTCTCCCGAGnTTA |
(TC)10 | 261 | 57 | JZ922158 |
| GenBank_Accn | Annotated sequence identifier | Annotation description |
| JZ919236 | sp|C0LGQ51|GSO1_ARATH | LRR receptor-like serine/threonine-protein kinase GSO1 |
| JZ922644 | sp|C0LGP4|Y3475_ARATH | Probable LRR receptor-like serine/threonine-protein kinase At3g47570 |
| JZ920215 | sp|O64780|Y1614_ARATH | G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 |
| JZ920265 | sp|O64780|Y1614_ARATH | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61400 |
| JZ917839 | sp|Q9LFG1|Y3359_ARATH | Putative leucine-rich repeat receptor-like serine/threonine-protein kinase At3g53590 |
| JZ921696 | sp|C0LGS21|Y4361_ARATH | Probable LRR receptor-like serine/threonine-protein kinase At4g36180 |
| JZ920273 | sp|C0LGP4|Y3475_ARATH | Probable LRR receptor-like serine/threonine-protein kinase At3g47570 |
| JZ921196 | sp|C0LGP4|Y3475_ARATH | Probable LRR receptor-like serine/threonine-protein kinase At3g47570 |
| JZ922387 | sp|C0LGH3|Y5614_ARATH | Probable LRR receptor-like serine/threonine-protein kinase At1g56140 |
| JZ917721 | sp|O64825|LYK4_ARATH | LysM domain receptor-like kinase 4 |
| JZ919870\JZ920485\JZ922291 | sp|Q9M2S4|LRKS4_ARATH | L-type lectin-domain containing receptor kinase S.4 |
| JZ918074\JZ921455\JZ918973 \JZ917791\JZ919688\JZ920169 |
sp|Q9LYX1|LRK82_ARATH | L-type lectin-domain containing receptor kinase Ⅷ.2 |
| JZ918026 | sp|O494451LRK72_ARATH | Probable L-type lectin-domain containing receptor kinase Ⅶ.2 |
| JZ919127 | sp|Q9LT96|Y5977_ARATH | Probable leucine-rich repeat receptor-like protein kinase At5g49770 |
| JZ919147 | sp|O22938|Y2182_ARATH | Leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 |
This study is the first to successfully construct high quality cDNA library using RNA derived from coralloid roots of C. debaoensis. We have obtained 2984 unigenes, including 641 sequences with no nucleotide similarity with public databases. These sequences are an important addition to existing databases. We have identified highly expressed genes (mainly stress-responsive genes and antimicrobial genes). A total number of 94 SSR loci were detected, of which 20 loci were successfully amplified in C. debaoensis. These SSR markers can be used for various population and conservation genetics studies of C. debaoensis and other cycads. The cDNA library also provides an excellent resource for discovering genes involved in signaling in symbiosis and defense. A total of 22 ESTs that encoded putative receptor-like protein kinase (RLK) genes were identified.
Author contributionsConceived and designed the experiments: Yunhua Wang, Nan Li, Ting Chen; Performed the experiments: Yunhua Wang, Ting Chen; Analyzed the data: Yunhua Wang, Yiqing Gong; Wrote the paper: Yunhua Wang.
AcknowledgmentsThis work was supported by the Grant (201522) from Shenzhen Urban Management.
Antolínllovera M. Ried M.K., Binder A., Parniske M., 2012. Receptor kinase signaling pathways in plant-microbe interactions. Annu. Rev. Phytopathol, 50(50), 451.
|
||
Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G., 2000. Gene ontology:tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet, 25(1), 25-29.
DOI:10.1038/75556 |
||
Birol I., Raymond A., Jackman S.D., Pleasance S., Coope R., Taylor G.A., Yuen M.M., Keeling C.I., Brand D., Vandervalk B.P., Kirk H., Pandoh P., Moore R.A., Zhao Y., Mungall A.J., Jaquish B., Yanchuk A., Ritland C., Boyle B., Bousquet J., Ritland K., Mackay J., Bohlmann J., Jones S.J., 2013. Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data. Bioinformatics, 29(12), 1492-1497.
DOI:10.1093/bioinformatics/btt178 |
||
Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., Pilbout S., Schneider M., 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res, 31(1), 365-370.
|
||
Brenner E.D., Katari M.S., Stevenson D.W., Rudd S.A., Douglas A.W., Moss W.N., Twigg R.W., Runko S.J., Stellari G.M., McCombie W.R., Coruzzi G.M., 2005. EST analysis in Ginkgo biloba:an assessment of conserved developmental regulators and gymnosperm specific genes. BMC Genom, 6, 143.
DOI:10.1186/1471-2164-6-143 |
||
Brenner E.D., Stevenson D.W., McCombie R.W., Katari M.S., Rudd S.A., Mayer K.F., Palenchar P.M., Runko S.J., Twigg R.W., Dai G., Martienssen R.A., Benfey P.N., Coruzzi G.M., 2003. Expressed sequence tag analysis in Cycas, the most primitive living seed plant. Genome Biol, 4(12), R78.
DOI:10.1186/gb-2003-4-12-r78 |
||
Chen J.R., Zhong Y.C., 1997. A new cycad from China. Acta Phytotaxon. Sin, 35(6), 571.
|
||
Gao Z.F., Barry A.T., 1989. A review of fossil cycad megasporophylls, with new evidence of Crossozamia pomel and its associated leaves from the lower permian of Taiyuan, China. Rev. Palaeobot. Palynol, 60(3), 205-223.
|
||
Hill K.D., Stevenson D.W., Osborne R., 2004. The world list of cycads. Bot. Rev, 70(2), 274-298.
DOI:10.1663/0006-8101(2004)070[0274:TWLOC]2.0.CO;2 |
||
Ju L.P., Kuo C.C., Chao Y.S., Cheng Y.P., Gong X., Chiang Y.C., 2011. Microsatellite primers in the native perennial cycad Cycas taitungensis (Cycadaceae). Am. J. Bot, 98(4), e84-86.
DOI:10.3732/ajb.1000504 |
||
Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y., 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res, 36(Database issue), D480-D484.
|
||
Lindblad P., Costa J.L., 2002. The cyanobacterial:cycad symbiosis. Biol. Environ, 102B(1), 31-33.
|
||
Liu P., Goh C.J., Loh C.S., Pua E.C., 2002. Differential expression and characterization of three metallothionein-like genes in Cavendish banana (Musa acuminata). Physiol. Plantarum, 114(2), 241-250.
DOI:10.1034/j.1399-3054.2002.1140210.x |
||
Liu P.L., Liang D., Yuan H., Gao S.M., Meng Y., 2017. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol, 17(1), 47.
DOI:10.1186/s12862-017-0891-5 |
||
Loconte H., Stevenson D.W., 1990. Cladistics of the Spermatophyta. Brittonia, 42(3), 197-211.
DOI:10.2307/2807216 |
||
Ma, X.Y., Jian, S.G., Wu, M., Liu, N., 2003. The population characters and conservation of cycas debaoensis Y. C. Zhong et C. J. Chen. Guihaia 23 (2), 123-126.
|
||
Mann I.K., Wegrzyn J.L., Rajora O.P., 2013. Generation, functional annotation and comparative analysis of black spruce (Picea mariana) ESTs:an important conifer genomic resource. BMC Genom, 14, 702.
DOI:10.1186/1471-2164-14-702 |
||
Martínez L.C.A. Artabe A.E.E., Bodnar J., 2012. A new cycad stem from the Cretaceous in Argentina and its phylogenetic relationships with other Cycadales. Trends Ecol. Evol, 170(3), 436-458.
|
||
Nixon K.C., Crepet W.L., Stevenson D., Friis E.M., 1994. A reevaluation of seed plant phylogeny. Ann. Mo. Bot. Gard, 81(3), 484-533.
DOI:10.2307/2399901 |
||
Norstog K., Nicholls T.J., 1997. The Biology of the Cycads. Cornell University Press, New York. |
||
Powell W., Machray G.C., Provan J., 1996. Polymorphism revealed by simple sequence repeats. Trends Plant Sci, 1(7), 215-222.
DOI:10.1016/S1360-1385(96)86898-0 |
||
Prashant S., Laurent Z., 2013. Lectin receptor kinases in plant innate immunity. Front. Plant Sci, 4(4), 124.
|
||
Rai A.N., Söderbäck E. Bergman B., 2000. Cyanobacterium-plant symbioses. New Phytol, 147(3), 449-481.
DOI:10.1046/j.1469-8137.2000.00720.x |
||
Ranade S.S., Lin Y.C., Zuccolo A., Van de Peer Y., Garcia-Gil Mdel R., 2014. Comparative in silico analysis of EST-SSRs in angiosperm and gymnosperm tree genera. BMC Plant Biol, 14, 220.
DOI:10.1186/s12870-014-0220-8 |
||
Rungis D., Berube Y., Zhang J., Ralph S., Ritland C.E., Ellis B.E., Douglas C., Bohlmann J., Ritland K., 2004. Robust simple sequence repeat markers for spruce (Picea spp.) from expressed sequence tags. Theor. Appl. Genet, 109(6), 1283-1294.
DOI:10.1007/s00122-004-1742-5 |
||
Shiu S.H., Bleecker A.B., 2003. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol, 132(2), 530.
DOI:10.1104/pp.103.021964 |
||
Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H., 2004. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell, 16(5), 1220-1234.
DOI:10.1105/tpc.020834 |
||
Soares M.B., Bonaldo M.F., Jelene P., Su L., Lawton L., Efstratiadis A., 1994. Construction and characterization of a normalized cDNA library. Proc Natl Acad Sci USA, 91(20), 9228-9232.
DOI:10.1073/pnas.91.20.9228 |
||
Soltis D.E., Soltis P.S., Zanis M.J., 2002. Phylogeny of seed plants based on evidence from eight genes. Am. J. Bot, 89(10), 1670-1681.
DOI:10.3732/ajb.89.10.1670 |
||
Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V., 2000. The COG database:a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res, 28(1), 33-36.
DOI:10.1093/nar/28.1.33 |
||
Treutlein J., Wink M., 2002. Molecular phylogeny of cycads inferred from rbcL sequences. Naturwissenschaften, 89(5), 221-225.
DOI:10.1007/s00114-002-0308-0 |
||
Vaid N., Macovei A., Tuteja N., 2013. Knights in action:lectin receptor-like kinases in plant development and stress responses. Mol. Plant, 6(5), 1405-1418.
DOI:10.1093/mp/sst033 |
||
Vessey J.K., Pawlowski K., Bergman B., 2005. Root-based N2-fixing symbioses:Legumes, Actinorhizal plants, Parasponia sp. and cycads. Plant Soil, 274(1), 51-78.
|
||
Victoria F.C., da Maia L.C., de Oliveira A.C., 2011. In silico comparative analysis of SSR markers in plants. BMC Plant Biol, 11, 15.
DOI:10.1186/1471-2229-11-15 |
||
von Stackelberg M., Rensing S.A., Reski R., 2006. Identification of genic moss SSR markers and a comparative analysis of twenty-four algal and plant gene indices reveal species-specific rather than group-specific characteristics of microsatellites. BMC Plant Biol, 6, 9.
DOI:10.1186/1471-2229-6-9 |
||
Yang Y., Li Y., Li L.F., Ge X.J., Gong X., 2008. Isolation and characterization of microsatellite markers for Cycas debaoensis Y. C. Zhong et C. J. Chen (Cycadaceae). Mol. Ecol. Resour, 8(4), 913-915.
|
||
Zhou G.K., Xu Y.F., Liu J.Y., 2005. Characterization of a rice class Ⅱ metallothionein gene:tissue expression patterns and induction in response to abiotic factors. J. Plant Physiol, 162(6), 686-696.
DOI:10.1016/j.jplph.2004.11.006 |
||
Zonneveld B.J.M., 2012. Genome sizes for all genera of Cycadales. Plant Biol, 14(1), 253-256.
|



