b. University of Chinese Academy of Sciences, Beijing, 100049, China;
c. Food Crops Research Institute, Yunnan Academy of Agricultural Sciences(YAAS), Kunming, 650200, China
Rice (Oryza sativa) is one of the most important cereal crops and is a staple food in many countries worldwide. With the rapid increase in the global population and the emergence of a food crisis, increasing rice yields has become the focus of rice research and breeding programs (Khush, 2005). Plant height is an important rice trait that directly affects the yield and lodging resistance. Dwarf plants show enhanced lodging resistance, but an excessively dwarfed plant leads to inadequate growth and a tendency for densely overlapping leaves, thus reducing rice yield. However, extremely tall plants are susceptible to lodging which also reduces crop yield and quality (Zhang et a., 2017b). The 'Green Revolution' and the breeding of hybrid rice cultivars have significantly improved rice yield by utilization of semi-dwarf gene resources (Hedden, 2003; Khush, 1999). Therefore, the discovery and utilization of semi-dwarf genes are of considerable importance for the improvement of rice yield.
In recent years, many genes the modulate plant height, including more than 60 recessive dwarf genes and 10 recessive semi-dwarf genes, have been identified or cloned (Zhang et a., 2014). DWARF 18 (D18) is a dwarf gene that encodes OsGA3ox2, which controls rice plant height by participating in gibberellins (GA) biosynthesis (Tong et a., 2014; Iwamoto et a., 2011; Itoh et a., 2001). The dwarf gene gibberellin insensitive dwarf 1 (gid1) encodes a soluble gibberellin receptor that affects plant height by mediating GA signaling transduction in rice (Ayano et a., 2014; Tanaka et a., 2006; Ueguchi-Tanaka et a., 2005). Brassinosteroid-deficient dwarf 1 (brd1) encodes a key enzyme involved in rice brassinolide (BR) biosynthesis, and the mutant for which is characterized by a severely dwarfed stem, curved and deformed leaves, crown root, small ear shape and grain size (Mori et a., 2002; Hong et a., 2002). The gene D61, which encodes a rice BR receptor kinase involved in BR signaling transduction, reduces plant height (Zhang et a., 2016; Nakamura et a., 2006; Yamamuro et a., 2000). DWARF KAMIKAWABUNWAI TILLERING (HTD2, also known as D88 or D14), a component of the strigolactones (SLs) signaling pathway, encodes an esterase that is involved in inhibition of branching and negatively regulates the number of tillers (Kagiyama et a., 2013; Liu et a., 2009). WEALTHY FARMER'S PANICLE (WFP, also known as IPA1) encodes a transcription factor that can directly bind to the promoter of the negative regulator FINE CULM 1 (FC1), which controls the growth of tiller lateral buds and thus inhibits tillering. WFP also affects plant height and panicle length by directly regulating the important gene DENSE AND ERECT PANICLE 1 (DEP1), thereby determining the spike type (Zhang et a., 2017a; Lu et a., 2013; Miura et a., 2010).
In contrast to recessive genes, few dominant dwarf genes have been reported. In rice, only a few dominant or semi-dominant dwarf genes, including D53, Ssi1, Sdd(t), Dx, TID1, LB4D, Slr-d, D-h, SBI, Dd7, Sdt97, have been reported (Liu et a., 2008, 2018; Zhao et a., 2018b; Piao et a., 2014; Liang et a., 2011; Asano et a., 2009; Miura et a., 2009; Qin et a., 2008; Tong et a., 2007; Wei et a., 2006; Sunohara et a., 2003). However, dominant dwarf genes have special advantages in crop breeding (Qin et a., 2008). For instance, unless both parents carry the same recessive semidwarf gene, recessive genes cannot be used in hybrid rice cultivars. Dominant dwarf alleles, in contrast, can be used when only one parent carries the dominant allele. Thus, the germplasm of the other parent can be widely chosen, sharply reducing time, cost, and labor in breeding work (Qin et a., 2008; Liang et a., 2004). Therefore, it is important to discover and utilize novel dwarf genes, especially dominant genes for breeding dwarf varieties.
The dwarf rice germplasms widely used in commercial production in China include 'Aijiaonante', 'Aizaizhan', 'Dee-geo-woogen', which genetic analysis has shown all carry the recessive gene semi-dwarf 1 (sd1) (Chen et a., 2013). The frequent use of limited dwarfism sources may disadvantage the diversification of rice varieties, hinder the genetic improvement process, and cause a 'bottleneck' effect in the genetic background available for developing new rice varieties (Matsuo et a., 1997; Luh, 1980; Hargrove and Cabanilla, 1979). Thus, more useful dwarfism sources are required for rice breeding.
In the present study, we found a unique upland landrace 'Kaowenghan' (KWH) from Yunnan province, which unlike other upland rice landraces shows a semi-dwarf phenotype. Genetic analysis indicated that the semi-dwarf phenotype is semidominant. To identify a potentially novel dominant dwarf gene for rice breeding, we crossed KWH donor male parents with DJY1 to generate homozygous semi-dwarf IL-87 and a mapping population. QTL mapping and breeding value evaluation indicate that we identified a novel semi-dwarf gene which may play an important role in the study of the genetic mechanisms of dominant dwarf and breeding utilization in rice.
1. Materials and methods 1.1. Plant materialsDJY1 is a japonica rice cultivar, and KWH is a unique semi-dwarf landrace of japonica upland rice; both are grown in Yunnan province, China. Progeny of the F1 were obtained by crossing DJY1 with KWH; the F1 individuals were consecutively backcrossed to DJY1 as male and recurrent parent until BC2F1; semi-dwarf IL-87 were developed from the BC2F4 population. The BC3F2 population (2015H3E483), which comprised 68 plants, were developed for genetic analysis and preliminary QTL mapping by crossing DJY1 (male parent) and the semi-dwarf IL-87 (female parent). The BC3F3 population (2017J1E87), including 396 individuals developed from the heterozygous BC3F2 plants, were planted for further QTL mapping. To confirm the effect of QTL, we also developed QTL homozygous NIL(NIL-2) from the BC3F3 populations for phenotypic assessment.
For breeding value evaluation, F1 progeny were produced by crossing the NIL-2 with 20 core rice cultivars obtained from the International Rice Research Institute (IRRI). These cultivars are genetically diverse and actively used in international breeding programs on account of their wide range of agronomic attributes, and belong to three genetic background groups: Japonica, Aus and Indica (McNally et a., 2009) (Table S1). The plant height of F1 plants was investigated.
All materials were grown in a paddy field at an experimental station at the Xishuangbanna Tropical Botanical Garden, located in Jinghong, Yunnan Province, China. All F1 and parents were sequentially planted in three rows for each material. DJY1, KWH, IL-87 and NIL-2 were planted in a randomized complete block design with three repeats, each block contained 4 rows with 10 plants per row. All the material was planted within a 27 cm × 15 cm plant space. Agronomic traits were examined for eight randomly selected plants from the center of the block. Field management, including irrigation, fertilization, and pest control followed standard agricultural practices.
1.2. Phenotypic measurementsPlant height (cm) was measured from the ground to the tip of the tallest panicle (awns excluded) at full maturity (Würschum et a., 2015). Panicle length (cm) was measured from the base of the panicle to the tip of panicle (Zhao et a., 2018a). The main tiller of each plant was selected to measure the internode length (cm); n1 was measured from the base of the panicle to the first node below; n2 was measured from the first to the second node, and so on (Wang et a., 2017). All F1, parents, and individuals of mapping populations were measured for plant height; DJY1 and IL-87 were measured for panicle length and internode length. The yieldrelated traits, including tiller number, effective tiller number, number of primary branches, number of secondary branches, grain number per panicle, setting percentage and 1000-grain weight (g), were all investigated in DJY1 and NIL-2 at the mature stage. Fully filled grains were chosen randomly from each plant for measurement of 1000-grain weight.
1.3. Molecular markers and linkage mapGenomic DNA was extracted from fresh leaves of individual plants and the parents following the method of Edwards et a. (1991). Simple sequence repeat (SSR) markers were selected from the published molecular map of rice (McCouch et a., 2002). The SSR analysis was conducted in accordance with the method of Wu and Tanksley (1993). The genetic linkage map was constructed using QTL IciMapping version 3.2 software (Institute of Crop Science Chinese Academy of Agricultural Sciences, CAAS) with a minimum LOD score of 2.5.
1.4. Data analysisSPSS version 18 in IBM was used to analyze data. The frequency distribution of plant height in the mapping populations was analyzed. The LSD multiple comparative analysis and the Student's t-test were used to analyze those data. The threshold of α = 0.05 was considered to be statistically significant, and values of P < 0.01 were considered to be highly significant. The QTL controlling plant height was identified by interval mapping analysis using the QTL IciMapping software package.
2. Results 2.1. Phenotypic characteristics of IL-87To identify phenotypic characteristics of semi-dwarf IL-87, KWH, and DJY1, plant height was measured at the maturity stage (Fig. 1A). There was a significant reduction in plant height of IL-87 (71.97 cm) compared to DJY1 (103.09 cm); specifically, IL-87 plants were 70% of the height of DJY1 plants (Fig. 1A and C). To further determine the changes in plant heights of IL-87, internode length and panicle length of IL-87 and DJY1 plants were measured at maturity. Internode length in IL-87 was significantly shorter than in DJY1 (Fig. 1B and E). In IL-87 plants the average length of each successive internode from the tip to the ground level 18.67, 10.43, 6.24, and 2.05 cm respectively. In contrast, in DJY1 plants, the successive internode lengths were 33.15, 17.65, 11.39, and 4.74 respectively. Average panicle length of IL-87 and DJY1 plants were 18.39 cm and 20.85 cm respectively (Fig. 1B and E). These results revealed that IL-87 has a semi-dwarf phenotype.
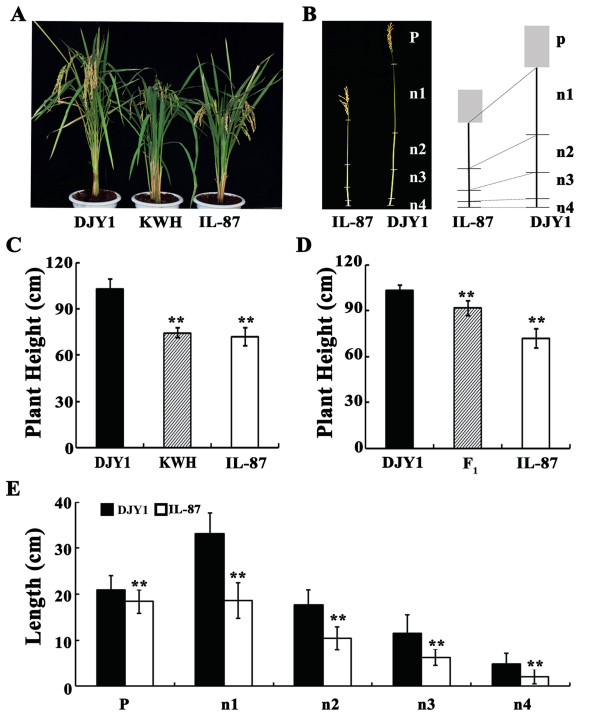
|
| Fig. 1 Phenotypic characteristics of IL-87 and F1 progeny (A) Representative whole plant photos of DJY1, KWH, and IL-87 after panicle heading. (B) Comparison of the individual regions of the main stems in DJY1 and IL-87, P, panicle, n, internode. (C) Plant height of DJY1, KWH, and IL-87, as determined by measuring the main tillers. (D) Plant height of DJY1, IL-87, and F1, as determined by measuring the main tillers. (E) Length of the panicle and internodes, as determined using the main stems of DJY1 and IL-87, P panicle, n internode. n = 3, ** indicates extremely significant difference compared with DJY1 (P < 0.01); * indicates significant differences compared with DJY1 (P < 0.05). |
DJY1 was crossed with IL-87 to analyze the genetic model of the semi-dwarf phenotype. The plant height of F1 progeny was significantly shorter than that of DJY1 but was taller than IL-87 (Fig. 1D), showing an intermediate plant height. These results indicated that the semi-dwarf trait was a semi-dominant trait. The frequency distribution of plant height of the BC3F2 population (2015H3E483) showed a continuous and trimodal distribution similar to that of the two parents and the intermediate types (Fig. 2A); the BC3F3 population (2017J1E87) also showed a similar distribution (Fig. 2B). These observations suggested that the semi-dwarf trait was controlled by a major QTL.
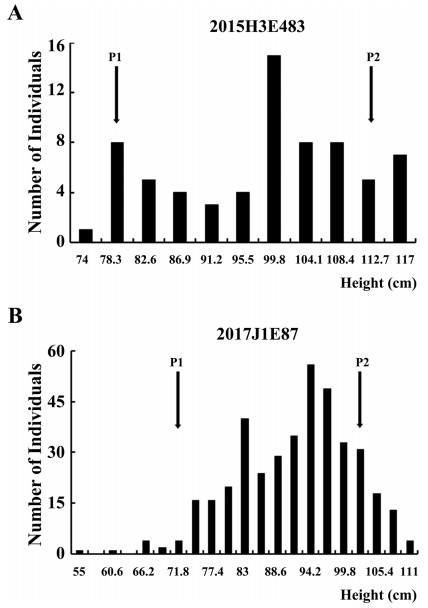
|
| Fig. 2 Frequency distribution of plant height in the mapping populations (A) Distribution of plant height in the BC3F2 population (2015H3E483). (B) Distribution of plant height in the BC3F3 population (2017J1E87). P1 represents the male parent (KWH), P2 represents the female parent (DJY1). |
A total of 452 SSR markers, relatively uniformly distributed throughout the rice genomes, were used to assess polymorphism between IL-87 and DJY1. Fifteen SSR markers were polymorphic and were distributed on chromosomes 1, 4, 10, and 11 in IL-87, which was indicative of genomic introgression from KWH. The genotype of 68 individuals from the BC3F2 population (2015H3E483) was investigated using the 15 polymorphic SSR markers. QTL analysis showed that a QTL between RM486 and RM12057 on chromosome 1 was detected, and explained 64.39% of the phenotypic variance in plant height, and the QTL was designated qDH1 (Fig. 3A; Table 1; Table S2).

|
| Fig. 3 Mapping of QTL for the semi-dwarf trait (A) The results of QTL mapping based on the BC3F2 population (2015H3E483). (B) The results of QTL mapping based on the BC3F3 population (2017J1E87). (C) Plant height of DJY1, NIL-2, and IL-87, as determined by measuring the main tillers. n= 3, ** indicates extremely significant difference compared with DJY1 (P < 0.01). |
| Populati | QTL | Flanking | Trait | LOD | Variance explained (%) | Additive effect |
| 2015H3E4 | qDH1 | RM486-RM | height | 11.9117 | 64.3906 | 10.4982 |
| 2017J1E87 | qDH1 | RM6696-R | height | 34.1495 | 35.7837 | 6.3784 |
To further localize the target QTL, a larger BC3F3 population (2017J1E87) containing 396 individuals derived from the original BC3F2 population (2015H3E483) was constructed by three heterozygous recombinant plants between RM486 and RM12057. Sixtytwo SSR markers were designed in the interval between RM486 and RM12057. Among them, 5 markers were polymorphic between DJY1 and IL-87 and used for QTL analysis of 396 individuals in the BC3F3 population (2017J1E87). The QTL qDH1 was further restricted to a 5.2 cM region flanked by RM6696 and RM12047 (Fig. 3B; Table S2).
To confirm that qDH1 was the major QTL controlling the semidwarf phenotype, we used the BC3F3 mapping population to develop a NIL-2 line that was homozygous for qDH1 at the chromosomal segment between RM6696 and RM12047. The plant heights of NIL-2 and DJY1 were assessed at maturity. The plant height of NIL-2 was significantly reduced compared with that of DJY1, with no significant difference detected between NIL-2 and IL-87 (Fig. 3C). These results illustrate that qDH1 is the major QTL that controlled plant height.
2.4. Yield of DJY1 and NIL-2To investigate whether the reduction in plant height had a side effect on yield, several yield-related traits were investigated between DJY1 and NIL-2 at the maturity stage. The number of grains per panicle and secondary branches of NIL-2 (57.9 and 6.10 respectively) were significantly reduced compared with those of DJY1 (83.7 and 12.04 respectively). The average 1000-grain weight of the NIL-2 (25.61 g) was also significantly lower than that of DJY1 (30.52 g). However, the number of tillers and effective tillers were significantly increased in the NIL-2 (15.2 and 13.2, respectively) compared with those in DJY1 (9.2 and 8.6, respectively). No significant differences in the number of primary branches and setting percentage were detected. The yield of a single plant of NIL-2 (15.62 g) was less than that of DJY1 (20.13 g), with no statistically significant difference observed (Table 2). These results suggest that the negative effects of qDH1 on rice yield are negligible.
| Traits | DJY1 | NIL-2 | P-value |
| Number of gain per panicle | 83.70±19.06 | 57.90±9.11* | 0.0211 |
| Setting percentage | 79.44%±8.91% | 81.01%±8.29% | 0.7788 |
| Number of tillers | 9.20±1.48 | 15.4.66* | 0.0252 |
| Number of effective tillers | 8.60±1.67 | 13.20±2.58* | 0.0102 |
| 1000-grain weight (g) | 30.52±1.72 | 25.61±0.46** | 0.0001 |
| Number of primary branches | 8.57±1.22 | 7.43±0.54 | 0.0858 |
| Number of secondary branches | 12.04±4.02 | 6.10±1.97* | 0.0141 |
| Theoretical yield of single plant (g) | 20.13±3.49 | 15.62±3.33 | 0.0701 |
| N = 3, **P < 0.01 by Student's t-test; *P < 0.05 by Student's t-test. | |||
To further estimate the effect of qDH1 on rice height, NIL-2 was crossed with 20 core cultivars obtained from the IRRI, and 18 F1 progenies were raised (two crosses failed). All F1 progeny were planted under the same conditions in our experimental station and phenotyped for plant height. Among the 18 crosses, the plant height of F1 progeny in nine crosses decreased, whereas the plant height of the F1 progeny in the remaining nine cross groups was unchanged. No significant difference in plant height was observed between the F1 progenies and the parent 'IR64-21', 'M 202', 'Minghui 63' (MH63), 'N 22', 'Nipponbare' (Npb), 'Sadu-Cho' (Sadu), 'Shan-Huang-Zhan-2' (SHZ2), 'Zhenshan 97B' (ZS97B) and 'Cypress' (Cyp) (Fig. 4A and C). Plant height of F1 progeny was highly significantly reduced compared with that of the parent 'Azucena' (Azu), 'Dom-Sufid' (Doms), 'FR13 A', 'Li-Jiang-Xin-Tuan-Hei-Gu' (LTH), and 'Pokkali's' (Pok), by about 30%, 20%, 18%, 33%, and 37%, respectively (Fig. 4A and B; Table S3). The F1 progenies of 'Dular', 'Moroberekan' (Moro), 'Tainung 67' (TNG67), and 'Aswina' (Asw) were significantly shorter than the parent by 27%, 10%, 11%, and 21%, respectively (Fig. 4A and B; Table S3). Among them, Azu, Moro, LTH, TNG67 and Doms belonged to the Japonica group, 'FR13 A' and 'Dular' belonged to the Aus group, Asw and Pok were attributed to Indica group (Table S1). These results indicate that homozygous NIL-2 can potentially reduce plant height in different types of rice cultivars from different genetic background.
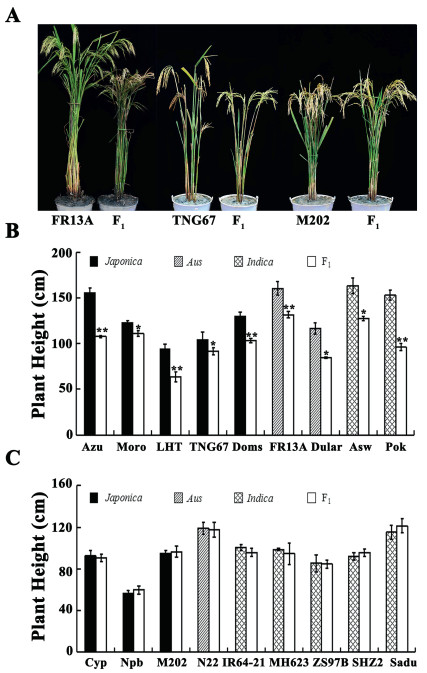
|
| Fig. 4 Evaluation of breeding value (A) Representative plant height phenotype. (B) (C) Height analysis of each parent and F1 generation. n = 3, ** indicates extremely significant differences between F1 and Parents (P < 0.01); * indicates significant differences between F1 and parent (P < 0.05). |
Plant height is an important target of crop breeding and is positively correlated with yield within a certain range. A suitable plant height is not only beneficial for the ideal plant type, but also plays an important role in improving the yield potential of rice and the photosynthetic efficiency of the upper leaves (Luo et a., 2012). Since the use of the sd1 gene in the 'Green Revolution' of rice, an increasing number of plant height genes have been identified and cloned. However, many of them cannot be used for breeding due to unwanted phenotypes such as extreme dwarfism, sterility, and short grain (Sakamoto et a., 2004). Dominant dwarf genes are of great value and widely used in crop breeding (Liang et a., 2011). The 'Green Revolution' genes of wheat, Rht1 and Rht2, are dominant dwarf genes, have been successfully used in wheat breeding (Kurkiev et a., 2007; Gale et a., 1985). In this study, we used the japonica cultivar DJY1 and a KWH donor to develop the nearisogenic line NIL-2, which has the semi-dominant dwarf QTL qDH1. The semi-dominant semi-dwarf QTL qDH1 was localized between the markers RM6696 and RM12047 on chromosome 1 and we found that qDH1 reduces rice plant height with few side effects on yield.
In recent decades, a large number of plant height QTLs have been detected in the rice genome. Bao et a. (2009) used populations derived from residual heterozygous lines to detect QTLs for plant height in rice, and localized the QTL qph6-1 on chromosome 6 to a physical distance of about 51.7 KB. The QTL ph1 was identified on chromosome 1 of rice and cloned, the gene was reported highly correlated with plant height (Ishimaru et a., 2004). HEADING DATE 7 (GHD7), which controls the number of single panicles, plant height, and the timing of heading of rice, has been cloned. The gene encodes a CCT domain-containing protein and is mainly regulated by photoperiod (Xue et a., 2008). In the present study, we used different generations of the same genetic population to narrow down the QTL locus qDH1 for plant height between the markers RM6696 and RM12047 on chromosome 1. These results provide a sound foundation for further fine mapping, cloning, and functional analysis of the locus.
The use of dwarf genes is important for the breeding of hybrid rice. Although breeders are constantly seeking to create novel dwarf germplasm by a variety of methods, few dwarf genes have been exploited for rice breeding. For example, the dwarf germplasm of japonica, which is mainly derived from 'Nonken 58' and 'Balil1a', is rarely used for breeding because of poor economic characteristics (Chen et a., 2013). We found that the major QTL qDH1 induces reduction of plant height in DJY1, but has no negative effects on yield (Fig. 1B; Table 2). Hybridization of NIL-2 with 20 core rice cultivars belonging to three genetic background groups (Table S1), and assessment of the plant height of the F1 progeny showed that qDH1 could reduce the plant height of nine cultivars from different genetic background. Because plant height is a phenotype controlled by multiple genes (Ni et a., 2009; Jaiswal et a., 2002), the interactions between these genes determine plant height differences (Wade, 2001). This may explain why plant height was affected in only half the cultivars examined.
The well-characterized semi-dwarf gene sd1 was also located between the markers RM6696 and RM12047 on chromosome 1. sd1 is a recessive gene located on the long arm of chromosome 1 and reduces rice plant height with few side effects on yield (Hedden, 2003). The QTL qDH1 also presents a similar phenotype. Thus, we speculate that qDH1 might be the dominant allele of sd1. However, in our study, the QTL qDH1 was semi-dominant. If QTL qDH1 is an allele of sd1, it will be important for studying the molecular mechanisms by which different alleles change from recessive to dominant.
AcknowledgementsThis research was partially funded by grants from National Natural Science Foundation of China (31360330) and Chinese Academy of Science (XDA08020203). We thank Robert McKenzie, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Appendix A. Supplementary dataSupplementary data related to this article can be found at https://doi.org/10.1016/j.pld.2018.09.001.
Asano K., Hirano K., Ueguchi-Tanaka M., et al., 2009. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol. Genet. Genom, 281, 223-231.
DOI:10.1007/s00438-008-0406-6 |
||
Ayano M., Kani T., Kojima M., et al., 2014. Gibberellin biosynthesis and signal transduction is essential for internode elongation in deepwater rice. Plant Cell Environ, 37, 2313-2324.
|
||
Bao Q.J., Fan Y.Y., Yu W.D., 2009. Fine mapping of QTL qPH6-1 for plant height on the short arm of rice chromosome 6. Chin. J. Rice Sci, 23, 470-474.
|
||
Chen F., Lin H.Z., Zhou Q.X., et al., 2013. Research progress of dwarf genes in rice. Shandong Agric. Sci, 45, 127-133.
|
||
Edwards K., Johnstone C., Thompson C., 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res, 19, 1349.
DOI:10.1093/nar/19.6.1349 |
||
Gale M.D., Youssefian S., Russell G.E., 1985. Dwarfing Genes in Wheat. Elsevier Inc, London.. Elsevier Inc, London. |
||
Hargrove T.R., Cabanilla V.L., 1979. The impact of semidwarf varieties on Asian ricebreeding programs. Bioscience, 29, 731-735.
DOI:10.2307/1307667 |
||
Hedden P., 2003. The genes of the green revolution. Trends Genet, 09, 5-9.
|
||
Hong Z., Ueguchi-Tanaka M., Shimizu-Sato S., et al., 2002. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J, 32, 495-508.
DOI:10.1046/j.1365-313X.2002.01438.x |
||
Ishimaru K., Ono K., Kashiwagi T., 2004. Identification of a new gene controlling plant height in rice using the candidate-gene strategy. Planta, 218, 388-395.
DOI:10.1007/s00425-003-1119-z |
||
Itoh H., Ueguchi-Tanaka M., Sentoku N., 2001. Cloning and functional analysis of two gibberellin 3b-hydroxylase genes that are differently expressed during the growth of rice. P.N.A.S, 98, 09-14.
|
||
Iwamoto M., Kiyota S., Hanada A., et al., 2011. The multiple contributions of phytochromes to the control of internode elongation in rice. Plant Physiol, 157, 1187-1195.
DOI:10.1104/pp.111.184861 |
||
Jaiswal P., Ware D., Ni J., et al., 2002. Gramene:development and integration of trait and gene ontologies for rice. Comp. Funct. Genom, 3, 132-136.
DOI:10.1002/cfg.156 |
||
Kagiyama M., Hirano Y., Mori T., et al., 2013. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Gene Cell, 18, 147-160.
DOI:10.1111/gtc.2013.18.issue-2 |
||
Khush G.S., 1999. Green revolution:preparing for the 21st century. Genome, 42, 646-655.
DOI:10.1139/g99-044 |
||
Khush G.S., 2005. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol, 59, 1-6.
|
||
Kurkiev K.U., 2007. Introgression of the reduced height gene Rht10 from the wheat cultivar Ai-bian 1 into the triticale genotyp. Genetika, 43, 1269-1272.
|
||
Liang F., Xin X.Y., Hu Z.J., et al., 2011. Genetic analysis and fine mapping of a novel semidominant dwarfing gene LB4D in rice. J. Integr. Plant Biol, 53, 312-323.
DOI:10.1111/j.1744-7909.2011.01031.x |
||
Liang G.H., Cao X.Y., Sui J.M., et al., 2004. Fine mapping of a semidwarf gene sd-g in indica rice (Oryza sativa L.). Chin. Sci. Bull, 49, 900-904.
|
||
Liu B.M., Wu Y.J., Fu X.D., et al., 2008. Characterizations and molecular mapping of a novel dominant semi-dwarf gene Sdd (t) in rice (Oryza sativa). Plant Breed, 127, 125-130.
DOI:10.1111/j.1439-0523.2007.01444.x |
||
Liu C., Zheng S., Gui J.S., et al., 2018. Shortened basal internodes encodes a gibberellin 2-oxidase and contributes to lodging resistance in rice. Mol. Plant, 11, 288-299.
DOI:10.1016/j.molp.2017.12.004 |
||
Liu W.Z., Wu C., Fu Y.P., et al., 2009. Identification and characterization of HTD2:a novel gene negatively regulating tiller bud outgrowth in rice. Planta, 230, 649-658.
DOI:10.1007/s00425-009-0975-6 |
||
Lu Z.F., Yu H., Xiong G.S., et al., 2013. Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell, 25, 3743-3759.
DOI:10.1105/tpc.113.113639 |
||
Luh, B.S., 1980. Rice:Production and Utilization. AVI Publishing Co., Inc, Westport, CT.
|
||
Luo J., Shao G.N., Wei X.J., et al., 2012. Genetic analysis of a QTL qPH3 for plant height in rice. Chin. J. Rice Sci, 26, 417-442.
|
||
Matsuo, T., Futsuhara, Y., Kikuchi, F., et al., 1997. Science of the Rice Plant. 3. Genetics. Supplementary Volume: Indices. Food and Agriculture Policy Research Center, Tokyo.
|
||
McCouch S.R., Teytelman L., Xu Y., et al., 2002. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.) (supplement). DNA Res, 09, 199-207.
|
||
McNally K.L., Childs K.L., Bohnert R., et al., 2009. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. P.N.A.S. USA, 106, 12273-12278.
DOI:10.1073/pnas.0900992106 |
||
Miura K., Wu J., SunoharaU H., et al., 2009. High-resolution mapping revealed a 1. 3-Mbp genomic inversion in Ssi1, a dominant semidwarf gene in rice (Oryza sativa). Plant Breed, 128, 63-69.
DOI:10.1111/pbr.2009.128.issue-1 |
||
Miura K., Ikeda M., Matsubara A., et al., 2010. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet, 42, 545-549.
DOI:10.1038/ng.592 |
||
Mori M., Nomura T., Ooka H., et al., 2002. Isolation and characterization of a rice dwarf mutant with a defect in brassinosteroid biosynthesis. Plant Physiol, 130, 1152-1161.
DOI:10.1104/pp.007179 |
||
Nakamura A., Fujioka S., Sunohara H., et al., 2006. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol, 140, 580-590.
DOI:10.1104/pp.105.072330 |
||
Ni, J.J., Pujar, A., Youens-Clark, K., et al., 2009. Gramene QTL database: development, content and applications. Database J. Biol. Databases. Curation 1, bap005.
|
||
Piao R.H., Chu S.H., Jiang W.Z., et al., 2014. Isolation and characterization of a dominant dwarf gene, D-h, in rice. PLoS One 09, e86210.
|
||
Qin R.Z., Qiu Y., Cheng Z.J., et al., 2008. Genetic analysis of a novel dominant rice dwarf mutant 986083D. Euphytica, 160, 379-387.
DOI:10.1007/s10681-007-9548-6 |
||
Sakamoto T., Miura K., Itoh H., et al., 2004. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol, 135, 1642-1653.
DOI:10.1104/pp.103.028308 |
||
Sunohara H., Kitano H., 2003. New dwarf mutant controlled by a dominant gene, Twisted dwarf1. Rice Genet. Newsl, 20, 20-21.
|
||
Tanaka N., Matsuoka M., Kitano H., et al., 2006. Gid1, a gibberellin-insensitive dwarf mutant, shows altered regulation of probenazole-inducible protein(PBZ1) in response to cold stress and pathogen attack Plant. Plant Cell Environ, 29, 619-631.
DOI:10.1111/pce.2006.29.issue-4 |
||
Tong H.N., Xiao Y.H., Liu D.P., et al., 2014. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell, 26, 4376-4393.
DOI:10.1105/tpc.114.132092 |
||
Tong J.P., Liu X.J., Zhang S.Y., et al., 2007. Identification, genetic characterization, GA response and molecular mapping of Sdt97:a dominant mutant gene conferring semi-dwarfism in rice (Oryza sativa L.). Genet. Res, 89, 221-230.
|
||
Ueguchi-Tanaka M., Ashikari M., Nakajima M., et al., 2005. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature, 437, 693-698.
DOI:10.1038/nature04028 |
||
Wade M.J., 2001. Epistasis, complex traits, and mapping genes. Genetica, 112-113, 59-69.
DOI:10.1023/A:1013316611768 |
||
Wang J.G., Gang S., Yang L., et al., 2017. Markers associated with culm length and elongated internode length in, japonica, rice. Crop Sci, 57, 2329-2344.
DOI:10.2135/cropsci2016.06.0533 |
||
Wei L.R., Xu J.C., Li X.B., et al., 2006. Genetic analysis and mapping of the dominant dwarfing gene D-53 in rice. J. Integr. Plant Biol, 48, 447-452.
DOI:10.1111/jipb.2006.48.issue-4 |
||
Wu K.S., Tanksley S.D., 1993. Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol. Gen. Genet, 241, 225-235.
|
||
Würschum T. Langer S.M., Longin C.F.H., 2015. Genetic control of plant height in European winter wheat cultivars. Theor. Appl. Genet, 128, 865.
DOI:10.1007/s00122-015-2476-2 |
||
Xue W.Y., Xing Y.Z., Weng X.Y., et al., 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet, 40, 761-767.
DOI:10.1038/ng.143 |
||
Yamamuro C., Ihara Y., Wu X., et al., 2000. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell, 12, 1591-1606.
DOI:10.1105/tpc.12.9.1591 |
||
Zhang B.W., Wang X.L., Zhao Z.Y., et al., 2016. OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol, 170, 1149-1161.
DOI:10.1104/pp.15.01668 |
||
Zhang, J., Liu, X.Q., Li, S.Y., et al., 2014. The rice semi-dwarf mutant sd37, caused by a mutation in CYP96B4, plays an important role in the fine-tuning of plant growth. PLoS One 09, e88068.
|
||
Zhang, L., Yu, H., Ma, B., et al., 2017a. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 08, 14789.
|
||
Zhang, Y.X., Yu, C.S., Lin, J.Z., et al., 2017b. OsMPH1 regulates plant height and improves grain yield in rice. PLoS One 12, e0180825.
|
||
Zhao, C.F., Zhao, Q.Y., Zhao, L., et al., 2018a. Characterization and fine mapping of qPE12, a new locus controlling rice panicle exsertion. Euphytica 214, 47.
|
||
Zhao Z.G., Zhang C., Liu X., et al., 2018b. Genetic analysis and fine mapping of a dominant dwarfness gene from wild rice (Oryza barthii). Plant Breed, 137, 50-59.
DOI:10.1111/pbr.2018.137.issue-1 |



