b. Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, 650201, China;
c. University of Chinese Academy of Sciences, Beijing, 100049, China
Burmanniaceae are currently placed in the monocot order Dioscoreales, which molecular data indicates consists of Nartheciaceae, Burmanniaceae, and Dioscoreaceae (Merckx et al., 2006). However, the phylogenetic relationships between these families remains poorly understood. Burmannia is the largest genus within the family Burmanniacea, and consists of about 60 herbaceous species which use a range of trophic strategies, from autotrophy, hemi-mycohetertrophy to holo-mycoheterotrophy (Jonker, 1938; Merckx et al., 2006; Bolin et al., 2017). The genus is mainly distributed in pantropical regions, although some species can extend into warm temperate regions (Zhang and Saunders, 2000). Burmannia disticha L., is an annual chlorophyllous herbs that grows up to 12.5-70 cm tall. Stems of B. disticha are green and cauline leaves are linear. The plant commonly grows in wet thickets or grasslands and is distributed mainly in southern and southeastern Asia, and southern and south-western China (Zhang, 2001).
Whole plastid genome sequencing of heterotrophs has become increasingly common. Since the first plastid genome of a nonphotosynthetic plant was sequenced in 1992 (Epifagus virginiana; Wolfe et al., 1992), researchers have increasingly focused on plastid evolution. For instance, researchers have attempted to determine which plastid genes have been conserved in mycoheterotrophic land plant taxa by examining the plastomes of five families of monocot angiosperms (Petrosaviaceae, Orchidaceae, Corsiaceae, Triuridaceae, Thismiaceae) (Delannoy et al., 2011; Logacheva et al., 2011, 2014; Barrett and Davis, 2012; Barrett et al., 2014; Mennes et al., 2015; Schelkunov et al., 2015; Bodin et al., 2016; Lim et al., 2016).
Burmanniaceae is a small mycoheterotrophic monocot family with nearly 130 species described and in which loss of chlorophyll has occurred on at least eight independent occasions (Merckx et al., 2008). Thus, to fully clarify the evolutionary pathways from autotrophy to full mycoheterotrophy, data representing other mycoheterotrophic monocot families regarding the loss of photosynthetic activity are needed. Furthermore, previous studies have indicated that more Burmanniaceae DNA sequences are needed to clarify the phylogenetic placement of the family within Dioscoreales. Plastid phylogenomics has recently become an effective way to elucidate the difficult evolutionary relationships in plants (Mennes et al., 2013, 2015; Barrett et al., 2014; Logacheva et al., 2014; Schelkunov et al., 2015; Bodin et al., 2016; Lim et al., 2016) due to its moderate nucleotide substitution rate and suitable genome size.
In this study, we use Illumina technology and genome skimming sequencing data to characterize the complete plastome of the normal photosynthetically plant B. disticha. Specifically, we describe B. disticha plastid genome size and gene content. To explore areas of the plastid genome that might be informative in resolving Burmannia phylogeny, we compare the B. disticha genome to seven Dioscoreales genomes. This study reports the first plastid genome sequence from Burmanniaceae, and provides a reference plastid genome for plants in Dioscoreales outside of Dioscoreaceae and Nartheciaceae. Finally, this study is the first step in a more extensive comparison of plastid genome diversity within Burmannia.
2. Materials and methods 2.1. Plant material, DNA extraction, and library preparationPlant material from B. disticha was collected in the field at Xiajinchang, Malipo County, Yunnan Province, China on October 28, 2013. The leaves were dried in silica gel and stored at room temperature for DNA extraction. Approximately 100 mg of leaf tissue was used for total genomic DNA extraction using a modified cetyltrimethyl ammonium bromide (CTAB) method (Doyle and Doyle, 1987; Rai et al., 2003). The quality of the obtained DNA was assessed by 1.0% agarose gel electrophoresis. Illumina shortinsert, paired-end (PE) libraries with average insert size of 500 bp were constructed for B. disticha, generating approximately 700 Mb clean reads of 90 bp on an Illumina HiSeq platform.
2.2. De novo contig assembly, plastome annotation, and plastome reconstructionThe genomic reads of B. disticha were assembled de novo using the SOAPdenovoV.2.0 (Luo et al., 2012) with a variety of kmer length (from 41 to 81). The resulting contigs/scaffolds were mapped to seven plastid genomes of Dioscoreales to identify potential plastid origin fragments. The process was accomplished by blasting plastid gene sequences of Aletris fauriei (GenBank accession NC_033412.1), Aletris spicata (NC_033411.1), Dioscorea elephantipes (NC_009601.1), Dioscorea rotundata (KJ490011.1), Dioscorea villosa (NC_034686.1), Dioscorea zingiberensis (NC_027090.1) and Metanarthecium luteoviride (NC_029214.1) against the assembled contig/ scaffolds pool using BLAST 2.2.26 (Altschul et al., 1990) with default settings. The aligned contig/scaffolds larger than 1000 bp were further connected by overlapped sequences, guided by their mapped locations in the reference plastid genomes. Finally, we mapped the raw sequencing reads to the assembled plastid genome sequences by Bowtie 2 (Langmead and Salzberg, 2012) to validate the assembly. The consensus sequences were annotated using the online tool DOGMA (Wyman et al., 2004) at the default settings. To complete annotation and check the gene predictions suggested by DOGMA, two additional methods were used: (1) aligning the Dioscoreales genes (as described above) using Geneious tool "MAFFT Alignment" [Version 10.1.2, Biomatters Limited; Kearse et al., 2012] and (2) annotating plastomes with the online tool CpGAVAS (Liu et al., 2012). The annotation results obtained were manually checked and corrected as necessary. In addition, we used tRNAscanSE (Schattner et al., 2005) to form more precise candidate transfer RNA (tRNA) genes. The circular plastome maps were generated in the online tool OGDRAW (Lohse et al., 2013). Finally, we determined the exact position of IR copies in sequences by aligning plastome sequences to themselves using BLASTN. A blast match was considered to describe IR if its length was more than 500 bp with identity more than 95%, and two sequences constituting the match were reverse complement to each other.
2.3. Phylogenetic trees reconstructionWe used 78 protein-coding genes from 8 complete plastomes of Dioscoreales to reconstruct the phylogenetic tree (Carludovica palmata as outgroup). C. palmata, belonging to the monocot order Pandanales, is a close, photosynthetic relative of Dioscoreales (Lam et al., 2015, 2016). Protein-coding genes were aligned by ClustalW in MEGA v 4.0 (Tamura et al., 2007), and the phylogenetic tree for a concatenated alignment was calculated in RAxML v 8.2 (Stamatakis, 2006) with a GTR+G model for the rapid Bootstrap (BS) algorithm, that was combined with the search for the best scoring maximumlikelihood (ML) tree (1000 bootstrap replicates).
3. Results and discussion 3.1. Size and structure of the B. disticha plastid genomeIllumina sequencing produced 7, 559, 832 paired-end reads, among which 698, 426 reads (9.24% out of total) could be mapped to the assembled plastid genome of B. disticha with an average sequencing depth of 399.1×. The plastid genome was assembled as a circular sequence of 157, 480 bp (GenBank accession number MG792012; Fig. 1). B. disticha has the typical quadripartite structure of plant plastid genomes, with an 81, 231-bp large single copy (LSC) region, a 13, 017-bp small single copy (SSC) region, two inverted repeat (IR) regions of 31, 616 bp, and exemplifies a typical angiosperm plastome arrangement. The overall GC content of B. disticha plastome is 34.9%. However, the distribution of nucleotides is uneven across the genome, and the IRs are more GC rich (39.5%) than the LSC (32.3%) and SSC (28.8%) regions. Approximately 120-130 unique genes are retained in the plastomes of photosynthetic land plants, and the plastid genome size is commonly about 120-170 kb (Ruhlman and Jansen, 2014). The plastome of B. disticha contains 111 genes including 78 protein-coding genes, 4 ribosomal RNA (rRNA) genes, and 29 tRNA genes (Fig. 1; Table 1). Of these, 12 genes are located in the small single copy region and 20 are duplicated in the IR. Coding regions comprise 61.3% of the B. disticha plastid genome. The mean length of intergenic regions and the longest intergenic region are 398 bp and 1918 bp, respectively, and the ratio of genic:intergenic DNA is 1.58:1.
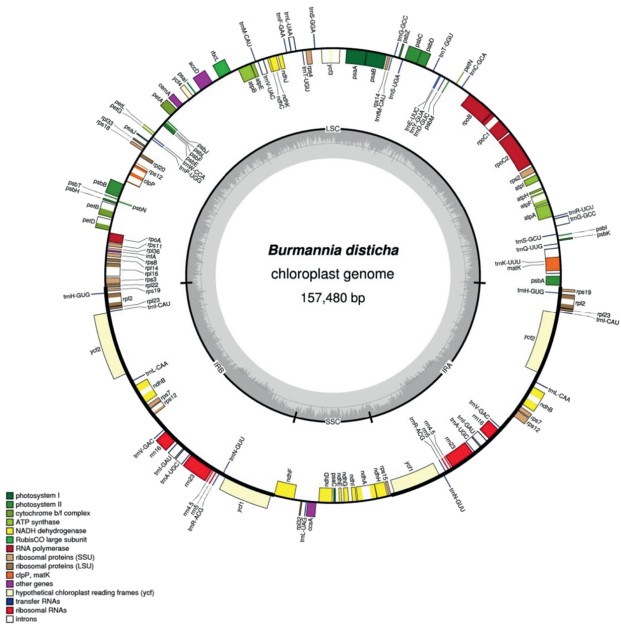
|
| Fig. 1 Circular plastome map of Burmannia disticha. Genes located inside the circle are transcribed clockwise; those outside are transcribed counterclockwise. The gray circle marks the GC content with the inner circle marking a 50% threshold. Thick branches indicate IR copies |
| Category | Genes |
| Photosynthesis | psaA, psaB, psaC, psal, psaJ psbA, psbB, psbC, psbD, psbE, psbF, psbH psbl, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ atpA, atpB, atpE, atpFa, atpH, atpl petA, petB a, petDa, petG, petL, petN rbcL ycf3b, ycf4 ndhAa, ndhBa(×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhl, ndhJ, ndhK |
| Ribosomal | rpl2a(×2), rpl14, rpl16a, rpl20, rpl22, rpl23(×2), rpl32, |
| proteins | rpl33, rpl36 rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12 b(×2), rps14, rps15, rps18, rps19(×2) |
| RNA polymerase | rpoA, rpoB, rpoC1a, rpoC2 |
| rDNAs | rrn4.5 (×2), rrn5(×2), rrn16(×2), rrn23(×2) |
| tRNAs | trnA_UGCa(×2), trnC_GCA, trnD_GUC, trnE_UUC, trnF_GAA, trnG_GCCb, trnfM_CAU, trnH_GUG(×2), trnI_CAU (×2), trnI_GAUa(×2), trnK_UUUa trnL_UAAa, trnL_CAA(×2), trnL_UAG, trnM_CAU, trnN_GUU(×2), trnP_UGG, trnQ_UUG, trnR_ACG(×2), trnR_UCU, trnS_GCU, trnS_GGA, trnS_UGA trnT_GGU, trnT_UGU, trnV_GAC(×2), trnV_UACa, trnW_CCA, trnY_GUA |
| Other proteincoding genes | accD, clpPb, infA, matK, ccsA, ycf1(×2), ycf2(×2), cemA |
| a Indicates genes with one intron. b Indicates genes with two introns; (×2) indicates duplicated genes in the IR regions. | |
Seven Dioscoreales species have plastid genomes smaller than that of B. disticha (Table 2). D. elephantipes has the smallest plastid genome (152, 609 bp) and A. spicata has the largest (154, 999 bp). The length of their LSC region varies from 81, 231 bp (B. disticha) to 85, 601 bp (D. rotundata), that of the SSC region from 13, 017 bp (B. disticha) to 19, 038 bp (D. zingiberensis), and their IR region from 25, 476 bp (D. rotundata) to 31, 616 bp (B. disticha) (Table 2). The overall GC content ranges from 37.2% to 37.5% among them, however it has been reduced to 34.9% in B. disticha. The GC content in the LSC and SSC of B. disticha are 32.3% and 28.8%, which is lower than those of the seven related plastomes, including Aletris (35.5% and 31.3%, respectively), Dioscorea (35.1% and 31.1%, respectively), and M. luteoviride (35.2% and 31.2%, respectively). With respect to the GC content in the IR, Dioscorea (43.0%) is slightly higher than M. luteoviride (42.9%) and Aletris (42.85%), but lower in B. disticha (39.5%).
| B. disticha | A.fauriei | A. spica | D. eleph | D. rotun | D. villo | D. zingi | M. luteoviride | |
| Genome size | 157, 480 | 154, 440 | 154, 999 | 152, 609 | 155, 406 | 153, 919 | 153, 970 | 153, 777 |
| LSC size | 81, 231 | 83, 501 | 83, 511 | 82, 777 | 85, 601 | 83, 865 | 83, 950 | 82, 882 |
| SSC size | 13, 017 | 18, 183 | 18, 160 | 18, 814 | 18, 840 | 18, 902 | 19, 038 | 17, 777 |
| IR size | 31, 616 | 26, 378 | 26, 664 | 25, 509 | 25, 476 | 25, 576 | 25, 491 | 26, 559 |
| No. protein coding | 78 | 79 | 79 | 78 | 78 | 78 | 78 | 78 |
| No.rRNA | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| No. tRNA | 29 | 29 | 29 | 29 | 29 | 29 | 29 | 29 |
| No. pseudogenes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total GC (%) | 34.9 | 37.5 | 37.5 | 37.2 | 37.2 | 37.2 | 37.2 | 37.4 |
| GC (%) in LSC | 32.3 | 35.5 | 35.4 | 34.9 | 35.2 | 35 | 35.1 | 35.2 |
| GC (%) in SSC | 28.8 | 31.3 | 31.3 | 31.2 | 30.9 | 31.2 | 31.2 | 31.2 |
| GC (%) in IR | 39.5 | 42.9 | 42.8 | 43 | 43 | 43 | 43 | 42.9 |
The length of the IR has increased approximately 18.6-24.1% in B. disticha compared with seven Dioscoreales species. The expansion of the IR at the 3'LSC boundary to include trnH_GUG is typical for monocots and also occurs in some early diverging angiosperms (Wang et al., 2008). However, the borders in these species are not the same. Both A. fauriei and D. zingiberensis have canonical IRs that contain trnH_GUG and parts of the 5'-end of ycf1 (D. zingiberensis; 286 bp) (Fig. 2). The LSC/IRb junction (JLB) of the remaining six Dioscoreales species (IR-expanded) is located in rps19, making 2-279 bp of the 5'-end of this gene extend into IR. By contrast, a 540-bp IR expansion into LSC is found in B. disticha, making its JLB occur within rps19, the rps19-rpl22 intergenic region and 93 bp of the 5'-end of rpl22 extend into the IR. Lam et al. (2015) also demonstrated a further expansion of the IR at the 3'LSC boundary to include the rps19 gene in the plastid genome of C. palmata. The IRa/LSC border in all plastomes except that of B. disticha is located downstream from the trnH_GUG gene, by convention located where plastome numbering begins. However, in B. disticha the border is located downstream from the rps19 gene, placing the rps19 gene in the IRa region. Additionally, in seven related Dioscoreales species the SSC/ IR border is located in the ycf1 gene with 286-1675 bp duplicated in the IR regions. In B. disticha, the IR/SSC junction occurs within the rps15-ycf1 intergenic region, resulting in the duplication of the whole ycf1 gene sequence in IRs.
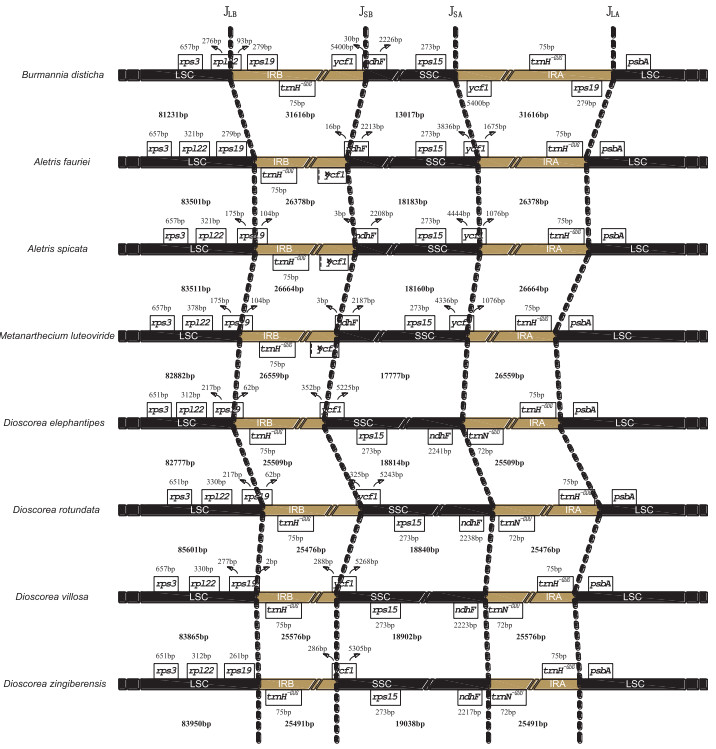
|
| Fig. 2 Comparison of the IR-SC boundaries among eight Dioscoreales plastomes. The JLA, JLB, JSA and JSB refer to junctions of LSC/IRA, LSC/IRB, SSC/IRA, and SSC/IRB, respectively. Ψ indicates pseudogene |
The plastome of Aletris has the most number of potentially functional genes with a total of 112, including 79 protein-coding genes, 4 ribosomal RNA (rRNA) genes, and 29 tRNA genes, whereas other Dioscoreales species, including B. disticha, contain 111 genes (Table 2). The majority of the variation in plastome genes involves the ribosome protein gene rps16. All species of Dioscoreales have lost the rps16 gene that is present in Aletris. In Aletris, a functional rps16 gene (246 bp) is located in the 2281-bp-long intergenic region between trnK_UUU and trnQ_UUG. Furthermore, the plastomes of seven related species lack a functional ycf1 gene, which has instead become a pseudogenewith onlyabout 1000 bp. In B. disticha, the two ycf1 genes in the duplicated IR region have intact reading frames and appear to be functional. Our overall results indicate that the plastid gene content of B. disticha is very similar to that of those closely related species in the order Dioscoreales.
While most start codons conformed to the commonly used canonical ATG, several atypical start codons were detected in the plastome of B. disticha, such as ACG for rpl2 and GTG for rps19. The ACG for rpl2 is presumably converted to the canonical ATG codon by RNA editing, a feature commonly found in all monocots, whether photosynthetic or not (Schelkunov et al., 2015).
3.3. Phylogenetic analysesThe final concatenated data matrix of 78 protein-coding genes was 68, 911 bp with 55, 060 (79.9%) identical sites. Phylogenetic analysis showed that B. disticha was strongly supported as the sister group to a clade that included D. elephantipes, D. rotundata, D. villosa, and D. zingiberensis (Fig. 3). Despite the potential drawbacks of phylogenies derived from plastid genomes (see Davis et al., 2014), our results are consistent with previous studies based on both mitochondrial and nuclear data which showed that Burmanniaceae is close to Dioscoreaceae, and therefore a member of Dioscoreales (Angiosperm Phylogeny Group, 1998, Angiosperm Phylogeny Group, 2003; Caddick et al., 2002a, b; Soltis et al., 2000; Davis et al., 2004; Merckx et al., 2006). Well-supported relationships in Dioscoreales include a small clade comprising M. luteoviride, A. fauriei, and A. spicata, a sister-group relationship between D. elephantipes and D. rotundata, and between D. villosa and D. zingiberensis (Fig. 3). In this study, we were unable to retrieve sufficient representative Burmanniaceae data to compare our phylogeny to those of previous studies on this family. While further taxon sampling is necessary in the future, our study provides evidence that plastome sequences may offer a new approach to resolving the phylogenetic position of Burmannia.

|
| Fig. 3 ML phylogenetic trees based on nine complete plastid genome sequences. The tree was estimated with RAxML (Stamatakis, 2006), applying the rapid bootstrapping algorithm (1000 bootstrap replicates). Bootstrap values are plotted above nodes. The bars indicate nucleotide substitutions per site |
We thank Dr. An-Dan Zhu (Kunming Institute of Botany) for the circular drawing editing and valuable comments; This study was supported by the Large-scale Scientific Facilities of the Chinese Academy of Sciences (Grant No: 2017-LSFGBOWS-02) and the Youth Innovation Promotion Association of Chinese Academy of Sciences (2015321).
Altschul S.F., Gish W., Miller W., et al., 1990. A basic local alignment search tool. J. Mol. Biol, 215(3), 403-410.
DOI:10.1016/S0022-2836(05)80360-2 |
||
Angiosperm Phylogeny Group, 2003. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants:APG Ⅱ. Bot. J. Linn. Soc, 141, 399-436.
DOI:10.1046/j.1095-8339.2003.t01-1-00158.x |
||
Angiosperm Phylogeny Group, 1998. An ordinal classification for the families of flowering plants. Ann. Mo. Bot. Gard, 4, 531-553.
|
||
Barrett C.F., Davis J.I., 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot, 99, 1513-1523.
DOI:10.3732/ajb.1200256 |
||
Barrett C.F., Freudenstein J.V., Li J., et al., 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol, 31(12), 3095-3112.
DOI:10.1093/molbev/msu252 |
||
Bolin J.F., Tennakoon K.U., Majid M.B.A., et al., 2017. Isotopic evidence of partial mycoheterotrophy in Burmannia coelestis (Burmanniaceae). Plant Spec. Biol, 32, 74-80.
DOI:10.1111/psbi.2017.32.issue-1 |
||
Bodin S.S., Kim J.S., Kim J.H., 2016. Phylogenetic inferences and the evolution of plastid DNA in Campynemataceae and the mycoheterotrophic Corsia dispar D.L Jones & B. Gray (Corsiaceae). Plant Mol. Biol. Rep, 34, 192-210.
DOI:10.1007/s11105-015-0914-6 |
||
Caddick L.R., Rudall P.J., Wilkin P., et al., 2002a. Phylogenetics of Dioscoreales based on combined analyses of morphological and molecular data. Bot. J. Linn. Soc, 138, 123-144.
DOI:10.1046/j.1095-8339.2002.138002123.x |
||
Caddick L.R., Wilkin P., Rudall P.J., et al., 2002b. Yams reclassified:a recircumscription of Dioscoreaceae and Dioscoreales. Taxon, 51, 103-114.
DOI:10.2307/1554967 |
||
Davis C.C., Xi Z.X., Mathews S., 2014. Plastid phylogenomics and green plant phylogeny:almost full circle but not quite there. BMC Biol, 12(1), 11.
DOI:10.1186/1741-7007-12-11 |
||
Davis J.I., Stevenson D.W., Petersen G., et al., 2004. A phylogeny of the monocots, as inferred from rbcL and atpA sequence variation, and a comparison of methods for calculating jackknife and bootstrap values. Syst. Biol, 29, 467-510.
|
||
Delannoy E., Fujii S., Francs-Small C.D.C., et al., 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol, 28, 2077-2086.
DOI:10.1093/molbev/msr028 |
||
Doyle J.J., Doyle J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull, 19, 11-15.
|
||
Jonker, F., 1938. A Monograph of the Burmanniaceae, vol. 51. Meeded Bot. Mus. Herb. Rijks Univ. Utrecht, Utrecht, pp. 1-279.
|
||
Kearse M., Moir R., Wilson A., et al., 2012. Geneious Basic:an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647-1649.
DOI:10.1093/bioinformatics/bts199 |
||
Lam V.K.Y., Merckx V.S.F.T., Graham S.W., 2016. A few-gene plastid phylogenetic framework for mycoheterotrophic monocots. Am. J. Bot, 103(4), 692-708.
DOI:10.3732/ajb.1500412 |
||
Lam V.K.Y., Gomez M.S., Graham S.W., 2015. The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biol. Evol, 7, 2220-2236.
DOI:10.1093/gbe/evv134 |
||
Langmead B., Salzberg S.L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Meth, 9, 357-359.
DOI:10.1038/nmeth.1923 |
||
Lim G.S., Barrett C.F., Pang C.C., et al., 2016. Drastic reduction of plastome size in the mycoheterotrophic Thismia tentaculata relative to that of its autotrophic relative Tacca chantrieri. Am. J. Bot, 103, 1-9.
DOI:10.3732/ajb.1500441 |
||
Liu C., Shi L.C., Zhu Y.J., et al., 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom, 13, 715.
DOI:10.1186/1471-2164-13-715 |
||
Logacheva M.D., Schelkunov M.I., Nuraliev M.S., et al., 2014. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol. Evol, 6(1), 238-246.
DOI:10.1093/gbe/evu001 |
||
Logacheva M.D., Schelkunov M.I., Penin A.A., 2011. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol. Evol, 3, 1296-1303.
DOI:10.1093/gbe/evr102 |
||
Lohse M., Drechsel O., Kahlau S., et al., 2013. Organellar Genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression datasets. Nucleic Acids Res, 41, 575-581.
DOI:10.1093/nar/gks1075 |
||
Luo R., Liu B., Xie Y., et al., 2012. SOAPdenovo2:an empirically improved memoryefficient short-read de novo assembler. Giga Sci, 1, 18.
DOI:10.1186/2047-217X-1-18 |
||
Mennes C.B., Lam V.K.Y., Rudall P.J., et al., 2015. Ancient Gondwana break-up explains the distribution of the mycoheterotrophic family Corsiaceae (Liliales). J. Biogeogr, 42(6), 1123-1136.
DOI:10.1111/jbi.12486 |
||
Mennes C.B., Smets E.F., Moses S.N., et al., 2013. New insights in the long-debated evolutionary history of Triuridaceae (Pandanales). Mol. Phylogenet. Evol, 69, 994-1004.
DOI:10.1016/j.ympev.2013.05.031 |
||
Merckx V., Chatrou L.W., Lemaire B., et al., 2008. Diversification of myco-heterotrophic angiosperms:evidence from Burmanniaceae. BMC Evol. Biol, 8(1), 178.
DOI:10.1186/1471-2148-8-178 |
||
Merckx V., Schols P., Maas-Van D.K.H., et al., 2006. Phylogeny and evolution of Burmanniaceae (Dioscoreales) based on nuclear and mitochondrial data. Am. J. Bot, 93(11), 1684-1698.
DOI:10.3732/ajb.93.11.1684 |
||
Rai H.S., O'Brien H.E., Reeves P.A., et al., 2003. Inference of higher-order relationships in the cycads from a large chloroplast data set. Mol. Phylogenet. Evol, 29, 350-359.
DOI:10.1016/S1055-7903(03)00131-3 |
||
Ruhlman, T., Jansen, R.K., 2014. The plastid genomes of flowering plants. In: Maliga, P. (Ed.), Chloroplast Biotechnology. Humana Press, New York, pp. 33-38.
|
||
Schattner P., Brooks A.N., Lowe T.M., 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res, 33, 686-689.
DOI:10.1093/nar/gki366 |
||
Schelkunov M.I., Shtratnikova V.Y., Nuraliev M.S., et al., 2015. Exploring the limits for reduction of plastid genomes:a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol. Evol, 7(4), 1179-1191.
DOI:10.1093/gbe/evv019 |
||
Soltis D.E., Soltis P.S., Chase M.W., et al., 2000. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc, 133, 381-461.
DOI:10.1111/boj.2000.133.issue-4 |
||
Stamatakis A., 2006. RAxML-Ⅵ-HPC:maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 11, 2688-2690.
|
||
Tamura K., Dudley J., Nei M., et al., 2007. Tamura, K., Dudley, J., Nei, M., et al., 2007. MEGA4:molecular evolutionary genetics analysis (mega) software version 4.0. Mol. Biol. Evol, 24, 1596-1599.
DOI:10.1093/molbev/msm092 |
||
Wang R.J., Cheng C.L., Chang C.C., et al., 2008. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol, 8, 36.
DOI:10.1186/1471-2148-8-36 |
||
Wolfe K.H., Morden C.W., Palmer J.D., 1992. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. U. S. A, 89(22), 10648-10652.
DOI:10.1073/pnas.89.22.10648 |
||
Wyman S.K., Jansen R.K., Boore J.L., 2004. Automatic annotation of organellar genomes with DOGM. Bioinformatics, 20, 3252-3255.
DOI:10.1093/bioinformatics/bth352 |
||
Zhang D.X., 2001. Phylogenetic reconstruction of Burmannia L. (Burmanniaceae):a preliminary study. Acta Phytotaxon. Sin, 39(3), 203-223.
|
||
Zhang D., Saunders R.M.K., 2000. Systematics of the Burmannia coelestis complex(Burmanniaceae). Nord. J. Bot, 20, 385-394.
DOI:10.1111/j.1756-1051.2000.tb01578.x |



