b. University of Chinese Academy of Sciences, Beijing 100049, China
Flora is the plant life occurring in a particular region, generally the naturally growing or native plant life, and it is a natural synthesis of family, genus, and species of plant life (Wu et al., 2006). Floristic geography is the study of the past and present distribution of plants, and provides important insight on their origin and diversification in time and space (Wang, 1992). Sources of floristic data include published national and regional texts that summarize taxonomic relationships, provide discussion of taxa, and give brief geographical and ecological data pertinent to the taxa of the area covered (Morin et al., 1989). Based on the floristic data of the Flora Reipublicae Popularis Sinicae (FRPS) and other available regional floras of China, Wu analyzed the distribution patterns of over 3000 genera of seed plants in China and established a scheme of classification of distribution patterns or areal-types of genera of seed plants in China (Wu, 1965, 1991), in which 15 areal-types were recognized. As floristic data accumulated with the completion of the FRPS and progress of the Flora of China (FOC), the scheme was modified and elucidated, and enlarged to include distribution patterns or areal-types of families of seed plants of the world, of which 18 types were recognized (Wu et al., 2003, 2006). The scheme of classification of areal-types or distribution patterns of families and genera of seed plants has been widely used in analyzing national and regional floras of China at various levels and proved helpful in understanding biogeographical issues, such as floristic divisions, endemism and disjunctive distributions (e.g., Wu, 1979, 1983; Li and Li, 1997; Wu et al., 2005, 2010; Li et al., 2007).
However, the species composition of regional plant assemblages is driven by the interplay between evolutionary processes, including speciation, extinction, dispersal, and ongoing ecological processes (Ricklefs, 2007; Xing and Ree, 2017). A floristic assemblage can only comprise plants whose environmental tolerances allow species to maintain a population (Qian et al., 2017a), given both the abiotic and biotic conditions in that region (Fraterrigo et al., 2014; Qian and Sandel, 2017a). Meanwhile, the ability of a species to persist in a particular set of ecological environments is constrained by its evolutionary history (Ricklefs, 1987; Qian and Sandel, 2017b). Then, analyses of the composition and assembly of floristic assemblages that only focus on present day and local-scale ecological interactions are incomplete and will not be able to successfully explain the primary reasons why plant species composition varies across regions (Swenson, 2011a, 2013). Phylogenetics is the study of evolutionary relationships among species and higher taxonomic ranks of organisms, which when calibrated with the dimension of time can provide important information on patterns of inferred genetic similarity in time and space (Pennington et al., 2006; Donoghue, 2008). Therefore, integrating phylogenetic information into ecology provides a promising way to explore the ecological, biogeographical, and evolutionary processes that drive plant assemblies at multiple spatial scales (Webb et al., 2002; Graham and Fine, 2008).
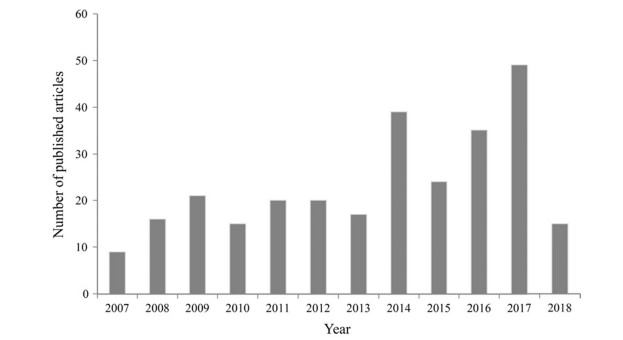
Swenson and Umaña (2014) introduced the term 'phylofloristics' to define a phylogenetically enabled analysis of the species composition of regional plant assemblages. The aim of this analysis is to explore the spatio-temporal patterns of floristic assemblages and uncover the evolutionary and biogeographical history that potentially explains the assembly processes shaping regional species pools by integrating phylogeny into traditional floristic study. The analytical approach uses conceptual and analytical advances from studies that have sought to quantify phylogenetic alpha and beta diversity of community assemblies (e.g., Webb, 2000; Schweiger et al., 2008; Swenson, 2011b) with the primary difference being that phylofloristics focuses on the entire flora of regional plant assemblages. Analogous to taxonomic diversity, which measures species composition in a floristic region, phylogenetic diversity (PD) is a measure of diversity to quantify phylogenetic composition of floristic assemblages (Forest et al., 2007; Chao et al., 2014; Lu et al., 2018). Phylogenetic diversity is calculated as the sum of all phylogenetic branch lengths connecting taxa in a defined region (Faith, 1992). In additional to the measure of phylogenetic diversity, with associated statistical tests, a series of metrics have been proposed to assess the spatial phylogenetics of regional plant assemblages and explore their origin, diversification and formation. For example, net relatedness index (NRI) or nearest taxon index (NTI) can be used as a measure of phylogenetic relatedness among coexisting species within a region (Webb, 2000; Kembel and Hubbell, 2006), mean pairwise phylogenetic index (Dpw) or mean nearest phylogenetic neighbor index (Dnn) can be used to quantify the spatial turnover of phylogenetic composition between sites (Webb et al., 2008), and relative phylogenetic diversity (RPD) and relative phylogenetic endemism (RPE) can be used to quantify distinctions between centres of neo- and paleoendemism within a region (Mishler et al., 2014). Since an early attempt to perform a phylogenetic analysis of regional plant assemblages (Forest et al., 2007), the use of phylogenetic information to investigate evolutionary features of floristic assemblages has blossomed (Fig. 1).
|
| Fig. 1 Results of a search in the Web of Science (accessed on 28 May, 2018) using the key words 'phylogenetic diversity' and 'floristic'. |
Phylogenetic structure is used to quantify the average degree of phylogenetic relatedness among coexisting species within a floristic region, which reflects the evolutionary character of floristic assemblages (Ricklefs, 2006; Cadotte et al., 2010; Qian et al., 2017b). Phylogenetic clustering indicates that species in this region are more closely related than expected, whereas phylogenetic overdispersion or evenness indicates that species are more distantly related than expected (Webb et al., 2002). By analyzing a comprehensive data set including all floristic taxa with their phylogenetic relationships, one can explore the patterns of phylogenetic relatedness of plant assemblages within a particular floristic region. For example, Qian et al. (2013) explored the latitudinal gradient patterns in phylogenetic relatedness of angiosperm trees in North America. They found that tree species in regional assemblages tend to be more phylogenetically related (clustered) in regions at higher latitudes with lower temperatures in North America. This finding is consistent with the prediction of the tropical niche conservation hypothesis that species in colder or drier climates should on average be more closely related to each other (more phylogenetically clustered) than those in warmer or wetter climates (Latham and Ricklefs, 1993; Wiens and Donoghue, 2004). Similar to the latitudinal diversity gradient, the patterns of phylogenetic relatedness of floristic assemblages were revealed in the elevational diversity gradient. For example, by measuring net related index and net nearest taxon index, Li et al. (2014) analyzed the phylogenetic structure of each elevational belt of alpine plants in the Hengduan Mountains of southwest China. They found that alpine plants tended to show phylogenetic overdispersion at low elevational belts and phylogenetic clustering at high elevational belts. They concluded that phylogenetic structure was influenced by an environmental filter, interspecies interactions, and rapid speciation during the uplift of the Qinghai-Tibet Plateau.
Phylogenetic structure of regional plant assemblages is determined by environmental conditions and biogeographical history. Incorporating phylogenetic information into the study of spatial turnover in species composition allows researchers to explore the role of ecological and evolutionary processes in structuring floras. For example, Qian et al. (2013b) investigated the phylogenetic beta diversity of floristic assemblages across North America north of Mexico, and found that geographical and environmental distances together explained on average about 76% of the variation in phylogenetic beta diversity with environmental distance explaining more variation in phylogenetic similarity of floristic assemblages between latitudinal bands. Also, Swenson and Umaña (2014) studied phylogenetic turnover of vascular plants among 18 islands in the Lesser Antillean of Central America. Their results show a major influence of environmental heterogeneity on the assembly of the island floras and that spatial distance is not a significant predictor of compositional dissimilarity of plant species between islands.
2. patial phylogenetics of floristic assemblagesUnderstanding spatial patterns of biodiversity, such as identifying areas with highly concentrated richness or endemism, has long been the desired objective of biodiversity assessments. However, previous studies have most commonly focused on species alone, an approach that can provide valuable insights but gives an incomplete picture of diversity since it fails to incorporate their evolutionary relationships among species (e.g., Myers et al., 2000; López-Pujol et al., 2011). Based on the concepts of phylogenetic diversity (PD) and phylogenetic endemism (PE), two derived metrics called relative phylogenetic diversity (RPD) and relative phylogenetic endemism (RPE) were designed to examine the distribution of branch lengths in an area (Mishler et al., 2014). Mishler et al. (2014) also developed a randomization test for PE and RPE called categorical analysis of neo- and paleo-endemism (CANAPE). This test is capable of distinguishing areas containing significant concentrations of rare long branches (paleo-endemism), rare short branches (neo-endemism), or mixed endemism (e.g., Heenan et al., 2017; Thornhill et al., 2017). These measures allow one to better evaluate the spatial patterns of diversity and endemism from an evolutionary perspective by combining spatial information from herbarium collections and DNA-based phylogenies. For example, using a genus-level phylogeny for 87% of the Chilean vascular flora and detailed geographical locality information, Scherson et al. (2017) explored the distribution patterns of evolutionary features of the Chilean vascular flora by calculating PD, PE, RPD, RPE and performing analyses of neo- and paleo-endemism. Their results indicated that PD patterns were consistent with known centers of high taxon richness and the Chilean biodiversity hotspot. In addition, the south of the country showed areas of significantly high RPD and a concentration of paleo-endemism, and the north showed areas of significantly low PD and RPD, and a concentration of neoendemism.
In addition to simply identifying areas of taxon richness and endemism, incorporating the phylogeny facilitates investigations of potential evolutionary and ecological causes (e.g., centers of divergence or competitive exclusion) of significant concentrations of diversity and endemism. For example, using a phylogeny sampling 90% of Australia's angiosperm genera, and 3.4 million georeferenced specimens, Thornhill et al. (2016) investigated the spatial patterns of diversity and endemism of the Australian flora and the evolutionary and ecological processes that have created them. They found that the Wet Tropics and Cape York flora show the highest observed taxonomic richness and PD for angiosperms. However, the PD randomization test revealed significantly low PD in this area, which indicates flora establishments in this area are biased toward clades that are adapted for tropical conditions. The high RPD score shows that, although the plants in this area are phylogenetically clustered, they also have longer than expected branches, and may represent relict taxa. The CANAPE results identify the area as a super-endemic region, which gives support to the idea that the tropical flora is extremely range-restricted and long-branched. In addition, the arid interior flora showed low observed richness for angiosperms. The significantly low PD suggests phylogenetic clustering in this region, an indication that only certain clades are adapted to arid conditions. Significantly low RPD suggests that branches are shorter than expected and may indicate recent radiations in the region.
3. Origin and diversification of floristic assemblagesA central theme in floristic study is the origin and diversification of regional plant assemblages in time and space (Wang, 1992; Mittelbach and Schemske, 2015). At the spatial scale, it addresses the evolution of differences in species richness and composition among continents. At the time-scale, it deals with plant diversification to geological time for continental differentiation. Previous study on the evolutionary history of floristic assemblages has typically been explored using specific taxa as exemplars (e.g., Richardson et al., 2001; Klak et al., 2004; Pillon, 2012). Recently, dated phylogenies and detailed distribution data from selected clades or entire flora were used to address the origin and diversification of floristic assemblages. For example, Lu et al. (2018) investigated the spatio-temporal divergence patterns of the Chinese flora using a dated phylogeny of 92% of the angiosperm genera for the region and detailed spatial distribution data. They found that 66% of the angiosperm genera in China did not originate until the early Miocene 23 million years ago (Mya). The flora of eastern China bears a signature of older divergence (mean divergence times of 22.04-25.39 Mya) and the flora in western China shows more recent divergence (mean divergence times of 15.29-18.86 Mya). Linder (2014) explored the evolution of African plant diversity by analysing the evolutionary history of a selection of African clades. He argued that the current plant diversity in Africa can be interpreted as the result of the continuous adding of new floras. The region entered the Cenozoic with the oldest Lowland forest Flora. During the Paleocene, the Arid Flora and the Austro-temperate Floras were added. The doming and volcanism of the Neogene resulted in the adding of the Tropic-montane and Tropic-alpine Floras. The Savanna Flora may have been the result of a biotic event - the evolution and spread of grassland. Verboom et al. (2009) used the dated phylogenies of multiple plant groups to explore the origin and diversification of succulent karoo flora and fynbos flora in southern African. They found that all succulent karoo lineages were less than 17.5 Mya old, which was largely consistent with suggestions that this biome was the product of recent radiation, probably triggered by climatic deterioration since the late Miocene. In contrast, fynbos lineages showed a broader age distribution, with some lineages originating in the Oligocene, but most being more recent.
Tracing the patterns of distribution and dispersal of selected plant lineages from a particular floristic region can help to better understand the biogeographical origins of flora. For example, Chen et al. (2018) explored the biogeographical origins and historical evolution of the East Asian Flora using molecular phylogenetic data from 213 clades as well as relevant fossil information on the seed plants through meta-analysis. Their results suggest a multiple biogeographical origin of the East Asian Flora including Laurasian floras and Gondwanan floras. Moreover, the East Asian Flora does exhibit a close affinity to the floras in the Northern Hemisphere, which include North America, Arctic/Boreal, Tethyan areas, and Tropical Asia. Based on the compiled data for 85 dated phylogenetic clades of vascular plants, Crisp and Cook (2013) reviewed the evolution of Australian flora and the principal forces that transformed the ancestral Gondwanan rainforest flora to the presentday island-continent flora through the Cenozoic. They found that today's vegetation is a mix of ancient radiations that have persisted in Australia through dramatic climate change since before the breakup of Gondwana (38 clades, 45%), and more recent lineages whose ancestors arrived by trans-oceanic dispersal (41 clades, 48%). Moreover, the temperate flora in Australia had a Gondwanan origin, whereas tropical taxa are hypothesized to have arrived by dispersal from Asia as the Australian plate drifted north into tropical latitudes.
4. Phylogenetic delimitation of floristic regionsA central aim in floristic study is to classify groups of organisms into meaningful geographical units at different scales for the purpose of better understanding patterns of biodiversity (Mackey et al., 2008; Kreft and Jetz, 2010). Traditional attempts to delineate floristic regions typically based on the similarity of floristic assemblages and the degree of endemism (e.g., Takhtajan, 1986; Olson et al., 2001; Zhang et al., 2016). However, such analyses treat taxa as independent units, often ignoring the rich context that phylogenetic relationships can provide, and therefore do not reflect the varied evolutionary histories of floristic regions (Donoghue, 2008; Cavender-Bares et al., 2016). By including phylogenetic relationships in a traditional floristic analysis, a phylogenetic distance-based biogeographical delimitation of floristic regions can be generated. For example, Slik et al. (2018) presented a classification of the world's tropical forests based on phylogenetic similarity of floristic composition. They identified five principal floristic regions: Indo-Pacific forest region, subtropical forest region, African forest region, American forest region, and dry forests region. Their results do not support the traditional neo-versus paleotropical forest division, but African and American forests are grouped, reflecting their former western Gondwanan connection, while Indo-Pacific forests range from eastern Africa and Madagascar to Australia and the Pacific. A northern-hemisphere subtropical forest region was identified with representatives in Asia and America, providing support for a link between Asian and American northernhemisphere forests. Additionally, global dry forest regions were identified as a single tropical biome, with representatives in America, Africa, Madagascar, and India. Also, Li et al. (2015b) proposed a system of floristic regions within a global biodiversity hotspot region (Yunnan of China) by combining data on the distributions and phylogenetic relationships of 1983 genera of native seed plants. They identified eight distinct floristic regions in Yunnan, which were grouped into two larger northern and southern geographical units. The proposed eight floristic regions are in broad agreement with previously published floristic divisions of Yunnan based on the qualitative evidence by expert and modelled distribution of woody plant species. Moreover, across the Yunnan region they examined, the central Yunnan region showed the lowest level of spatial turnover in phylogenetic relationships and taxonomic composition of the floristic assemblages. Daru et al. (2016) identified 15 phylogenetically distinct biogeographical units using a published time-calibrated phylogenetic tree for 1400 woody plant species along with their geographical distributions in southern Africa. Their phylogeny-based delimitation of southern Africa's woody vegetation broadly matches currently recognized phytogeographical classifications, but also highlights parts of the Namib Karoo and Greater Limpopo Transfrontier Park as distinct, but previously under-recognized biogeographical units.
5. Biodiversity conservation of phylogenetic dimensionBiodiversity conservation has become a central focus of research because species extinction risks resulting from global climate change, habitat fragmentation, exotic species invasion, and increased human population density have grown in recent decades (Strauss et al., 2006; Cardinale et al., 2012). Historically, biodiversity conservation has focused on using rarity, endemism, and distinctiveness to set priorities (Myers et al., 2000; Kier et al., 2009). However, in recent years, the evolutionary processes that maintain and generate biodiversity have also been recognized as important factors that need to be accounted for in conservation planning (Moritz, 2002; Mace and Purvis, 2008; Winter et al., 2013). Some have argued that preserving a site containing a few highly diverse, distantly related lineages may be more beneficial, ultimately, than preserving a site with a large number of closely related taxa (Lyashevska and Farnsworth, 2012; Diniz-Filho et al., 2013; Costion et al., 2015). This is important because two regions of equal species richness may be composed of species with either highly similar or distinct phylogenetic histories, namely if both regions have different lineage diversity (Webb et al., 2002). Moreover, the extinction of the most phylogenetically distinct species would result in the loss of a greater proportion of biological diversity (Faith, 2008; Thuiller et al., 2011), which may decrease the ability of the biota of a region to persist in the face of environmental change (González-Orozco et al., 2016). Thus, phylogenetic information is used to prioritize conservation of species assemblages that have a distinct evolutionary history relative to others in the region (Devictor et al., 2010; Tucker and Cadotte, 2013). For example, Forest et al. (2007) and Tucker et al. (2012) found that although the flora in the western part of the Cape Floristic Region of South Africa has twice the species richness as that of the eastern region, the western flora is less phylogenetically diverse, because the flora of the western part of the Cape is phylogenetically clustered and is made up of relatively closely related genera. In contrast, the flora of the eastern region is phylogenetically overdispersed relative to the western region, and contains genera that are, on average, less closely related to one another. The authors recommendation would not reject recently diversifying sites in the west as conservation targets, but to maximize the evolutionary potential of the flora to respond to environmental changes in the future, more attention should be paid to the suitable areas in the east because they contain high levels of phylogenetic diversity within the region (Forest et al., 2007). Also, Li et al. (2015a) explored how phylogenetic diversity, species richness, and phylogenetic community structure vary in seed plant communities along an elevational gradient in a relatively understudied high mountain region, the Dulong Valley, in southeastern Tibet, China. They found evergreen broad-leaved forests had the highest levels of species richness and phylogenetic diversity (Fig. 2). Moreover, evergreen broad-leaved forests were phylogenetically overdispersed, whereas other vegetation types tended to be phylogenetically clustered. Thus, they suggest that evergreen broad-leaved forests with high species richness and overdispersed phylogenetic structure should be a focus for biodiversity conservation within the Dulong Valley because these areas may help maximize the potential of this flora to respond to future global climate change.

|
| Fig. 2 Photos of selected living plants from evergreen broad-leaved forests in the Dulong Valley of southeastern Tibet, China. A. Dipentodon sinicus; B. Dobinea vulgaris; C. Euptelea pleiosperma; D. Helwingia japonica; E. Stachyurus himalaicus; F. Tetracentron sinense. |
Although the integration of phylogenetic information and floristic data is driving the discovery of complex patterns and new hypotheses for further study, we would like to emphasize limitations to these studies that future phylofloristic analyses should improve. Firstly, the phylogenetic trees have been largely unresolved within genera and this limits more detailed investigations into the biogeography and evolutionary history of floristic assemblages in this system. Secondly, detailed and accurate information regarding the geographic distribution of all plant life is still poorly known in areas with sparse specimen collections. These data limitations have hindered our ability to better understand spatial patterns of regional plant assemblages (e.g., identifying areas of taxon richness and endemism, biogeographical regionalization). Thirdly, plant functional traits provide information about adaptations to climate and environmental conditions, which can be used to explore species co-occurrence of floristic assemblages (Kraft et al., 2015; Wang et al., 2018). However, extensive trait information remains stored in publications and herbarium specimens, and the ability to extract these data is limited. The lack of trait information is an impediment to fully understanding floristic assembly. Finally, phylofloristic study demands the use of an integrative, multifaceted, big data approach, which incorporates bioinformatics, large-scale phylogeny reconstruction, use of digitized specimen records and fossil data, and complex ecological models. However, the development of the tool or program for these analyses is in many ways still in its infancy and generally is not yet as well developed as needed. These limitations will likely attract future phylofloristic studies. However, the removal of these obstacles will greatly help refine analyses and promote more focused investigations into the evolutionary and biogeographical imprint on flora assembly that can't be detected by analyzing species compositions alone.
AcknowledgementsThis study was supported by the National Natural Science Foundation of China (grant no. 31570212, 31770228, 31370243) and the Talent Project of Yunnan (grant no. 2015HB092). We are grateful to two anonymous reviewers for constructive comments in improving the manuscript. We also thank Professor Zhekun Zhou for invitation to contribute to this special issue.
Cadotte M.W., Davies T.J., Regetz J., Kembel S.W., Cleland E.E., Oakley T.H., 2010. Phylogenetic diversity metrics for ecological communities:integrating species richness, abundance and evolutionary history. Ecol. Lett, 13, 96-105.
DOI:10.1111/ele.2009.13.issue-1 |
||
Cardinale B.J., Duffy J.E., Gonzalez A., Hooper D.U., Perrings C., Venail P., Narwani A., Mace G.M., Tilman D., Wardle D.A., Kinzig A.P., Daily G.C., Loreau M., Grace J.B., Larigauderie A., Srivastava D.S., Naeem S., 2012. Biodiversity loss and its impact on humanity. Nature, 486, 59-67.
DOI:10.1038/nature11148 |
||
Cavender-Bares J., Ackerly D.D., Hobbie S.E., Townsend P.A., 2016. Evolutionary legacy effects on ecosystems:biogeographic origins, plant traits, and implications for management in the era of global change. Annu. Rev. Ecol. Evol. Syst, 47, 433-462.
DOI:10.1146/annurev-ecolsys-121415-032229 |
||
Chao A., Chiu C.H., Jost L., 2014. Unifying species diversity, phylogenetic diversity, functional diversity, and related similarity and differentiation measures through Hill numbers. Annu. Rev. Ecol. Evol. Syst, 45, 297-324.
DOI:10.1146/annurev-ecolsys-120213-091540 |
||
Chen Y.S., Deng T., Zhou Z., Sun H., 2018. Is the East Asian flora ancient or not? Natl. Sci. Rev, 0, 1-13.
|
||
Costion C.M., Edwards W., Ford A.J., Metcalfe D.J., Cross H.B., Harrington M.G., Richardson J.E., Hilbert D.W., Lowe A.J., Crayn D.M., 2015. Using phylogenetic diversity to identify ancient rain forest refugia and diversification zones in a biodiversity hotspot. Divers. Distrib, 21, 279-289.
DOI:10.1111/ddi.2015.21.issue-3 |
||
Crisp M.D., Cook L.G., 2013. How was the Australian flora assembled over the last 65 million years? A molecular phylogenetic perspective. Annu. Rev. Ecol. Evol.Syst, 44, 303-324.
DOI:10.1146/annurev-ecolsys-110512-135910 |
||
Daru B.H., van der Bank M., Maurin O., Yessoufou K., Schaefer H., Slingsby J.A., Davies T.J., 2016. A novel phylogenetic regionalization of phytogeographical zones of southern Africa reveals their hidden evolutionary affinities. J. Biogeogr, 43, 155-166.
DOI:10.1111/jbi.12619 |
||
Devictor V., Mouillot D., Meynard C., Jiguet F., Thuiller W., Mouquet N., 2010. Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity:the need for integrative conservation strategies in a changing world. Ecol. Lett, 13, 1030-1040.
|
||
Diniz-Filho J.A.F., Loyola R.D., Raia P., Mooers A.O., Bini L.M., 2013. Darwinian shortfalls in biodiversity conservation. Trends Ecol. Evol, 28, 689-695.
DOI:10.1016/j.tree.2013.09.003 |
||
Donoghue M.J., 2008. A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. U.S.A, 105, 11549-11555.
DOI:10.1073/pnas.0801962105 |
||
Faith D.P., 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv, 61, 1-10.
DOI:10.1016/0006-3207(92)91201-3 |
||
Faith D.P., 2008. Threatened species and the potential loss of phylogenetic diversity:conservation scenarios based on estimated extinction probabilities and phylogenetic risk analysis. Conserv. Biol, 22, 1461-1470.
DOI:10.1111/cbi.2008.22.issue-6 |
||
Forest F., Grenyer R., Rouget M., Davies T.J., Cowling R.M., Faith D.P., Balmford A., Manning J.C., Proches S., Van Der Bank M., Reeves G., Hedderson T.A.J., Savolainen V., 2007. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature, 445, 757-760.
DOI:10.1038/nature05587 |
||
Fraterrigo J.M., Wagner S., Warren R.J., 2014. Local-scale biotic interactions embedded in macroscale climate drivers suggest Eltonian noise hypothesis distribution patterns for an invasive grass. Ecol. Lett, 17, 1447-1454.
DOI:10.1111/ele.12352 |
||
González-Orozco C.E., Pollock L.J., Thornhill A.H., Mishler B.D., Knerr N., Laffan S.W., Miller J.T., Rosauer D.F., Faith D.P., Nipperess D.A., Kujala H., Linke S., Butt N., Külheim C., Crisp M.D., Gruber B., 2016. Phylogenetic approaches reveal biodiversity threats under climate change. Nat. Clim. Change, 6, 1110-1114.
DOI:10.1038/nclimate3126 |
||
Graham C.H., Fine P.V.A., 2008. Phylogenetic beta diversity:linking ecological and evolutionary processes across space in time. Ecol. Lett, 11, 1265-1277.
DOI:10.1111/ele.2008.11.issue-12 |
||
Heenan P.B., Millar T.R., Smissen R.D., McGlone M.S., Wilton A.D., 2017. Phylogenetic measures of neo-and paleo-endemism in the indigenous vascular flora of the New Zealand archipelago. Aust. Syst. Bot, 30, 124-133.
DOI:10.1071/SB17009 |
||
Kembel S.W., Hubbell S.P., 2006. The phylogenetic structure of a Neotropical forest tree community. Ecology, 87, S86-S99.
DOI:10.1890/0012-9658(2006)87[86:TPSOAN]2.0.CO;2 |
||
Kier G., Kreft H., Lee T.M., Jetz W., Ibisch P.L., Nowicki C., Mutke J., Barthlott W., 2009. A global assessment of endemism and species richness across island and mainland regions. Proc. Natl. Acad. Sci. U.S.A, 106, 9322-9327.
DOI:10.1073/pnas.0810306106 |
||
Klak C., Reeves G., Hedderson T., 2004. Unmatched tempo of evolution in Southern African semi-desert ice plants. Nature, 427, 63-65.
DOI:10.1038/nature02243 |
||
Kraft N.J.B., Godoy O., Levine J.M., 2015. Plant functional traits and the multidimensional nature of species coexistence. Proc. Natl. Acad. Sci. U.S.A, 112, 797-802.
DOI:10.1073/pnas.1413650112 |
||
Kreft H., Jetz W., 2010. A framework for delineating biogeographical regions based on species distributions. J. Biogeogr, 37, 2029-2053.
DOI:10.1111/jbi.2010.37.issue-11 |
||
Latham R.E., Ricklefs R.E., 1993. Global patterns of tree species richness in moist forest:energy-diversity theory does not account for variation in species richness. Oikos, 67, 325-333.
DOI:10.2307/3545479 |
||
Li R., Dao Z.L., Ji Y.H., Li H., 2007. A floristic study on the seed plants of the northern Gaoligong mountains in western Yunnan, China. Acta Bot. Yunnanica, 29, 601-615.
|
||
Li R., Kraft N.J.B., Yang J., Wang Y.H., 2015b. A phylogenetically informed delineation of floristic regions within a biodiversity hotspot in Yunnan, China. Sci.Rep, 5, 9396.
DOI:10.1038/srep09396 |
||
Li R., Kraft N.J.B., Yu H.Y., Li H., 2015a. Seed plant phylogenetic diversity and species richness in conservation planning within a global biodiversity hotspot in eastern Asia. Conserv. Biol, 29, 1552-1562.
DOI:10.1111/cobi.12586 |
||
Li X.H., Zhu X.X., Niu Y., Sun H., 2014. Phylogenetic clustering and overdispersion for alpine plants along elevational gradient in the Hengduan Mountains Region, southwest China. J. Systemat. Evol, 52, 280-288.
DOI:10.1111/jse.v52.3 |
||
Li X.W., Li J., 1997. The Tanaka-Kaiyong line-an important floristic line for the study of the flora of East Asia. Ann. Missouri Bot. Gard, 84, 888-892.
DOI:10.2307/2992033 |
||
Linder H.P., 2014. The evolution of African plant diversity. Front. Ecol. Evol, 2, 38.
|
||
López-Pujol J., Zhang F.M., Sun H.Q., Ying T.S., Ge S., 2011. Centres of plant endemism in China:places for survival or for speciation? J. Biogeogr, 38, 1267-1280.
DOI:10.1111/jbi.2011.38.issue-7 |
||
Lu L.M., Mao L.F., Yang T., Ye J.F., Liu B., Li H.L., Sun M., Miller J.T., Mathews S., Hu H.H., Niu Y.T., Peng D.X., Chen Y.H., Smith S.A., Chen M., Xiang K.L., Le C.T., Dang V.C., Lu A.M., Soltis P.S., Soltis D.E., Li J.H., Chen Z.D., 2018. Evolutionary history of the angiosperm flora of China. Nature, 554, 234-238.
DOI:10.1038/nature25485 |
||
Lyashevska O., Farnsworth K.D., 2012. How many dimensions of biodiversity do we need? Ecol. Indicat, 18, 485-492.
DOI:10.1016/j.ecolind.2011.12.016 |
||
Mace G.M., Purvis A., 2008. Evolutionary biology and practical conservation:bridging a widening gap. Mol. Ecol, 17, 9-19.
DOI:10.1111/mec.2008.17.issue-1 |
||
Mackey B.G., Berry S.L., Brown T., 2008. Reconciling approaches to biogeographic regionalization:a systematic and generic framework examined with a case study of the Australian continent. J. Biogeogr, 35, 213-229.
|
||
Mishler B.D., Knerr N., González-Orozco C.E., Thornhill A.H., Laffan S.W., Miller J.T., 2014. Phylogenetic measures of biodiversity and neo-and paleoendemism in Australian Acacia. Nat. Commun, 5, 4473.
DOI:10.1038/ncomms5473 |
||
Mittelbach G.G., Schemske D.W., 2015. Ecological and evolutionary perspectives on community assembly. Trends Ecol. Evol, 30, 241-247.
DOI:10.1016/j.tree.2015.02.008 |
||
Morin, N. R., Whetstone, R. D., Wilken, D., Tomlinson, K. L., 1989. Floristics for the 21st Century. Missouri Botanical Garden, St. Louis.
|
||
Moritz C., 2002. Strategies to protect biological diversity and the evolutionary process that sustain it. Syst. Biol, 51, 238-254.
DOI:10.1080/10635150252899752 |
||
Myers N., Mittermeier R.A., Mittermeier C.G., da Fonseca G.A.B., Kent J., 2000. Biodiversity hotspots for conservation priorities. Nature, 403, 853-858.
DOI:10.1038/35002501 |
||
Olson D.M., Dinerstein E., Wikramanayake E.D., Burgess N.D., Powell G.V.N., Underwood E.C., D'amico J.A., Itoua I., Strand H.E., Morrison J.C., Loucks C.J., Allnutt T.F., Ricketts T.H., Kura Y., Lamoreux J.F., Wettengel W.W., Hedao P., Kassem K.P., 2001. Terrestrial ecoregions of the world:a new map of life on earth. Bioscience, 51, 933-938.
DOI:10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2 |
||
Pennington R.T., Richardson J.E., Lavin M., 2006. Insights into the historical construction of species-rich biomes from dated plant phylogenies, neutral ecological theory and phylogenetic community structure. New Phytol, 172, 605-616.
DOI:10.1111/nph.2006.172.issue-4 |
||
Pillon Y., 2012. Time and tempo of diversification in the flora of New Caledonia. Bot.J. Linnean Soc, 170, 288-298.
DOI:10.1111/boj.2012.170.issue-3 |
||
Qian H., Jin Y., Ricklefs R.E., 2017a. Phylogenetic diversity anomaly in angiosperms between eastern Asia and eastern North America. Proc. Natl. Acad. Sci. U.S.A, 114, 11452-11457.
DOI:10.1073/pnas.1703985114 |
||
Qian H., Jin Y., Ricklefs R.E., 2017b. Patterns of phylogenetic relatedness of angiosperm woody plants across biomes and life-history stages. J. Biogeogr, 44, 1383-1392.
DOI:10.1111/jbi.2017.44.issue-6 |
||
Qian H., Sandel B., 2017a. Phylogenetic relatedness of native and exotic plants along climate gradients in California, USA. Divers. Distrib, 23, 1323-1333.
DOI:10.1111/ddi.2017.23.issue-11 |
||
Qian H., Sandel B., 2017b. Phylogenetic structure of regional angiosperm assemblages across latitudinal and climatic gradients in North America. Global Ecol.Biogeogr, 26, 1258-1269.
DOI:10.1111/geb.2017.26.issue-11 |
||
Qian H., Swenson N.G., Zhang J.L., 2013b. Phylogenetic beta diversity of angiosperms in North America. Global Ecol. Biogeogr, 22, 1152-1161.
DOI:10.1111/geb.2013.22.issue-10 |
||
Qian H., Zhang Y.J., Zhang J., Wang X.L., 2013a. Latitudinal gradients in phylogenetic relatedness of angiosperm trees in North America. Global Ecol. Biogeogr, 22, 1183-1191.
DOI:10.1111/geb.2013.22.issue-11 |
||
Richardson J.E., Pennington R.T., Pennington T.D., Hollingsworth P.M., 2001. Rapid diversification of a species-rich genus of Neotropical rain forest trees. Science, 293, 2242-2245.
DOI:10.1126/science.1061421 |
||
Ricklefs R.E., 1987. Community diversity:relative roles of local and regional processes. Science, 235, 167-171.
DOI:10.1126/science.235.4785.167 |
||
Ricklefs R.E., 2006. Evolutionary diversification and the origin of the diversityenvironment relationship. Ecology, 87, S3-S13.
DOI:10.1890/0012-9658(2006)87[3:EDATOO]2.0.CO;2 |
||
Ricklefs R.E., 2007. History and diversity:explorations at the intersection of ecology and evolution. Am. Nat, 170, S56-S70.
DOI:10.1086/519402 |
||
Scherson R.A., Thornhill A.H., Urbina-Casanova R., Freyman W.A., Pliscoff P.A., Mishler B.D., 2017. Spatial phylogenetics of the vascular flora of Chile. Mol.Phylogenet. Evol, 112, 88-95.
DOI:10.1016/j.ympev.2017.04.021 |
||
Schweiger O., Klotz S., Durka W., Kuhn I., 2008. A comparative test of phylogenetic diversity indices. Oecologia, 157, 485-495.
DOI:10.1007/s00442-008-1082-2 |
||
Slik J.W.F., Franklin J., Arroyo-Rodríguez V., Field R., Aguilar S., Aguirre N., Ahumada J., Aiba S.I., Alves L.F., et al., 2018. Phylogenetic classification of the world's tropical forests. Proc. Natl. Acad. Sci. U.S.A, 115, 1837-1842.
DOI:10.1073/pnas.1714977115 |
||
Strauss S.Y., Webb C.O., Salamin N., 2006. Exotic taxa less related to native species are more invasive. Proc. Natl. Acad. Sci. U.S.A, 103, 5841-5845.
DOI:10.1073/pnas.0508073103 |
||
Swenson N.G., 2011a. The role of evolutionary processes in producing biodiversity patterns, and the interrelationships between taxonomic, functional and phylogenetic biodiversity. Am. J. Bot, 98, 472-480.
DOI:10.3732/ajb.1000289 |
||
Swenson N.G., 2011b. Phylogenetic beta diversity metrics, trait evolution and inferring the functional beta diversity communities. PLoS One, 6, e21264.
DOI:10.1371/journal.pone.0021264 |
||
Swenson N.G., 2013. The assembly of tropical tree communities-the advances and shortcomings of phylogenetic and functional trait analyses. Ecography, 36, 264-276.
DOI:10.1111/j.1600-0587.2012.00121.x |
||
Swenson N.G., Umaña M.N., 2014. Phylofloristics:an example from the lesser antilles. J. Plant Ecol, 7, 166-175.
DOI:10.1093/jpe/rtt074 |
||
Takhtajan A., 1986. Floristic Regions of the World. University of California Press, Berkeley. |
||
Thornhill A.H., Baldwin B.G., Freyman W.A., Nosratinia S., Kling M.M., MoruetaHolme N., Madsen T.P., Ackerly D.D., Mishler B.D., 2017. Spatial phylogenetics of the native California flora. BMC Biol, 15, 96.
DOI:10.1186/s12915-017-0435-x |
||
Thornhill A.H., Mishler B.D., Knerr N.J., González-Orozco C.E., Costion C.M., Crayn D.M., Laffan S.W., Miller J.T., 2016. Continental-scale spatial phylogenetics of Australian angiosperms provides insights into ecology, evolution and conservation. J. Biogeogr, 43, 2085-2098.
DOI:10.1111/jbi.12797 |
||
Thuiller W., Lavergne S., Roquet C., Boulangeat I., Lafourcade B., Araujo M.B., 2011. Consequences of climate change on the tree of life in Europe. Nature, 470, 531-534.
DOI:10.1038/nature09705 |
||
Tucker C.M., Cadotte M.W., 2013. Unifying measures of biodiversity:understanding when richness and phylogenetic diversity should be congruent. Divers. Distrib, 19, 845-854.
DOI:10.1111/ddi.2013.19.issue-7 |
||
Tucker C.M., Cadotte M.W., Davies T.J., Rebelo A.G., 2012. Incorporating geographical and evolutionary rarity into conservation prioritization. Conserv.Biol, 26, 593-601.
DOI:10.1111/cobi.2012.26.issue-4 |
||
Verboom G.A., Archibald J.K., Bakker F.T., Bellstedt D.U., Conrad F., Dreyer L.L., Forest F., Galley C., Goldblatt P., Henning J.F., Mummenhoff K., Linder H.P., Muasya A.M., Oberlander K.C., Savolainen V., Snijmanm D.A., van der Niet T., Nowell T.L., 2009. Origin and diversification of the Greater Cape flora:ancient species repository, hot-bed of recent radiation, or both? Mol. Phylogenet. Evol, 51, 44-53.
DOI:10.1016/j.ympev.2008.01.037 |
||
Wang H., Harrison S.P., Prentice I.C., Yang Y., Bai F., Togashi H.F., Wang M., Zhou S., Ni J., 2018. The China Plant Trait Database:toward a comprehensive regional compilation of functional traits for land plants. Ecology, 99, 500.
DOI:10.1002/ecy.2091 |
||
Wang H.S., 1992. Floristic Geography. Science Press, Beijing. |
||
Webb C.O., 2000. Exploring the phylogenetic structure of ecological communities:an example for rain forest trees. Am. Nat, 156, 145-155.
DOI:10.1086/303378 |
||
Webb C.O., Ackerly D.D., Kembel S.W., 2008. Phylocom:software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics, 24, 2098-2100.
DOI:10.1093/bioinformatics/btn358 |
||
Webb C.O., Ackerly D.D., McPeek M.A., Donoghue M.J., 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Evol. Syst, 33, 475-505.
DOI:10.1146/annurev.ecolsys.33.010802.150448 |
||
Wiens J.J., Donoghue M.J., 2004. Historical biogeography, ecology, and species richness. Trends Ecol. Evol, 19, 639-644.
DOI:10.1016/j.tree.2004.09.011 |
||
Winter M., Devictor V., Schweiger O., 2013. Phylogenetic diversity and nature conservation:where are we? Trends Ecol. Evol, 28, 199-204.
|
||
Wu Z.Y., 1965. On the tropical affinities of Chinese flora. Chin. Sci. Bull, 1, 25-33.
|
||
Wu Z.Y., 1979. The regionalization of Chinese flora. Acta Bot. Yunnanica, 1, 1-22.
|
||
Wu Z.Y., 1983. On the significance of Pacific intercontinental discontinuity. Ann.Missouri Bot. Gard, 70, 577-590.
DOI:10.2307/2398977 |
||
Wu Z.Y., 1991. Areal-types of Chinese genera of seed plants. Acta Bot. Yunnanica(Ⅳ), 1-139.
|
||
Wu Z.Y., Sun H., Zhou Z.K., Li D.Z., Peng H., 2010. Floristics of Seed Plants from China. Science Press, Beijing. |
||
Wu Z.Y., Sun H., Zhou Z.K., Peng H., Li D.Z., 2005. Origin and differentiation of endemism in the flora of China. Acta Bot. Yunnanica, 27, 577-604.
|
||
Wu Z.Y., Zhou Z.K., Li D.Z., Peng H., Sun H., 2003. The areal-types of the world families of seed plants. Acta Bot. Yunnanica, 25, 245-257.
|
||
Wu Z.Y., Zhou Z.K., Sun H., Li D.Z., Peng H., 2006. The Areal-types of Seed Plants and Their Origin and Differentiation. Yunnan Science and Technology Press, Kunming. |
||
Xing Y.W., Ree R.H., 2017. Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A, 114, E3444-E3451.
DOI:10.1073/pnas.1616063114 |
||
Zhang D.C., Ye J.X., Sun H., 2016. Quantitative approaches to identify floristic units and centres of species endemism in the Qinghai-Tibetan plateau, south-western China. J. Biogeogr, 42, 2465-2476.
|



