b. University of Chinese Academy of Sciences, Beijing 100049, China
Cytosine DNA methylation represents a major molecular mechanism that provides critical genomic information and contributes to understand the molecular basis of phenotypic variation based on epigenetic modification in eukaryotic organisms, though the levels and patterns of cytosine DNA methylation appear to vary drastically among different organisms (Avramidou et al., 2015; Okano et al., 1999). Cytosine-5-methylation is the most common DNA modification, which transfers a methyl group from S-adenosyl-L-methionine to fifth carbon of cytosine residues, extensively occurring in eukaryote genomes (Finnegan et al., 2000). In plants, DNA methylation generally refers to three different nucleotide sequence contexts: CG, CHG and CHH methylation (where H = C, T or A) (Gruenbaum et al., 1981; Law and Jacobsen, 2010). Many studies have showed that DNA methylation might play indispensable roles in regulating diverse physiological processes during plant growth and development, including coordination of gene expression to adapt environmental changes (Zhang et al., 2016), involvement in chromatin organization (Chinnusamy and Zhu, 2009), and maintenance of genomic integrity (Wang et al., 2016). Recent studies have showed that environmental stimuli (including biotic and abiotic stresses) can induce changes of genomic DNA methylation (Dowen et al., 2012; Garg et al., 2015), and such environmentally induced methylation can be heritable, which may result in a re-shaping of phenotypes in next generations (Herman and Sultan, 2016). Particularly, the combination of genotype-specific environmental sensitivity, DNA methylation and transgenerational stability, and phenotypic impacts might be a primary candidate mechanism for adaptive evolution in plants (Alsdurf et al., 2013; Herman and Sultan, 2016; Zhang et al., 2013). Investigation of the inherent relationships of phenotypic and DNA methylation variations might be a good way to dissect this potential mechanism for plant adaptive evolution.
Castor bean (Ricinus communis L., Euphorbiaceae), a typical nonedible oilseed crop, displays rich phenotypic variations within the intra-species. Owing to its seed oils (made of high ricinoleic acids) are often used in aviation oil, lubricants, nylon, dyes, inks, soaps, adhesive, biodiesel and so on, with its high economic value. Castor bean is widely cultivated in tropical, sub-tropical and temperate countries, particularly India, China and Brazil (Akpan et al., 2006; Alexandrov and Karlov, 2016; Sujatha et al., 2008; Webster, 1994). Because of the limited domestication, wide seed dissemination in the ancient time, and the ease and rapidity with which it becomes established in margin lands, landraces of castor bean germplasm occur in the warm regions worldwide with rich phenotypic variations in its growth habit, color of foliage and stems, seed size and oil content (Wolf, 2000). Most types are large perennials that often develop into small trees in tropical or subtropical areas; however it is usually shorter and smaller and grown annually in areas prone to frost. It is obvious that castor bean exhibits great phenotypic diversity and phenotypic plasticity to environmental factors (Qiu et al., 2010). However, previous studies have showed that the genetic diversity of castor bean appears relatively low, based on AFLP, SSR and SNP studies (Allan et al., 2008; Foster et al., 2010; Qiu et al., 2010). The genetic or molecular basis of phenotypic variation and plasticity remains unknown. Epigenetic variations may create a potential power to drive the phenotypic diversity and plasticity in castor bean.
As mentioned above, DNA methylation is the main type of epigenetic regulation. The methylation-sensitive amplification polymorphism (MSAP) technique, a modified AFLP (amplified fragment length polymorphism) method, has been well developed for determining the global DNA methylation level without any prior knowledge genomic sequences (Reyna-López et al., 1997; Vos et al., 1995; Xu et al., 2000). Owing to the two methylation-sensitive restriction enzymes HpaII and MspI, which replaces restriction enzymes used in AFLP, HpaII and MspI can recognize unmethylated tetranucleotide sequence 5'-CCGG-3' loci, but be sensitive to different methylation contexts. Specifically, HpaII can recognize and cleave the hemi-methylated sequence (mCCGG, only one DNA strand methylated), whereas MspI can recognize and cleave fully methylated CCGG sequence (CmCGG, methylation of both DNA strands). Thus, different methylation status at the cytosines of the CCGG sites could be recognized by the different amplification products obtained from EcoRI–HpaII and EcoRI–MspI enzyme digestion files (Lee et al., 1994; Walder et al., 1983; Xu et al., 2000). Because of its advantages including low cost, operation easy, high sensitivity and high polymorphism, MSAP technique has become a powerful and highly informative tool and been extensively applied to detect global DNA methylation level in plants in last two decades. These studies were mainly focused on detecting the DNA methylation inheritance such as in maize and rice (Lauria et al., 2004; Xiong et al., 1999; Zhao et al., 2007); investigating the epigenetic diversity at the population level such as in mangrove plants, cocoa, white polar and violet (Herrera and Bazaga, 2010; López and Wilkinson, 2010; Liramedeiros et al., 2010; Ma et al., 2013); inspecting the global DNA methylation variation with biotic or abiotic stresses such as in rice, wheat, cotton and rapeseed (Dai et al., 2015; Hong et al., 2013; Marconi et al., 2013; Zhong et al., 2009). However, there were relatively few studies on investigating the inherent relationships between phenotypic diversity and DNA methylation variation within intra-species. In this study, the global DNA methylation variations among the sixty castor bean accessions, which exhibited rich phenotypic diversity and were collected worldwide, were investigated by using MSAP method in order to explore relationships between phenotypic diversity and DNA methylation variation. This study provides new data in understanding the formation of phenotypic diversity and its epigenetic basis in castor bean.
2. Material and methods 2.1. Sample collection and DNA extractionSixty worldwide accessions of castor bean were collected from the USDA National Plant Germplasm System (http://www.ars-grin.gov/npgs) and our collected landraces in China, which covered the 29 countries. The detailed information for each accession used in this study is listed in Table 1. Meanwhile, we measured the hundred-grain weight and oil content of castor bean seeds for each accession by minispec mq-one seed-analyzer (Bruker Optik GmbH, Germany).
| No | PI | Country | Oil content (%) | Hundred-grain weight (g) | No. | PI | Country | Oil cont–nt (%) | Hundred-grain weight (g) |
| 1 | 206515 | Jamaica (Ja) | 48.803 | 36.56 | 31 | 250027 | Iran (IR-2) | 52.813 | 21.36 |
| 2 | 208689 | Algeria (AG) | 49.188 | 33.16 | 32 | 250397 | Pakistan (PK) | 51.079 | 20.36 |
| 3 | 209326 | Virgin Islands (U.S.) (VI) | 48.313 | 19.02 | 33 | 250743 | Iran (IR-4) | 52.847 | 22.48 |
| 4 | 209436 | Panama (Pa) | 48.731 | 18.94 | 34 | 250876 | Iran (IR-5) | 52.974 | 19.20 |
| 5 | 215774 | Peru (Pe) | 49.636 | 28.82 | 35 | 274769 | South Africa (SA-1) | 51.100 | 24.44 |
| 6 | 219773 | Argentina (Ar) | 51.988 | 26.90 | 36 | 323589 | Portugal (Po-1) | 51.220 | 38.72 |
| 7 | 240674 | Uruguay (Ur) | 47.115 | 15.89 | 37 | 631156 | USA (USA) | 50.969 | 29.20 |
| 8 | 248502 | South Africa (SA) | 50.412 | 29.51 | 38 | 203661 | Paraguay (Pr-1) | 47.772 | 17.90 |
| 9 | 248929 | India (IN) | 44.169 | 22.77 | 39 | 208842 | Cuba (Cu-1) | 48.297 | 15.76 |
| 10 | 248935 | India (IN-1) | 44.268 | 22.10 | 40 | 219774 | Argentina (Ar-2) | 51.724 | 30.14 |
| 11 | 250881 | Iran (IR) | 44.083 | 21.96 | 41 | 248968 | India (IN-3) | 52.180 | 34.44 |
| 12 | 265509 | Colombia (Co) | 51.399 | 27.32 | 42 | 248441 | Zaire (ZA-1) | 46.759 | 26.92 |
| 13 | 271861 | Ecuador (Ec) | 50.004 | 31.20 | 43 | 250575 | Egypt (EG) | 50.772 | 20.16 |
| 14 | 298600 | Brazil (Br-1) | 51.142 | 26.64 | 44 | 258368 | Former Soviet Union (SU-1) | 52.340 | 29.24 |
| 15 | 167342 | Turkey (TR) | 49.812 | 40.23 | 45 | 274772 | South Africa (SA-2) | 49.031 | 19.92 |
| 16 | 176751 | Turkey (TR-1) | 50.624 | 14.77 | 46 | 181916 | Syria (SY) | 48.216 | 15.64 |
| 17 | 180335 | India (IN-5) | 46.156 | 15.33 | 47 | 201830 | Madagascar (MD) | 44.633 | 24.12 |
| 18 | 183468 | India (IN-6) | 48.271 | 16.40 | 48 | 212115 | Afghanistan (AF) | 50.161 | 24.92 |
| 19 | 202711 | Brazil (Br-2) | 49.035 | 19.81 | 49 | 223408 | Iran (IR-1) | 49.727 | 16.98 |
| 20 | 203126 | India (IN-7) | 51.246 | 32.01 | 50 | 240310 | Benin (BE) | 49.606 | 31.27 |
| 21 | 203128 | India (IN-8) | 51.406 | 31.96 | 51 | 248424 | Zaire (ZA-2) | 48.944 | 30.52 |
| 22 | 208839 | Cuba (Cu) | 48.758 | 15.56 | 52 | 253620 | Morocco (MA-1) | 49.769 | 33.12 |
| 23 | 209132 | Puerto Rico (PR) | 49.904 | 15.75 | 53 | – | Pakistan (PK-1) | – | – |
| 24 | 215770 | Peru (Pe-1) | 51.887 | 44.98 | 54 | – | Pakistan (PK-2) | – | – |
| 25 | 219772 | Argentina (Ar-1) | 52.046 | 27.96 | 55 | – | Pakistan (PK-3) | – | – |
| 26 | 240673 | Uruguay (Ur-1) | 60.622 | 17.25 | 56 | 257445 | South Africa (SA-3) | 50.187 | 31.20 |
| 27 | 241368 | Brazil (Br-3) | 51.005 | 28.62 | 57 | 176751 | Turkey (TR-4-1) | 50.624 | 14.77 |
| 28 | 243062 | Brazil (Br-4) | 49.153 | 19.57 | 58 | – | ZB306 (CN) | – | – |
| 29 | 248937 | India (IN-9) | 44.621 | 24.47 | 59 | – | ZB107 (CN-1) | – | – |
| 30 | 248938 | India (IN-10) | 53.173 | 27.66 | 60 | – | Hale (USA-1) | – | – |
| – Data missing. | |||||||||
All seeds were germinated at greenhouse and the leaves of three-leaf stage seedlings were collected. High quality genomic DNA of each individual was then extracted using the Plant Genomic DNA Kit (TianGen, Beijing, China). The DNA quality was confirmed by 0.8% agarose gel electrophoresis, and the DNA concentration was estimated by NanoDrop2000 (Thermo, USA).
2.2. Primer screening and MSAP analysisIn this study, MSAP technique was used to investigate epigenetic polymorphism of different castor bean germplasms (Reyna-Lđpez et al., 1997). Briefly, high-quality genomic DNA was digested with two sets of restriction enzyme combinations, EcoRI/MspI and EcoRI/HpaII. The digestion reaction mix contained 1 ml genomic DNA, 10 × buffer, 8 U EcoRI, 16 U MspI or HpaII (New England Biolabs, Beverly, MA, USA). The digestion reaction was carried out at 37 ℃ for 9 h and ended at 70 ℃ for 10 min. Subsequently, the 4 ml digested DNA fragments were ligated to the specific adapters in a reaction mix containing 5 pmol EcoRI adapter, 50 pmol MspI/HpaII adapter, 2 ml ligase buffer and 1 U T4 DNA ligase (New England Biolabs, Beverly, MA, USA) that was incubated at 16 ℃ for 10 h.
The 2 ml ligated DNA was used as template for pre-amplification in a reaction mix containing 2.5 ml 10 × PCR buffer, 2 ml dNTP (2.5 mmol/ml), 1.5 ml Mgcl2 (25 mmol/ml), 1 ml pre-selective primer (see Table 2), and 1 U of Taq Polymerase (Takara Biotech. Inc., Dalian). The PCR conditions were following profile: 94 ℃ for 3 min, 26 cycles at 94 ℃ for 30 s, 30 s annealing at 56 ℃ and 60 s extension at 72 ℃, final extension at 72 ℃ for 5 min. The diluted (40-fold) amplified products were used as the template for selective amplification. A total volume of 25 ml PCRs were carried out with 20 selective primer combinations (see Table 2). The touch-down PCR conditions were performed and reaction conditions were following profile: 94 ℃ for 3 min followed by 12 cycles of 94 ℃ for 30 s, 65 ℃ for 30 s (reduced 0.7 ℃ per cycle), and 72 ℃ for 1 min, and a subsequent 23 cycles of 94 ℃ for 30 s, 56 ℃ for 30 s, and 72 ℃ for 1 min. A fluorescence-based MSAP technique was used for fragment separation and detection by using an ABI3730 × l DNA analyzer (Applied Biosystems, Milano, IT). The fragments were scored and translated to binary data (0 for absence and 1 for presence) in the EcoRI/MspI and EcoRI/HpaII samples by GeneMarker 2.2.0 software. Meanwhile, for polymorphic primers, selective amplification products were separated on 6% polyacrylamide gels at 180 V for 1.5 h, and then the gels were stained with silver nitrate.
| Adapter/primer | Sequence |
| EcoRI adapter | 5'-CTCGTAGACTGCGTACC-3' |
| 5'-AATTGGTACGCAGTC-3' | |
| HpaII/MspI adapter | 5'-GATCATGAGTCCTGCT-3' |
| 5'-CGAGCAGGACTCATGA-3' | |
| E + A | 5'-GACTGCGTACCAATTCA-3' |
| HM + G | 5'-ATCATGAGTCCTGCTCGG-3' |
| H/M1 | 5'-ATCATGAGTCCTGCTCGGTCT-3' |
| H/M2 | 5'-ATCATGAGTCCTGCTCGGTCG-3' |
| H/M3 | 5'-ATCATGAGTCCTGCTCGGTCC-3' |
| H/M4 | 5'-ATCATGAGTCCTGCTCGGTTC-3' |
| H/M5 | 5'-ATCATGAGTCCTGCTCGGTTG-3' |
| H/M6 | 5'-ATCATGAGTCCTGCTCGGTTA-3' |
| H/M7 | 5'-ATCATGAGTCCTGCTCGGTGT-3' |
| H/M8 | 5'-ATCATGAGTCCTGCTCGGTGC-3' |
| H/M9 | 5'-ATCATGAGTCCTGCTCGGTAC-3' |
| E10 | 5'-GACTGCGTACCAATTCAAC-3' |
| E11 | 5'-GACTGCGTACCAATTCAAG-3' |
| E12 | 5'-GACTGCGTACCAATTCACA-3' |
| E13 | 5'-GACTGCGTACCAATTCACG-3' |
| E14 | 5'-GACTGCGTACCAATTCAGC-3' |
| E15 | 5'-GACTGCGTACCAATTCAGG-3' |
| E16 | 5'-GACTGCGTACCAATTCAGA-3' |
| E17 | 5'-GACTGCGTACCAATTCACC-3' |
According to different sensitivities to methylation of restriction enzyme HpaII and MspI to the CCGG sequence, the methylation states could be classified into following four basic types based on previous reports (Portis et al., 2004): Type Ⅰ pattern represents the non-methylation sites (E-H/E-M, 1/1), Type Ⅱ pattern represents the hemi-methylation sites (E-H/E-M, 1/0), Type Ⅲ pattern represents full-methylation sites (E-H/E-M, 0/1), and Type Ⅳ pattern represents the uninformative sites (E-H/E-M, 0/0).
For each primer, the well-resolved bands were scored as absence (0) and presence (1) with same molecular weight to form a binary matrix. Epigenetic diversity parameters, including the percentage of polymorphic loci (PPB), the expected heterozygosity (He), Shannon's Information Index (I), the effective number of alleles per locus (Ne) were estimated and analyzed by using the GenAlEx version 6.501 software (Rod and Petere, 2006). Based on the binary matrix, the Nei's genetic distance matrix was calculated by DPS (Data Processing System) software (Vision 7.05), and then the genetic distance matrix was transferred to MEGA6 software (Tamura et al., 2013) to construct the phylogenetic tree with neighbor-joining method.
To determine population differentiation, we used the STRUCTURE program which employs a Bayesian algorithm to calculate the number of populations. The number of the MCMC reps was set to 100, 000 and the length of the burn-in period was set to 50, 000 steps. In order to estimate the appropriate number of clusters (K), ΔK was calculated by the method of Evanno et al. (2005) with ten runs for each K ranging from K = 1 to K = 9.
2.4. Sequencing and annotation of polymorphic MSAP fragmentsSome bands showing differential DNA methylation level among castor bean accessions were selected and excised from polyacrylamide gel. The bands were rehydrated with 20 ml of sterile distilled water overnight at 4 ℃. The soluble DNA samples (about 2 ml) were used as PCR template for re-amplified by using the corresponding selective amplification primers and PCR conditions. Re-amplified products were checked by agarose gel electrophoresis, and were purified by using the Gel Extraction Kit (GENEray, China). The purified DNA fragments were then cloned into pMD19-T cloning vector (Takara Biotech. Inc., Dalian), and then transferred into Escherichia coli. Trans1-T1 (TransGen Biotech, China). Positive clones that were confirmed by colony PCR, were selected and sequenced. The obtained sequence information was BLASTN searched against the castor bean genome database including nuclear, mitochondrial and chloroplast genome in NCBI (http://www.ncbi.nlm.nih.gov/).
3. Results 3.1. The DNA methylation level and epigenetic diversity of castor bean germplasmsTo obtain the efficient and polymorphic primers, we initially screened PCR products for 8 random selected DNA samples using polyacrylamide gel electrophoresis. Initially, seventy-two EcoRI/ HpaII and EcoRI/MspI primer combinations were tested. The primer combinations which can produce arbitrary, clear and reproductive PCR bands were sort out for further analysis. Finally, twenty primer combinations were determined to test global DNA methylation variation of all accessions. Since the EcoRI primers were fluorescent-labeled, the PCR products were accurately analyzed with ABI3730 × l DNA analyzer. To exclude the noise peaks, the fragment size of PCR products out of the ranging from 50 bp to 500 bp was excluded for the further analysis.
In total, the 20 primer sets produced the reproducible PCR product fragments from the 60 accessions tested, ranging from 123 to 292 fragments among different landraces. The total number of bands ranged from small to large in order according to the size of bands, and these polymorphic fragments obtained from PCR products were combined into a binary matrix to indicate the presence (1) or absence (0) of the fragments in EcoRI/HpaII and EcoRI/MspI digestion profiles. According to the MSAP profiles, the total number of non-methylated sites (CCGG, double-strand; 1, 1), hemi-methylated sites (mCCGG, only one strand methylated; 1, 0) and fully methylated CCGG sites (CmCGG, double-strand methylated; 0, 1) were counted respectively. The relative total levels of non-methylation or methylation were calculated as the percentages non-methylation or methylation sites (includes the total number of hemi-methylated sites and fully methylated sites) at the total number of all bands. The total level of DNA methylation ranging from 3.80% to 34.31% of sixty castor bean accessions, of which the fully methylation level ranging from 3.80% to 32.31%. The methylation level of castor bean came from Cuba (Cu-1) was 3.80% and castor bean of Uruguay (Ur) was the highest (34.31%) in all castor bean germplasms. This indicated that DNA methylation level varies greatly among different landraces. In addition, our results showed that the fully methylated ratios (Type Ⅲ, methylation of the internal cytosines on both strands) were higher than the corresponding hemi-methylation levels (Type Ⅱ, hemi-methylation on the external cytosine) of all castor bean germplasms that we adopted.
In order to assess the epigenetic diversity of different castor bean germplasms, the binary matrix (zero and one) was subject to the estimation of the basic parameters including Na (No. of Different Alleles), Ne (No. of Effective Alleles), I (Shannon's Information Index), He (Expected Heterozygosity), uHe (Unbiased Expected Heterozygosity) and the PPB (the percentage of polymorphic loci) using the GenAlEx software. As a result, we found that the value of PPB was 69.07%, and the values of Na, Ne and uHe were 1.691, 1.395 and 0.244, respectively. The average I per locus was 0.366 ranging from 0.389 to 0.693, and the average of He value per locus was 0.242 ranging from 0.218 to 0.498.
3.2. Population structure analysisSTRUCTURE is a Bayesian and model-based algorithm which was used for clustering genetic data analysis. Given the number of clusters (K) and assuming Hardy–Weinberg and linkage equilibrium within clusters, STRUCTURE estimates allele frequencies in each cluster and population memberships for every individual (Hubisz et al., 2009). Here, the Structure software was used for estimating the appropriate number of clusters (K) and inferring the population structure. The admixture model was described by Evanno et al. (2005), the most optimal number of cluster for the whole dataset was determined by the highest ΔK value, whose corresponding the value of K was regarded as the optimal number of cluster. The result of structure analysis for the sixty samples yielded a highest ΔK value (ΔK = 333.625) which showed a clear peak at the K value of 2 (Fig. 1B). Thus, sixty castor bean germplasms were properly divided into two clusters, 33 castor bean accessions were included in the first cluster (Fig. 1C, the red color), collected from African, America and Asia across the main collection regions. The second cluster consisted of the 27 castor bean accessions (Fig. 1C, the green color), collected from the countries of America, Asia, Africa and Europe, similarly across the main collection regions.

|
| Fig. 1 The results of structure analysis. (A) The mean of the log likelihood of the given each K (10 replicates) and (B) the distribution of ΔK. (C) Results for two to four clusters as detected by STRUCTURE software, the values of K ranging from 2 to 4. |
Further, using the neighbor-joining criteria we constructed a phylogenetic tree based on the Nei's genetic distance matrix data. As showed in Fig. 2, the sixty accessions were clustered as two distinct clades (Clade Ⅰ and Clade Ⅱ). The Clade Ⅰ was comprised of 16 germplasms collected from Africa, South America, North America, and Asia; The Clade Ⅱ included 44 accessions collected across the collection regions. In comparison with the results obtained from the population structure analysis (Fig. 1), the two main groups were consistent though the number of members within each group were slight different. While inspecting the geographical distribution of each accession within the two groups, it was clear that there was no distinct geographic structure among castor bean.
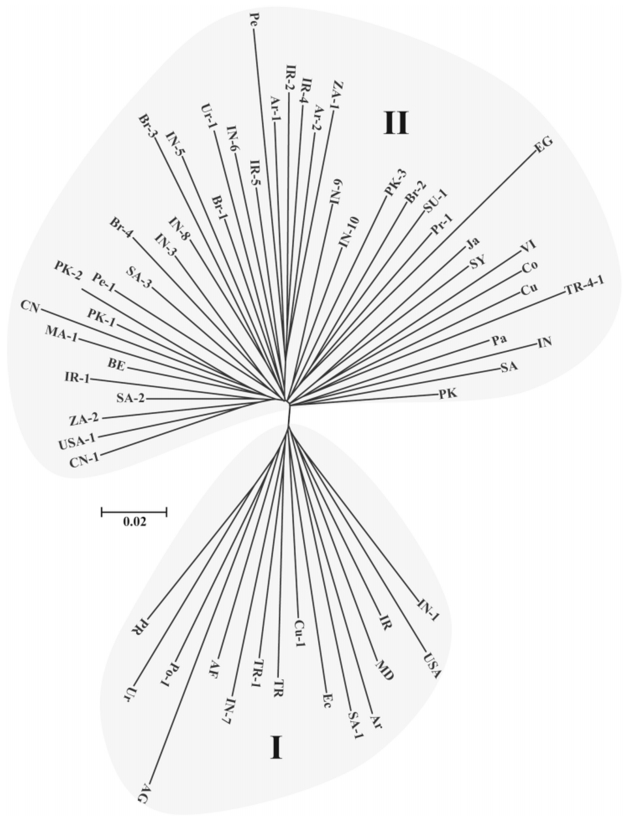
|
| Fig. 2 Phylogenetic tree of sixty castor bean accessions from MSAP data. |
To investigate which loci or genes on genome resulted in the methylation polymorphism, we randomly inspect more than 20 polymorphic fragments by separating the PCR polymorphic bands from the polyacrylamide gels and clone-sequencing to obtain the DNA sequences of polymorphic bands. In total, we successfully obtained 13 DNA sequences, ranging from 48 bp to 582 bp in length (in Table 3). After the BLASTN search in NCBI database, we found that the 13 DNA fragments were successfully annotated as four distinct regions of castor bean genomes. We denoted these DNA fragments as P1–P4 region as shown in Table 3. The P1 region was identified as a fragment located in downstream 600 bp of nad2 gene in the castor bean mitochondrion genome (HQ874649.1); the P2 region including ten DNA fragments was annotated as the ndhB gene (NADH dehydrogenase subunit 2) in chloroplast genome (JF937588.1), functionally involving photomorphogenesis (Catalá et al., 1997; Guéra et al., 2000); the P3 and P4 were identified as uncharacterized mRNAs with unknown function in nuclear genome. These results indicated that the polymorphic methylated loci occur widely in nuclear, chloroplast and mitochondrion genomes.
| Name | Primer combinations | ails and resu | Gene ID | Homology | E-value |
| P1 | E16/H6 | Size (bp) | HQ874649.1 | Ricinus communis mitochondrion genome | 0.0 |
| P2 | E11/H9 | 582 | XM_002530870.2 | Ricinus communis uncharacterized LOC8268543 (LOC8268543), mRNA | 9e–37 |
| P3 | E16/H9 | 89 | JF937588.1 | Ricinus communis chloroplast, complete genome | 0.0 |
| P4 | E16/H3 | 562 | XR_001535477.1 | Ricinus communis uncharacterized LOC107261565 (LOC107261565), transcript variant X2, ncRNA | 1e–13 |
DNA methylation is an ancient epigenetic modification that appears in the genomes of organisms throughout the eukaryotic phylogeny (Feng et al., 2010; Suzuki and Bird, 2008). Increasing evidence have shown that genomic DNA methylation contribute to heritable phenotypic variation (Das and Messing, 1994; Liu et al., 2004). As mentioned above, castor bean germplasms displayed rich phenotypic diversity, but previous studies had revealed that the genetic diversity among germplasms is relatively low (Allan et al., 2008; Foster et al., 2010; Qiu et al., 2010). Why castor bean exhibits the rich phenotypic diversity with the low genetic diversity remained uncertain. This study is the first report on natural epigenetic variation occurring in castor bean germplasms. Our results revealed that the genomic DNA methylation of castor bean exhibited a considerable variation among germplasms. About ten times divergence of DNA methylation level was identified between the lowest and the highest germplasms. Although these germplasms were collected from diverse nature environment, since the germplasms were collected worldwide and germinated and grown under the same environment (the greenhouse), the exhibited difference of genomic DNA methylation is supposed to be genetically evolved, not resulting from the environmental heterogeneity. Besides, leave samples used in this study were collected at the same developmental stage so that any developmental variation in methylation, to the extent that it occurs within leaves, would not confound our results to determine genotype-specific variation in methylation patterns. Although based on current study whether the divergent DNA methylation levels occurring among castor bean germplasms is a driving power to shape the rich phenotypic diversity in castor bean remains unclear yet, the considerable variation of genomic DNA methylation might be a potential clue to dissect the molecular basis of phenotypic variation among germplasms. Using the same MSAP methods, previous studies have shown that 30–41% of loci were methylated in Brassica oleraceae accessions (Salmon et al., 2008), 32% of loci were methylated in Gossypium hirsutum accessions (Keyte et al., 2006), and 35–43% of loci were methylated in Arabidopsis thaliana ecotypes (Cervera et al., 2003). The degree of cytosine methylation may be expected to be even higher since some methylated sites cannot be detected with the MSAP technique (Sáez-Laguna et al., 2014), because detection is restricted to the recognition sites of the isoschizomers used (Xiong et al., 1999).
In this study, based on the epigenetic differentiation the two main clusters were identified. Compared to the previous study that investigated the genetic diversity among castor bean germplasms using EST–SSR markers and resulted in five distinct clusters (Qiu et al., 2010), it seems clear that the epigenetic differentiation is not dependent on genetic differentiation among castor bean germplasms. Probably, this divergence of epigenetic and genetic variations might be due to the different molecular mechanisms underlying the epigenetic and genetic variations occur. Interestingly, Rivarola et al. (2011) identified two main genetic clusters based on partial chloroplast genomic SNPs in castor bean germplasms. However, because of the limited samples tested in the study of Rivarola et al. and the different sample strategy from this study we could not figure out the relationships of epigenetic differentiation and genetic variations in castor bean. Combining our current study and with other reports (Kanchanaketu et al., 2012; Shan et al., 2012), it seems to be unexpected to learn that genetic and epigenetic diversity is not always linked together. In addition, the previous studies based on genetic variations revealed that there were no distinct geographic groups in castor bean germplasms (Allan et al., 2008; Qiu et al., 2010). Also, the current results indicated that epigenetic differentiation did not display a distinct geographic structure. This result is understandable because most of castor bean germplasms were multi-introduced across several continents with human activities, although castor bean landraces did not cultivated in many regions.
In addition, thirteen methylated DNA fragments were isolated from polymorphic methylated loci among castor bean accessions, and were mapped into four distinct regions on castor bean genomes (including nucleus, mitochondrion and chloroplast) in this study. This might be related to tissue-specific DNA methylation because our tested samples are young leaf tissues. Interestingly, ten DNA fragments with different length were resided in the same region of chloroplast genome and were identified as the gene ndhB, suggesting that this region or the gene ndhB was sensitive to DNA methylation among natural population. Although there were studies that found the DNA hypomethylation of chloroplast genome in Arabidopsis (Leutwiler et al., 1984), the DNA methylation degrees of chloroplast genomes might be variable among different plants because of some former reports on DNA methylation variations of chloroplast genome in other plants (Kobayashi et al., 1990; Ohta et al., 1991; Takio et al., 1994). The other two methylated loci were identified from genome as uncharacterized mRNA and non-coding RNA. Although limited polymorphic methylated loci were here identified, it seems to imply that polymorphic methylated loci occur widely, involving nuclear, mitochondrion and chloroplast genomes.
AcknowledgmentsWe thank Dr. Yupeng Geng from Yunnan University for his support and Ms. Ruiwen Li from Yunnan University for her assistance with the polyacrylamide gel electrophoresis experiment. This work was jointly supported by Chinese National Key Technology R & D Program (2015BAD15B02) and National Natural Science Foundation of China (31661143002 and 31501034).
Appendix A. Supplementary dataSupplementary data related to this article can be found at http://dx.doi.org/10.1016/j.pld.2017.05.007.
Akpan U.G., Jimoh A., Mohammed A.D., 2006. Extraction, characterization and modification of castor seed oil. Leonardo J. Sci, 5, 39417-39424.
|
||
Alexandrov O.S., Karlov G.I., 2016. Molecular cytogenetic analysis and genomic organization of major DNA repeats in castor bean (Ricinus communis L.). Mol.Genet. Genomics, 291, 775-787.
DOI:10.1007/s00438-015-1145-0 |
||
Allan G., Williams A., Rabinowicz P.D., Chan A.P., Ravel J., Keim P., 2008. Worldwide genotyping of castor bean germplasm (Ricinus communis L.) using AFLPs and SSRs. Genet. Resour. Crop Evol, 55, 365-378.
DOI:10.1007/s10722-007-9244-3 |
||
Alsdurf J.D., Ripley T.J., Matzner S.L., Siemens D.H., 2013. Drought-induced transgenerational tradeoff between stress tolerance and defence: consequences for range limits?. AoB Plants, 5, plt038.
DOI:10.1093/aobpla/plt038 |
||
Avramidou E.V., Doulis A.G., Aravanopoulos F.A., 2015. Determination of epigenetic inheritance, genetic inheritance, and estimation of genome DNA methylation in a full-sib family of Cupressus sempervirens L. Gene, 562, 180-187.
DOI:10.1016/j.gene.2015.02.068 |
||
Catalá R., Sabater B., Guéra A., 1997. Expression of the plastid ndhF gene product in photosynthetic and non-photosynthetic tissues of developing barley seedlings. Plant Cell Physiol, 38, 1382-1388.
DOI:10.1093/oxfordjournals.pcp.a029133 |
||
Cervera M.T., Ruiz-García L., Martínez-Zapater J.M., 2003. Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol. Genet. Genomics, 268, 832-833.
|
||
Chinnusamy V., Zhu J.K., 2009. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol, 12.
|
||
Dai L.F., Chen Y.L., Luo X.D., Wen X.F., Cui F.L., Zhang F.T., Zhou Y., Xie J.K., 2015. Level and pattern of DNA methylation changes in rice cold tolerance introgression lines derived from Oryza rufipogon Griff. Euphytica, 205, 73-83.
DOI:10.1007/s10681-015-1389-0 |
||
Das O.P., Messing J., 1994. Variegated phenotype and developmental methylation changes of a maize allele originating from epimutation.. Genetics, 136, 1121-1141.
|
||
Dowen R.H., Pelizzola M., Schmitz R.J., Lister R., Dowen J.M., Nery J.R., Dixon J.E., Ecker J.R., 2012. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U. S. A, 109, E2183.
DOI:10.1073/pnas.1209329109 |
||
Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol, 14, 2611-2620.
DOI:10.1111/mec.2005.14.issue-8 |
||
Feng S., Cokus S.J., Zhang X., Chen P.Y., Bostick M., Goll M.G., Hetzel J., Jain J., Strauss S.H., Halpern M.E., 2010. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U. S. A, 107, 8689.
DOI:10.1073/pnas.1002720107 |
||
Finnegan E.J., Kovac K.A., Matzke M.A., Matzke A.J.M., 2000. Plant DNA methyltransferases. Plant Mol. Biol, 43, 189-201.
DOI:10.1023/A:1006427226972 |
||
Foster J.T., Allan G.J., Chan A.P., Rabinowicz P.D., Ravel J., Jackson P.J., Keim P., 2010. Single nucleotide polymorphisms for assessing genetic diversity in castor bean (Ricinus communis). BMC Plant Biol, 10, 13.
DOI:10.1186/1471-2229-10-13 |
||
Garg R., Chevala V.N., Shankar R., Jain M., 2015. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep, 5.
|
||
Gruenbaum Y., Navehmany T., Cedar H., Razin A., 1981. Sequence specificity of methylation in higher plant DNA. Nature, 292, 860-862.
DOI:10.1038/292860a0 |
||
Guéra A., de Nova P.G., Sabater B., 2000. Identification of the Ndh (NAD(P)H-plastoquinone-oxidoreductase) complex in etioplast membranes of barley:changes during photomorphogenesis of chloroplasts. Plant Cell Physiol, 41, 49-59.
DOI:10.1093/pcp/41.1.49 |
||
Herman J.J., Sultan S.E., 2016. DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc. Biol. Sci, 283.
|
||
Herrera C.M., Bazaga P., 2010. Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet Viola cazorlensis. New Phytol, 187, 867.
DOI:10.1111/j.1469-8137.2010.03298.x |
||
Hong H.F., Wei J., Li T.C., Zheng P.L., Ning G., Yong P.C., Yi L., 2013. DNA methylation alterations of upland cotton (Gossypium hirsutum) in response to cold stress. Acta Physiol. Plant, 35, 2445-2453.
DOI:10.1007/s11738-013-1278-x |
||
Hubisz M.J., Falush D., Stephens M., Pritchard J.K., 2009. Inferring weak population structure with the assistance of sample group information. Mol. Ecol.Resour, 9, 1322-1332.
DOI:10.1111/men.2009.9.issue-5 |
||
Kanchanaketu T., Sangduen N., Toojinda T., Hongtrakul V., 2012. Genetic diversity analysis of Jatropha curcas L.(Euphorbiaceae) based on methylation-sensitive amplification polymorphism. Genet. Mol. Res, 11, 944.
|
||
Keyte A.L., Percifield R., Liu B., Wendel J.F., 2006. Infraspecific DNA methylation polymorphism in cotton (Gossypium hirsutum L. ). J. Hered, 97, 444-450.
DOI:10.1093/jhered/esl023 |
||
Kobayashi H., Ngernprasirtsiri J., Akazawa T., 1990. Transcriptional regulation and DNA methylation in plastids during transitional conversion of chloroplasts to chromoplasts. EMBO J, 9, 307-313.
|
||
Lauria M., Rupe M., Guo M., Kranz E., Pirona R., Viotti A., Lund G., 2004. Extensive maternal DNA hypomethylation in the endosperm of Zea mays. Plant Cell, 16, 510-522.
DOI:10.1105/tpc.017780 |
||
Law J.A., Jacobsen S.E., 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet, 11, 204.
DOI:10.1038/nrg2719 |
||
Lee C., Itoh T., Maeda R., Suga T., 1994. Effect of site-specific methylation on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res, 22, 3640-3659.
DOI:10.1093/nar/22.17.3640 |
||
Leutwiler L.S., Hough-Evans B.R., Meyerowitz E.M., 1984. The DNA of Arabidopsis thaliana. Mol. Genet. Genomics, 194, 15-23.
DOI:10.1007/BF00383491 |
||
Liramedeiros C.F., Parisod C., Fernandes R.A., Mata C.S., Cardoso M.A., Ferreira P.C., 2010. Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE, 5, e10326.
DOI:10.1371/journal.pone.0010326 |
||
Liu J., He Y., Amasino R., Chen X., 2004. siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev, 18, 2873-2878.
DOI:10.1101/gad.1217304 |
||
López C.M.R., Wilkinson M.J., 2010. Progressive erosion of genetic and epigenetic variation in callus-derived cocoa (Theobroma cacao) plants. New Phytol, 186, 856.
DOI:10.1111/j.1469-8137.2010.03242.x |
||
Ma K., Song Y., Yang X., Zhang Z., Zhang D., 2013. Variation in genomic methylation in natural populations of Chinese white poplar. PLoS ONE, 8, e63977.
DOI:10.1371/journal.pone.0063977 |
||
Marconi G., Pace R., Traini A., Raggi L., Lutts S., Chiusano M., Guiducci M., Falcinelli M., Benincasa P., Albertini E., 2013. Use of MSAP markers to analyse the effects of salt stress on DNA methylation in rapeseed (Brassica napus var.oleifera). PLoS ONE, 8, e75597.
DOI:10.1371/journal.pone.0075597 |
||
Ohta N., Sato N., Kawano S., Kuroiwa T., 1991. Methylation of DNA in the chloroplasts and amyloplasts of the pea, Pisum sativum. Plant Sci, 78, 33-42.
DOI:10.1016/0168-9452(91)90159-6 |
||
Okano M., Bell D.W., Haber D.A., Li E., 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247-257.
DOI:10.1016/S0092-8674(00)81656-6 |
||
Portis E., Acquadro A., Comino C., Lanteri S., 2004. Analysis of DNA methylation during germination of pepper (Capsicum annuum L.) seeds using methylationsensitive amplification polymorphism (MSAP). Plant Sci, 166, 169-178.
DOI:10.1016/j.plantsci.2003.09.004 |
||
Qiu L., Yang C., Bo T., Yang J.B., Liu A., 2010. Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol, 10, 1-10.
DOI:10.1186/1471-2229-10-1 |
||
Reyna-Lzópez G.E., Simpson J., Ruiz-Herrera J., 1997. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol. Genet. Genomics, 253, 703-710.
DOI:10.1007/s004380050374 |
||
Rivarola M., Foster J.T., Chan A.P., Williams A.L., Rice D.W., Liu X., Melakeberhan A., Creasy H.H., Puiu D., Rosovitz M.J., 2011. Castor bean organelle genome sequencing and worldwide genetic diversity analysis. PLoS ONE, 6, e21743.
DOI:10.1371/journal.pone.0021743 |
||
Rod P., Petere S., 2006. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes, 6, 288-295.
DOI:10.1111/men.2006.6.issue-1 |
||
Sáez-Laguna E., Guevara M.Á., Díaz L.M., Sánchez-Gómez D., Collada C., Aranda I., Cervera M.T., 2014. Epigenetic variability in the genetically uniform forest tree species Pinus pinea L. PLoS ONE, 9, e103145.
DOI:10.1371/journal.pone.0103145 |
||
Salmon A., Clotault J., Jenczewski E., Chable V., Manzanaresdauleux M.J., 2008. Brassica oleracea displays a high level of DNA methylation polymorphism. Plant Sci, 174, 61-70.
DOI:10.1016/j.plantsci.2007.09.012 |
||
Shan X.H., Li Y.D., Liu X.M., Wu Y., Zhang M.Z., Guo W.L., Liu B., Yuan Y.P., 2012. Comparative analyses of genetic/epigenetic diversities and structures in a wild barley species (Hordeum brevisubulatum) using MSAP, SSAP and AFLP. Genet.Mol. Res, 11, 2749-2759.
DOI:10.4238/2012.August.17.2 |
||
Sujatha M., Reddy T.P., Mahasi M.J., 2008. Role of biotechnological interventions in the improvement of castor (Ricinus communis L.) and Jatropha curcas L. Biotechnol. Adv, 26, 424-435.
DOI:10.1016/j.biotechadv.2008.05.004 |
||
Suzuki M.M., Bird A., 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet, 9, 465.
|
||
Takio S., Satoh Y., Satoh T., 1994. Occurrence of DNA methylation in chloroplasts of the suspension cultured cells from a Liverwort, Marchantia paleacea var. diptera.J. Plant Physiol, 143, 173-177.
DOI:10.1016/S0176-1617(11)81682-2 |
||
Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., 2013. MEGA6: molecular evolutionary genetics analysis version 6. 0. Mol. Biol. Evol, 30, 2725.
DOI:10.1093/molbev/mst197 |
||
Vos P., Hogers R., Bleeker M., Reijans M., Lee T.V.D., Hornes M., Friters A., Pot J., Paleman J., Kuiper M., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res, 23, 4407.
DOI:10.1093/nar/23.21.4407 |
||
Walder R.Y., Langtimm C.J., Chatterjee R., Walder J.A., 1983. Cloning of the MspI modification enzyme.The site of modification and its effects on cleavage by MspⅠ and HpaⅡ. J. Biol. Chem, 258, 1235-1241.
|
||
Wang P., Gao C., Bian X., Zhao S., Zhao C., Xia H., Song H., Hou L., Wan S., Wang X., 2016. Genome-wide identification and comparative analysis of cytosine-5 DNA methyltransferase and demethylase families in wild and cultivated peanut. Front. Plant Sci, 7, 7.
|
||
Webster G.L., 1994. Synopsis of the genera and suprageneric taxa of Euphorbiaceae. Ann. Missouri Bot. Gard, 81, 33.
DOI:10.2307/2399909 |
||
Wolf J., 2000. Oilseed crops (2nd edn), Edited by EA Weiss Blackwell Science, Oxford, 2000. J. Sci. Food Agric, 80, 1573.
DOI:10.1002/(ISSN)1097-0010 |
||
Xiong L.Z., Xu C.G., Saghai Maroof M.A., Zhang Q., 1999. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Genet. Genomics, 261, 439.
DOI:10.1007/s004380050986 |
||
Xu M., Li X., Korban S.S., 2000. AFLP-Based detection of DNA methylation. Plant Mol. Biol. Rep, 18, 361-368.
DOI:10.1007/BF02825064 |
||
Zhang Y.Y., Fischer M., Colot V., Bossdorf O., 2013. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol, 197, 314-322.
DOI:10.1111/nph.12010 |
||
Zhang B., Tieman D.M., Jiao C., Xu Y., Chen K., Fe Z., Giovannoni J.J., Klee H.J., 2016. Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc. Natl. Acad. Sci. U.S.A, 113, 12580.
DOI:10.1073/pnas.1613910113 |
||
Zhao X., Chai Y., Liu B., 2007. Epigenetic inheritance and variation of DNA methylation level and pattern in maize intra-specific hybrids. Plant Sci, 172, 930-938.
DOI:10.1016/j.plantsci.2007.01.002 |
||
Zhong L., Xu Y., Wang J., 2009. DNA-methylation changes induced by salt stress in wheat Triticum aestivum. Afr. J. Biotechnol, 8, 6201-6207.
DOI:10.5897/AJB |



