b. Graduate University of Chinese Academy of Sciences, Beijing, 100049, China
The genus Elymus L., which includes as many as 150 species, is the largest and most widely distributed genus in the Triticeae tribe of family Poaceae (Löve, 1984). Elymus nutans Griseb., an allohexaploid (2n = 6x = 42) species with an StStYYHH genome, is widely distributed in Asia and grows on grassland, bush land, river banks, mountain slopes, and swales at altitudes from 1000 m up to 5000 m (Lu, 1993). As it is a high-quality forage crop and has high tolerance to various environmental stresses associated with high altitude, such as cold and drought, E. nutans is treated as a potential forage crop for farming in the QinghaieTibet Plateau (Lu and Nie, 2002). E. nutans shows great variability, and its genetic diversity has been measured by morphological characters (Zhang et al., 2009), ISSR analysis (Chen et al., 2009a), SRAP (Chen et al., 2009b), AFLP (Yan et al., 2010), gliadin (Miao et al., 2011), and SSR analysis (Chen et al., 2013). All of these studies found that the majority of the total variation occurred within populations rather than among populations. E. nutans is regarded as a self-pollinating plant. Selfpollinating plant species usually present lower genetic variation within populations. To explain these contradictory results in E. nutans, it has been suggested that E. nutans may combine a certain level of self-pollination rate and out-crossing (Miao et al., 2011), or that the population structure of E. nutans is strongly affected by excess gene flow caused by seed dispersal from grazing animals (Yan et al., 2010).
Chromosome rearrangements in E. nutans were first reported from the analysis of chromosome pairing in intraspecific hybrids (Lu, 1993). This revealed an increased number of multivalents at metaetaphase Ⅰ and the presence of chromatid bridges and fragments at anaetaphase Ⅰ, and suggested that chromosome rearrangements such as inversions or translocations occurred in different populations. A high frequency of karyotype variation has been observed in a domesticated population of E. nutans using sequential FISH and GISH (Dou et al., 2009). Four types of translocation were well characterized, and many chromosome variations due to repeat amplifications or deletions were detected. This suggested that possible sib-crossing or selection pressure might be responsible for the karyotype variation in this population.
Chromosome rearrangements have played an important role in the evolution and intraspecific diversity of polyploid Triticeae species. Intraspecific diversity accompanied by chromosomal aberrations and translocations were reported in both Triticum araraticum Jakubz. and T. dicoccoides Korn (Kawahara, 1986; Badaeva et al., 1994, 1995). Karyotype Ⅴariants and translocations were detected in 104 of 185 accessions in T. araraticum by C-banding, and karyotype variation diversity varied by geographical region. Each geographical region contained a unique spectrum of translocations (Badaeva et al., 1994). A high frequency of inter-genomic translocations was reported in the Triticeae species Kengyilia thoroldiana (Keng) J.L.Yang et al. (Wang et al., 2012), and in the Elymus tangutorum (Nevski) Hand.-Mazz (Yang et al., 2015). Species-specific chromosome arrangements have been detected in polyploid species of Triticeae (Liu et al., 1992; Jiang and Gill, 1994). Nucleocytoplasmic interaction (NCI) has been proposed to explain how chromosome rearrangement plays an important role in restoring fertility and nucleo-cytoplasmic compatibility, resulting in the genetic stabilization of newly formed hybrids and polyploids (Gill, 1991; Jiang and Gill, 1994). Furthermore, karyotype variations were reported in recently resynthesized and naturally formed allopolyploid species (Xiong et al., 2011; Chester et al., 2012), and in natural hybrids produced by polyploid homoploid hybridization (Lipman et al., 2013).
Fluorescence in situ hybridization (FISH) signals derived from a single repetitive DNA probe or a cocktail containing several DNA probes can provide a hybridization pattern that allows identification of all chromosomes within a species (Jiang and Gill, 2006). Genomic in situ hybridization (GISH) using the total genomic DNA of an analyzer (a species with a known genome) has been used to reveal the presence or absence of the analyzer genome in polyploid species (Ørgaard and Heslop-Harrison, 1994). Using FISH and GISH sequentially on the same cell allows for the identification of chromosomes by FISH patterns as well as the simultaneous allocation of the identified chromosomes to a certain genome in an allopolyploid species (Dou et al., 2009, 2011; 2012, 2013).
The pAs1 repetitive sequence (Afa-family) reportedly exists in a number of genomes of the tribe Triticeae (Nagaki et al., 1995). Afafamily sequences are usually formed by numerous blocks in several subtelomeric and interstitial chromosome regions, and therefore, have been extensively used as chromosome markers (Tsujimoto et al., 1997). The distribution of microsatellite AAG on the chromosomes of wheat, barley, and related species in Triticeae are well reported, and the distribution patterns corresponding to the Nband make them useful cytological markers for chromosome identification (Pedersen et al., 1996; Cuadrado and Schwarzacher, 1998). A fully resolved karyotype of E. nutans can be produced by FISH using differently labeled pAs1 and AAG tandem repeats followed by GISH using two labeled progenitor genomes (Dou et al., 2009).
In this study, sequential FISH and GISH were used to investigate wild E. nutans plants indigenous to the Qinghai Plateau. Our specific objectives were: (1) to detect the types and extent of karyotype variation in wild populations; (2) to learn whether specific karyotype Ⅴariants were related to geographical regions; (3) to further explore the possible reasons for the origin of the karyotype variation in E. nutans.
1. Materials and methods 1.1. Plant materialsEight natural populations of E. nutans were sampled in the Qinghai Plateau in 2010 and 2011, at an altitude range from 2600 to 3700 m. Twenty to thirty plants were randomly sampled in each population. Mature seeds were collected from each plant individually. Only the materials whose molecular karyotypes were exclusively captured are listed in Table 1. Additionally, two genome donor species (Pseudoroegneria stipifolia (Czern. ex Nevski) Löve and Hordeum bogdanii Wilensky) were used in this study (Table 1).
| Species (genome symbol) | Population | Identification No. | Source | Altitude (m) |
| Elymus nutans (StHY) | 1 | Nw1-5; Nw6-1; Nw6-5; | Qinghai Lake | 3200 |
| Nw12-8; Nw56-3; Nw56-4 | ||||
| 2 | Nw20-4; Nw45-2; Nw45-3 | Guide, Qinghai | 3560 | |
| 3 | Nw22-4; Nw22-5; Nw40-1 | Guinan, Qinghai | 3330 | |
| 4 | Nw28-1; Nw29-1; Nw30-4; | Tongde, Qinghai | 3510 | |
| 5 | Nw32-13; Nw32-19; | Tongde, Qinghai | 3700 | |
| Nw32-20 | ||||
| 6 | Nw59-3; nw59-4; Nw59-5 | Huzhu, Qinghai | 2600 | |
| 7 | Nw60-3; Nw60-4; Nw60-5 | Xinghai, Qinghai | 3600 | |
| 8 | KLS1-1; KLS1-2 Kls1-3 | Germu, Qinghai | 3800 | |
| Hordeum bogdanii (H) | - | - | Germu, Qinghai | 2800 |
| Pseudoroegneria stipifolia (St) | - | PI 313960 | FRRL, USA | - |
The seeds collected from the wild plants were germinated at room temperature. Root tips of 1e2 cm in length were pretreated in ice-cold water at 0-4 ℃ in a refrigerator (4 ℃) for 24 h, before being fixed in 3:1 (v/v) ethanol:glacial acetic acid. Chromosome spreads and slide preparation were performed as described in Dou et al. (2013).
1.3. Probe labelingTwo repetitive sequences, the satellite DNA pAs1 (including a repetitive sequence from Aegilops squarrosa (Rayburn and Gill, 1986)); and the microsatellite DNA (AAG)10, were labeled for use as FISH probes. pAs1 was labeled with tetramethyl-rhodamine-5-dUTP by a nick-translation method, and (AAG)10 was synthesized with a fluorescein-12-dUTP at the 3ʹ-end. Genomic DNA of H. bogdanⅡ and P. stipifolia for the GISH probes was labeled with fluorescein-12-dUTP and tetramethy1-rhodamine-5-dUTP, respectively, as described by Dou et al. (2009).
1.4. In situ hybridizationFISH and GISH experiments were carried out as described by Dou et al. (2009). Images were captured with a cooled CCD camera (Photometrics CoolSNAP) under a fluorescence microscope (Leica) and processed with the Meta Imaging system (Universal Imaging Corporation).
2. Results 2.1. Karyotype variation and chromosome variantsThe combined (AAG)10 and pAs1 FISH probes produced a distinct hybridization pattern on each chromosome.Sequential GISH allowed us to clearly classify all chromosomes into three subgenomes: H, St and Y. Sequential FISH and GISH permitted identification of individual chromosomes within each subgenome (Fig. 1). Well-marked chromosomes were arbitrarily designated as 1-7 by their arm ratio and the relative length (Fig. 2). In this designation, the numerals do not correspond with those of the homologous groups as used in the nomenclature system of the chromosomes of wheat and barley. Since some of the chromosomes exhibited polymorphisms among different individuals, the polymorphisms showing the greatest similarity in the distribution of hybridization signal, arm ratio and length were allocated to the same number. Thus, the polymorphisms of each chromosome and the karyotype of each individual were well described (Fig. 2 and Table 2).
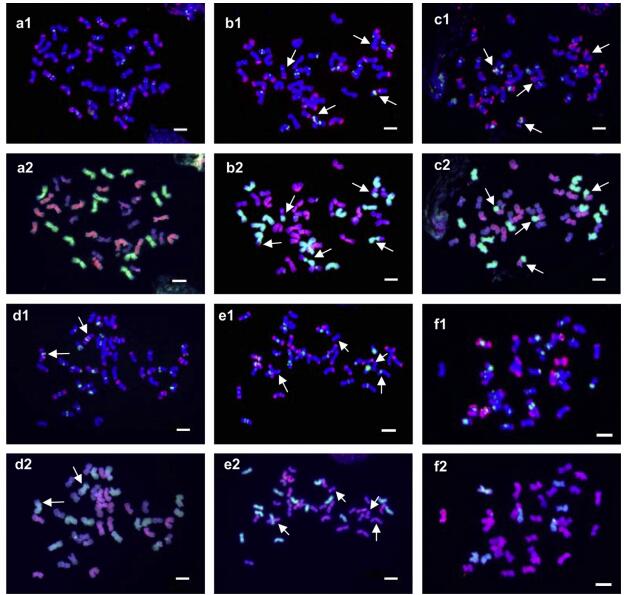
|
| Fig. 1 Sequential FISH and GISH patterns of representatives of E. nutans. a1, b1, c1, d1, e1, f1: FISH patterns probed with (AAG)10 (green) and pAs1 (red). a2, b2, c2, d2, e2, f2: Sequential GISH patterns probed with H. bogdanⅡ (green) and P. stipifolia (red). aee are E. nutans materials Nw45-2, Nw29-1, Nw6-1, Nw40-1, and 60-3, respectively, and f is a hybrid between E. nutans and an unknown StY Elymus species. Arrows indicate translocation chromosomes. Bar = 10 mm. |

|
| Fig. 2 Molecular karyotypes of 27 E. nutans samples. The patterns of most chromosomes were characterized by pAs1 (red) and (AAG)10 (green) probes. Different variants are annotated by different Roman letters. The translocated chromosomes are underlined, and indicated by numerals Ⅰ-Ⅵ. |
| Population | Samples | H genome | St genome | Y genome | ||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| 1 | Nw1-5 | a | a | a | b | a | a | b | a | b | a | a | a | a | a | b | a | i | a | e | a | a |
| Nw6-1 | b | a | a | a | b | cT | a | a | f | a | b | dT | a | a | d | e | h | d | e | e | a | |
| Nw6-5 | a | b | a | a | a | a | a | a | c | b | a | b | a | a | d | c | d | a | d | c | a | |
| Nw12-8 | b | a | b | a | b | cT | a | d | g | a | b | dT | a | a | g | f | d | f | e | a | c | |
| Nw56-3 | b | a | a | b | b | a | a | b | b | b | a | a | a | a | a | a | c | a | a | a | a | |
| Nw56-4 | b | a | a | a | a | a | a | a | b | b | a | a | a | a | a | b | c | a | a | b | ab | |
| 2 | Nw20-4 | b | a | b | b | b | a | a | a | b | a | d | a | a | a | h | g | b | b | a | b | a |
| Nw45-2 | a | a | b | a | a | a | a | a | a | a | a | a | a | a | a | a | a | a | a | a | a | |
| Nw45-3 | a | ab | c | a | a | a | a | a | b | a | a | b | a | a | b | b | b | b | a | a | a | |
| 3 | Nw22-4 | b | a | a | a | a | a | a | a | b | a | b | b | a | a | d | g | a | b | e | c | a |
| Nw22-5 | b | a | b | dT | a | a | a | a | h | a | c | b | cT | d | b | b | c | f | e | c | a | |
| Nw40-1 | b | a | a | b | b | bT | a | e | d | a | b | a | a | a | a | g | d | d | a | c | a | |
| 4 | Nw28-1 | a | a | a | a | a | a | a | a | c | a | a | c | a | a | d | c | d | c | d | c | a |
| Nw29-1 | b | a | ab | bcT | b | bT | a | a | bd | b | a | a | a | a | a | d | c | a | bT | a | a | |
| Nw30-4 | a | a | a | b | a | a | a | a | i | a | a | a | a | f | a | g | c | a | a | a | a | |
| 5 | Nw32-13 | a | a | a | a | b | a | a | a | b | a | a | b | a | a | a | g | c | a | d | b | a |
| Nw32-19 | a | a | a | b | a | a | b | b | bd | a | a | b | a | a | c | b | f | a | a | a | a | |
| Nw32-20 | b | a | a | b | a | a | b | c | d | a | a | a | a | a | ac | ab | ag | a | c | a | a | |
| 6 | Nw59-3 | a | a | ab | b | ab | bT | a | a | a | a | a | a | a | a | a | a | g | a | bT | a | a |
| Nw59-4 | a | a | b | b | b | a | a | a | b | a | a | c | a | a | b | a | g | b | c | b | a | |
| Nw59-6 | a | a | b | b | a | a | a | a | a | a | a | b | a | a | a | a | g | a | a | b | a | |
| 7 | Nw60-3 | c | a | a | a | b | a | a | e | b | a | d | eT | a | e | b | g | c | d | fT | f | a |
| Nw60-4 | a | a | a | b | a | bT | d | e | b | c | a | a | a | c | a | g | b | a | bT | a | a | |
| Nw60-5 | b | a | a | a | b | a | cT | c | b | a | b | a | b | b | e | e | e | d | d | d | a | |
| 8 | Kls1-1 | a | a | b | b | b | a | a | a | e | a | ab | a | a | c | f | b | b | e | d | c | a |
| Kls1-2 | c | a | a | a | b | a | a | e | b | a | d | b | a | a | b | g | c | d | e | f | a | |
| Kls1-3 | c | a | a | a | b | a | b | e | b | a | d | b | a | a | b | g | c | a | e | f | a | |
| No. of variants | 3 | 2 | 3 | 4 | 2 | 3 | 4 | 5 | 9 | 3 | 4 | 5 | 3 | 6 | 8 | 7 | 8 | 6 | 6 | 6 | 3 | |
| Total | 21 | 35 | 44 | |||||||||||||||||||
| Superscript "T" indicates the translocation chromosomes. | ||||||||||||||||||||||
The FISH/GISH results revealed a great variety of karyoTypes Ⅰn wild E. nutans plants. No identical karyotype was identified within or among populations. The extent of variation differed genome-togenome and chromosome-to-chromosome. A total of 100 chromosome variants were observed in the 27 samples. The Y genome showed the highest number of chromosome variants (44), whereas the H genome had the least (21). Chromosomes 1Y and 3Y exhibited the most polymorphic variants (8), while chromosomes 2H and 5H presented the least (2). The majority of plants carried the pattern in the homozygous state, although some were in the heterozygous state (22.2%, six of 27 plants; Table 2).
Most of the variants were characterized by the absence or presence of additional hybridization signals due to repeats, duplications, or deletions. However, distinct inter-genomic translocations were identified in nine of the 27 (33.3%) plants (Fig. 2 and Table 2). The translocations were classified into six types by the FISH/GISH patterns. Types Ⅰ, Ⅱ, Ⅲ, and Ⅳ were reciprocal translocations, whereas type Ⅴ and type Ⅵ represented a terminal fragment translocation lacking a counterpart (Fig. 2; 4Hc, 7Hc). Types Ⅰ and Ⅲ were a translocation between the H and Y genomes and a translocation between the H and St genomes, respectively, with breakpoints in the intercalary regions of the chromosomes (Fig. 2; 6Hb and 5Yb, 4Hd and 6Stc). Both Type Ⅱ and type Ⅳ were Robertsonian-like translocations with breakpoints in the centromeric regions of the chromosomes. Type Ⅱ was a translocation between H and St (Fig. 2; 6Hc and 5Std). Type Ⅳ was a translocation between St and Y (Fig. 2; 6Ste and 6Yf). Types Ⅰ and Ⅱ were more frequent than the others. Four (14.8%) and two (7.4%) of the 27 samples carried type Ⅰ and type Ⅱ translocations, respectively. The other types were observed in only one sample each.
2.2. Geographic distribution of inter-genomic translocationsGiven that high-frequency inter-genomic rearrangements may be associated with ecological adaptation, we analyzed the geographic distributions of type Ⅰ and Ⅱ translocations further. Type Ⅰ was found in populations 4, 6 and 7 (Table 2). Populations 4, and 7 are located in the southern part of the Qinghai Plateau, while population 6 is located in the eastern part. Type Ⅱ was only identified in population 1 around Qinghai Lake, which is located in the northwestern part of Qinghai Plateau. GISH analysis was used to screen other individuals in population 1 and another population in the northern part of Qilian. This demonstrated that a few type Ⅱ individuals were present in population 1. However, neither type Ⅰ nor type Ⅱ were detected in the Qilian population (data not shown). Moreover, no translocations were detected in the western region (population 8). Thus, the results suggest that the type Ⅰ translocation is widely distributed in the southeastern regions of Qinghai Plateau, whereas type Ⅱ is strictly distributed around Qinghai Lake.
3. DiscussionHere we found that wild populations of E. nutans exhibited a high frequency of karyotype variation. These findings are consistent with previous studies on the level of variation in domesticated populations of E. nutans (Dou et al., 2009). The chromosome variations were characterized as repeat deletions and amplifications, and inter-genomic translocations using a sequential FISH/GISH technique. Because chromosome inversion and intra-genomic translocation are difficult to detect using this technique, the karyotype variations might still include other chromosome structure aberrations such as inversions and intra-genomic translocations.
In domesticated populations of E nutans, the St genome has been shown to have the highest number of chromosome variants, while the Y genome has the lowest number of chromosome variants (Dou et al., 2009). However, in our study, the Y genome showed the highest number of chromosome variants, whereas the H genome had the least. In total, four inter-genomic translocations were identified in the domesticated population. Detailed comparisons of the FISH patterns of the translocation shoed the FISH pattern of translocation type Ⅱ in the domesticated population is identical to that in Elymus dahuricus (Dou et al., 2009). This strongly suggests seed mixing in the domesticated population. We found that the FISH pattern of translocation type Ⅰ in wild populations was identical to the translocation type Ⅰ pattern in the domesticated population. Translocation type Ⅰ was frequently detected in southeastern regions of Qinghai Plateau. This implies that the domesticated population originated from a southeastern wild population rather than from populations in the surrounding regions of Qinghai Lake as the seed supplier declared. Due to seed mixing and the uncertain origin of the domesticated population, the present study provides a more reliable assessment of karyotype variation in E. nutans indigenous to the Qinghai Plateau.
Karyotype variations have been reported in recently resynthesized and naturally formed allopolyploidy species (Xiong et al., 2011; Chester et al., 2012), and in natural hybrids produced by polyploidehomoploid hybridization (Lipman et al., 2013). Aneuploidy, inter-and intra-genomic arrangements, and repeat loss have been commonly detected in the early generations. Molecular phylogeny has demonstrated that reticulate evolution has occurred extensively in Elymus, and interspecific hybridization frequently occurred between Elymus species (Sun, 2014). High seed sets have previously been shown to result from inter-specific hybridizations between E. nutans and 13 tetraploid and four hexaploid Elymus species, especially in combinations with E. nutans as the pollinator (Lu, 1993). In our study, we identified natural inter-specific hybrids between E. nutans and an unknown Elymus tetraploid species (2n = 28, StStYY) in the offspring of two of the 27 investigated plants (Fig. 1f). Additionally, cytogenetic analysis of the secondary roots of a few sterile plants revealed a hybrid between E. nutans and E. dahuricus (data not shown). Taken together, this evidence demonstrates that E. nutans has high crossing ability with related species. Though F1 hybrids between E. nutans and other species may be partially or completely male sterile, opportunities for fertility restoration could be greatly increased by back-crossing because of the perennial habits of the hybrids. Thus, it can be inferred that the high frequency karyotype variation in E. nutans might be induced by interspecific hybridization. The St and Y genome chromosomes were revealed to be much more polymorphic than the H genome chromosomes in this study. Many Elymus species are sympatrically distributed with E. nutans in the QinghaieTibet Plateau (Lu et al., 1987, 1990, 1999). Single nuclear gene sequence analysis has shown diversification in the St and the Y genome in sympatrically distributed StY, StYH, and StYP Elymus species (Fan et al., 2013). This may further support the idea that the high variation in the St and Y genomes of E. nutans may be strongly affected by introgressive hybridization with these species.
High frequency inter-genomic translocations have been reported in K. thoroldiana indigenous to the QinghaieTibet plateau, and a significant correlation between chromosome translocations and environmental factors was revealed (Wang et al., 2012). It has been suggested that intergenomic rearrangements are associated with environmental factors and genetic differentiation of a single basic genome should be considered as an equally important genetic processes during ecotype evolution (Wang et al., 2012). Moreover, Yang et al. (2015) reported that various types of translocations are present in populations of E. tangutorum from different habitats or altitudes, and inferred that intergenomic rearrangements of E. tangutorum might be influenced by environmental factors. Though six inter-genomic translocation types were revealed in our study, only two had a high frequency. Apart from translocation type Ⅰ, type Ⅲ, Ⅳ, Ⅴ, and Ⅵ were restricted to special populations and type Ⅱ was restricted to a special region. This implies that the distribution of specific translocation types might be associated with unique environmental factors as suggested by Wang et al. (2012) and Yang et al. (2015). Since random populations and limited samples of each population were investigated in this study, more data is still needed to corroborate the correlation between intergenomic rearrangements and environmental factors in E. nutans.
AcknowledgmentsThis research was supported by the National Natural Science Foundation of Qinghai Province (2015-ZJ-903) and the National Natural Science Foundation of China (31072075).
| Badaeva E.D., Badaev N.S., Gill B.S., Filatenko A.A., 1994. Intraspecific karyotype divergence in Triticum araraticum (Poaceae). Plant Syst. Evol, 192, 117-145. DOI:10.1007/BF00985912 | ||
| Badaeva E.D., Jiang J., Gill B.S., 1995. Detection of intergenomic translocations with centromeric and noncentromeric breakpoints in Triticum araraticum: mechanism of origin and adaptive significance. Genome, 38, 976-981. DOI:10.1139/g95-128 | ||
| Chen S.Y., Ma X., Zhang X.Q., Chen Z.H., 2009a. Genetic variation and geographical divergence in Elymus nutans Griseb. (Poaceae: Triticeae) from west China. Biochem. Syst. Ecol, 37, 716-722. | ||
| Chen S.Y., Zhang X.Q., Ma X., Huang L.K., 2013. Assessment of genetic diversity and differentiation of Elymus nutans indigenous to QinghaieTibet Plateau using simple sequence repeats markers. Can. J. Plant Sci, 93, 1089-1096. DOI:10.4141/cjps2013-062 | ||
| Chen Z.H., Miao J.M., Zhong J.C., Ma X., Chen S.Y., Zhang X.Q., 2009b. Genetic diversity of wild Elymus nutans germplasm detected by SRAP markers. Acta Prataculturae Sin, 18(5), 192-200. | ||
| Chester M., Gallagher J.P., Symonds V.V., Cruz da Silva A.V., Mavrodiev E.V., Leitch A.R., et al., 2012. Extensive chromosomal variation in a recently formed natural allopolyploid species, tragopogon miscellus (asteraceae). Proc. Natl. Acad. Sci. U. S. A, 109(4), 1176-1181. DOI:10.1073/pnas.1112041109 | ||
| Cuadrado A., Schwarzacher T., 1998. The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma, 107(8), 587-594. DOI:10.1007/s004120050345 | ||
| Dou Q.W., Lei Y.T., Li X.M., Mott I., Wang R.R.-C., 2012. Characterization of alien chromosomes in backcross derivatives of Triticum aestivum × Elymus rectisetus hybrids by using molecular markers and sequential multicolor FISH/GISH. Genome, 55, 337-347. DOI:10.1139/g2012-018 | ||
| Dou Q.W., Chen Z.G., Liu Y.A., Tsujimoto H., 2009. High frequency of karyotype variation revealed by sequential FISH and GISH in plateau perennial grass forage Elymus nutans. Breed. Sci, 59, 651-656. DOI:10.1270/jsbbs.59.651 | ||
| Dou Q.W., Wang R.R.-C., Lei Y.T., Yu F., Li Y., Wang H.Q., Chen Z.G., 2013. Genome analysis of 7 Kengyilia species with FISH and GISH. Genome, 56, 641-649. DOI:10.1139/gen-2013-0113 | ||
| Dou Q.W., Zhang T.L., Tsujimoto H., 2011. Physical mapping of repetitive sequences and genomg analysis in six Elymus species by in situ hybridization. J. Syst. Evol, 49(4), 347-352. DOI:10.1111/jse.2011.49.issue-4 | ||
| Fan X., Sha L.N., Dong Z.Z., Zhang H.Q., Kang H.Y., Wang Y., et al., 2013. Phylogenetic relationships and Y genome origin in Elymus l. sensu lato (Triticeae; poaceae) based on single-copy nuclear Acc1 and Pgk1 gene sequences. Mol. Phylogenetics Evol, 69(3), 919-928. | ||
| Gill, B. S. , 1991. Nucleo-cytoplasmic interaction (NCI) hypothesis of genome evolution and speciation in polyploid plants. In: Sasakuma, T. , Kinoshita, T. (Eds. ), Proceedings of Dr. H. Kihara Memorial International Symposium on Cytoplasmic Engineering in Wheat, pp. 48-53. | ||
| Jiang J., Gill B.S., 1994. Different species-specific chromosome translocations in Triticum timopheevii and T. turgidum support the diphyletic origin of polyploid wheat. Chromosome Res, 2(1), 59-64. | ||
| Jiang J., Gill B.S., 2006. Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome, 49, 1057-1068. DOI:10.1139/g06-076 | ||
| Kawahara T., 1986. Difference in structural variability of genomes in Triticum and Aegilops. Wheat Inf. Serv, 63, 43. | ||
| Lipman M.J., Chester M., Soltis P.S., Soltis D.E., 2013. Natural hybrids between Tragopogon mirus and T. miscellus (asteraceae): a new perspective on karyotypic changes following hybridization at the polyploid level. Am. J. Bot, 100(10), 2016-2022. | ||
| Liu C.J., Atkinson M.D., Chinoy C.N., Devos K.M., Gale M.D., 1992. Nonhomoeologous translocations between group 4, 5 and 7 chromosomes within wheat and rye. Theor. Appl. Genet, 83(3), 305-312. | ||
| Löve A., 1984. Conspectus of the Triticeae. Feddes Repert, 95, 425-521. | ||
| Lu B.R., 1993. Meiotic studies of Elymus nutans and E. jacquemontii (Poaceae: Triticeae) and their hybrids with Pseudoroegneria spicata and seventeen Elymus species. Plant Syst. Evol, 186, 193-212. DOI:10.1007/BF00940798 | ||
| Lu G.P., Nie B., 2002. Field evaluation of Elymus nutans under alpine grassland conditions. Pratacultural Sci, 19, 13-15. | ||
| Lu B.R., Yan Ji, Ya ng, J. L., 1990. Cytological observations on Triticeae materials from Xinjiang, Qinghai and Sichuan. Plant Divers, 12(01), 1-3. | ||
| Lu S.L., Sun Y.H., Liu S.W., Yang Y.C., Wu Z.L., Guo B.Z., et al., 1987. Flora Reipublicae Popularis Sinicae. Science Press, Beijing. | ||
| Lu, S. L. , Liu, S. W. , Wu, Z. L. , He, T. N. , Zhou, L. H. , et al. , 1999. Flora Qinghaiica, vol. 4. Qinghai People's Publishing Press, Xining. | ||
| Miao J.M., Zhang X.Q., Chen S.Y., Ma X., Chen Z.H., et al., 2011. Gliadin analysis of Elymus nutans Griseb. from the Qinghai-Tibetan Plateau and Xinjiang, China. Grassl. Sci, 57, 127-134. | ||
| Nagaki K., Tsujimoto H., Isono K., Sasakuma T., 1995. Molecular characterization of a tandem repeat, Afa-family, and distribution among Triticeae. Genome, 38, 479-486. DOI:10.1139/g95-063 | ||
| Ørgaard M., Heslop-Harrison J.S., 1994. Investigation of genome relationships between Leymus, Psathyrostachys and Hordeum inferred from genomic DNA: DNA in situ hybridization. Ann. Bot, 73, 195-203. DOI:10.1006/anbo.1994.1023 | ||
| Pedersen C., Rasmussen S.K., Linde-Laursen I., 1996. Genome and chromosome identification in cultivated barley and related species of the Triticeae (Poaceae) by in situ hybridization with the GAA-satellite sequence. Genome, 39(1), 93-104. DOI:10.1139/g96-013 | ||
| Rayburn A.L., Gill B.S., 1986. Isolation of a D-genome specific repeated DNA sequence from Aegilops squarrosa. Plant Mol. Biol. Rep, 4(2), 102-109. DOI:10.1007/BF02732107 | ||
| Sun G., 2014. Molecular phylogeny revealed complex evolutionary process in Elymus species. J. Syst. Evol, 52(6), 706-711. DOI:10.1111/jse.v52.6 | ||
| Tsujimoto H., Mukai Y., Akagawa K., Nagaki K., Fujigaki J., Yamamoto M., Sasakuma T., 1997. Identification of individual barley chromosomes based on repetitive sequences: conservative distribution of Afa-family repetitive sequences on the chromosomes of barley and wheat. Genes Genet. Syst, 72(5), 303-309. DOI:10.1266/ggs.72.303 | ||
| Wang Q., Liu H., Gao A., Yang X., Liu W., Li X., et al., 2012. Intergenomic rearrangements after polyploidization of kengyilia thoroldiana (poaceae: Triticeae) affected by environmental factors. Plos One, 7(2), e31033. DOI:10.1371/journal.pone.0031033 | ||
| Xiong Z.Y., Gaeta R.T., Pires J.C., 2011. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. U. S. A, 108, 7908-7913. DOI:10.1073/pnas.1014138108 | ||
| Yan X.B., Guo Y.X., Liu F.Y., Zhao C., Liu Q.L., Lu B.R., 2010. Population structure affected by excess gene flow in self pollinating Elymus nutans and E. burchanbuddae (Triticeae: Poaceae). Popul. Ecol, 52, 233-241. DOI:10.1007/s10144-009-0169-x | ||
| Yang C.R., Zhang H.Q., Zhao F.Q., Liu X.Y., Fan X., Sha L.N., et al., 2015. Genome constitution of Elymus tangutorum (poaceae: triticeae) inferred from meiotic pairing behavior and genomic in situ hybridization. J. Syst. Evol, 53(6), 529-534. DOI:10.1111/jse.v53.6 | ||
| Zhang J.B., Bai S.Q., Zhang X.Q., Ma X., Yan J.J., Zhang C.B., You M.H., 2009. Study on ear characters of Elymus nutans Griseb. in the northwestern plateau of Sichuan province. J. Sichuan Univ. Nat. Sci. Ed, 46, 1505-1509. |



