b. Yunnan Key Laboratory for Wild Plant Resources, Kunming 650201, PR China;
c. Key Laboratory of Economic Plants and Biotechnology, Kunming Institute of Botany, Kunming, PR China
Plant photosynthesis, which occurs in plastids, is the primary energy source for life on Earth. However, some plants have partially lost their photosynthetic ability and become mixotrophic (MX), able to use both autotrophic and heterotrophic strategies to derive water and nutrients. Among angiosperms, mixotrophs exist in two groups: semiparasitic (e.g. genus Cuscuta) and partial mycoheterotrophic (MH) plants. The MH plants obtain carbon not only from their photosynthetic activity, but also from mycorrhizal fungi. This is a plant-fungus relationship which has evolved convergently in Orchidaceae and Ericaceae (Selosse and Roy, 2009; Tedersoo et al., 2007). Mixotrophs deploy a dual nutritional strategy which may have evolved repeatedly from various autotrophic (AH) plant lineages. However, the detailed evolutionary processes that lead to MH remain unclear (Bidartondo, 2005; Cameron et al., 2006; Julou et al., 2005; Tedersoo et al., 2007; Zimmer et al., 2007). Motomura et al. (2010) suggested that MH evolved after the establishment of MX rather than through direct shifts from AH to MH.
Heterotrophs have highly divergent morphology and DNA sequences (Barkman et al., 2007; Delannoy et al., 2011; Lemaire et al., 2011; Moore et al., 2010; Nickrent et al., 1998), thus precise phylogenetic relationships of heterotrophic plants to their respective autotrophic relatives have been notoriously difficult to ascertain (Stefanović and Olmstead, 2004). Plastid genome (plastome) of flowering plants is generally conserved in size, structure, gene content, and synteny (Gao et al., 2010; Wicke et al., 2011). However, owing to relaxation of strong functional constraints normally associated with photosynthesis, plastid genomes of heterotrophs exhibit a wide range of evolutionary degradation (Gao et al., 2010; Li et al., 2013; Wicke et al., 2011, 2013). To date, only several MH plant plastomes have been entirely sequenced (Delannoy et al., 2011; Li et al., 2013; Logacheva et al., 2011; Wicke et al., 2013; Wickett et al., 2008). In contrast to the highly reduced genome of fully MH plants (Gruzdev et al., 2016; Li et al., 2013; Wicke et al., 2013), the plastid genomes of Neottia Guett. and Aneura Dumort. have retained many genes related to photosynthesis. This may be attributed to a presumably recent switch of this species to mycoheterotrophy (Wickett et al., 2008).
Ericaceae is known to have formed symbiotic relationships with fungi. The tribe Pyroleae consists of mixotrophic species (with the exception of the fully MH Pyrola aphylla), while tribes Pteroporeae and Monotropeae consist of fully MH plants (Kron et al., 2002). All other tribes of the Ericaceae are, as far as is known, fully autotrophic. Although analysis of chloroplast DNA has contributed significantly to our understanding of Ericaceae evolution, most of the conclusions drawn thus far are based on nucleotide sequence segments (Kron, 1997; Kron et al., 2002). Extensive sampling and southern hybridization analysis across Ericaceae showed that mixotrophic taxa tend to retain most plastid genes relating to photosynthetic functions but vary regarding the gene content (Braukmann and Stefanović, 2012). In general, the plastome has a low rate of nucleotide substitution, which facilitates comparisons of variation in a wide range of plant taxa (Drouin et al., 2008; Gaut et al., 2011). Therefore, comparative analyses of the plastomes of autotrophs, mixotrophs and full heterotrophs, would shed new light on the evolution of plastome genomes in Ericaceae. With the loss of photosynthetic ability, genes required for the photosynthetic apparatus are predicted to undergo random mutations under relaxed natural selection (Funk et al., 2007; Logacheva et al., 2011; McNeal et al., 2007; Wolfe et al., 1992). Although some chloroplast genes have been retained and remain functional in several holoparasitic plants (Bungard, 2004), many holoparasites have reduced plastid genomes due to excessive gene loss and increased substitution rates in the remaining genes (Gruzdev et al., 2016; Li et al., 2013; Molina et al., 2014; Wicke et al., 2013; Wolfe et al., 1992). Examining nonsynonymous over synonymous substitution (dN/dS) ratios can be used to determine whether genes are evolving under purifying selection (Logacheva et al., 2011; McNeal et al., 2007).
In order to test if photosynthetic genes remain the most highly conserved in the plastid genome and whether relaxation of functional constraint precedes gene losses both before and after the evolution of heterotrophy in Ericaceae, we sequenced two plastomes of partial MH plants in the tribe Pyroleae (Pyrola decorata and Chimaphila japonica) which may be highly modified. The goal of this study was to provide a broad picture of chloroplast gene diversification in Pyroleae, and an understanding of the role of mixotrophs in the long history of Ericaceae evolution.
2. MethodsPlant material from P. decorata and C. japonica were collected from Qiongzhusi of Kunming (25°0'45"N, 102°42'04"E; altitude 2100 m), Yunnan Province, China. The plastid DNA was isolated from 100 g of fresh leaves using an improved extraction method that includes high ionic strength buffer at low pH (3.6) buffer (Triboush et al., 1998). Chloroplast DNA was sequenced with Illumina's Genome Analyzer at Beijing Genomics Institute (BGI) in Shenzhen, China. Subsequently, SOAP de novo were used to assemble the sequence reads of chloroplast genomes (Li et al., 2009). Eight small gaps were closed by PCR using Qiagen Taq Polymerase. Both plastid DNA and genomic DNA were used for PCR. Primers used for PCR were designed using available information from both ends of the gap. All primers were ordered from Sangon Biotech (Shanghai, China). PCR products were analyzed by electrophoresis on 1% agarose gel and then sequenced using standard Sanger protocols on ABI 3730 instruments. The thermal cycling program was as follows: 80 ℃ for 5 min; then 33 cycles of (95 ℃ for 45 s; 42-52 ℃ for 45 s; 65 ℃, 1 min), followed by 65 ℃ for 5 min.
Annotation of the sequenced genome was performed using DOGMA (Wyman et al., 2004), complemented with manual adjustment for start and stop codons and for intron/exon boundaries. Pairwise estimates of nonsynonymous substitution (dN) and synonymous substitution (dS) rates (dN/dS) were calculated for P. decorata, C. japonica, Rhododendron simsii (Moore et al., 2010), as well as Nicotiana tabacum L., when necessary (Kunnimalaiyaan and Nielsen, 1997), with the ratio calculated relative to Panax ginseng (Kim and Lee, 2004) as the outgroup. Non-triplet indels of pseudogenes were also adjusted and calculated. Plastomes from 41 seed plants were chosen and downloaded from GenBank (Supplement Table 1). The 43 taxa were divided into three groups: fully heterotrophic plants (Group Ⅰ), partial heterotrophic plants (Group Ⅱ, including two newly sequenced plastomes in this study) and autotrophic plants (Group Ⅲ) (Supplementary Table 1). All 79 protein-coding chloroplast genes (excluding ycf15, function presently unknown) were extracted from each genome and translated into amino acid sequences. Subsequently, they were classified in ten functional gene groups (Supplementary Table 2). Amino acid sequences for each gene were aligned by MUSCLE using MEGA 5 (Tamura et al., 2011) with default settings, verified by visual inspection, and used as guides to align the nucleotide sequences.
For dN/dS calculations, all protein-coding plastid genes were included in the analysis. The amino acids were only used in the alignment process; all analyses were done on nucleotide sequences. RNA editing sites were not considered. Standard errors were computed under the Kumar method using MEGA 5 (Tamura et al., 2011) for each gene and for classes of genes that together encode subunits of larger proteins. Wilcoxon rank sum tests performed with Kruskal Wallis Test method to estimate p-values between gene groups using SPSS software.
3. Results 3.1. Structure and gene content of the plastomeIllumina paired end (85 bp) sequencing produced 366 Mb of data for C. japonica and 470 Mb for P. decorata (Table 1). We aligned paired-end reads of each species to the published plastome of P. ginseng (Kim and Lee, 2004), which was chosen as a reference genome. In total, 527, 015 and 413, 293 paired-end reads were mapped to the reference genome for C. japonica and P. decorata, respectively. After de novo assembly with minor changes, we obtained more than 80% of the whole plastome. Gaps in the two plastomes were then filled by PCR sequencing. At last, we obtained 97% of the C. japonica plastome containing three gaps, and 94% of the P. decorata plastome with 17 gaps. Though less than 2000 bp (except ycf1 region), the unfinished gaps contain repeated sequences and lower GC content, so they were difficult to sequence using normal procedures. Although incomplete, the sequence data were sufficient for evolutionary analysis.
| Chimaphila | Pyrola | |
| Raw data (Mb) | 470.45 | 366.35 |
| Total reads (90 bp per read) | 3188238 | 3188238 |
| Aligned reads | 527015 | 413293 |
| Aligned % | 16.53 | 12.96 |
| Number of contigs | 42 | 94 |
| Average contig length (bp) | 3061 | 203 |
| Size of largest contig (bp) | 15751 | 640 |
| Contig N50 (bp) | 11365 | 221 |
| Number of scaffolds | 37 | 52 |
| Average scaffold length (bp) | 3478 | 1769 |
| Size of largest scaffold (bp) | 18085 | 16373 |
| Scaffold N50 (bp) | 15350 | 3039 |
| Overall average read depth (estimated value) | 329.38x | 295.2x |
| Number of gaps | 17 | 118 |
| Average gap length (bp) | 1123 | 402 |
| Size of largest gap (bp) | 1452 | 2237 |
| Sequence GC% | 35.94 | 36.21 |
The plastomes of both MX plants are about 140 kb, which is larger than most published holoparasites, including Cistanche deserticola Ma (~102 kb) (Li et al., 2013), Cuscuta reflexa Roxb. and C. exaltata Engelm. (~121 kb and 125 kb, respectively) (Funk et al., 2007; McNeal et al., 2007), as well as Monotropa hypopitys L. (34.8 kb) (Gruzdev et al., 2016), but smaller than the complete plastome of the autotroph N. tabacum (Kunnimalaiyaan and Nielsen, 1997). Similar results were reported for Corallorhiza Gagnebin (Barrett et al., 2014) and Orobanchaceae (Wicke et al., 2013). The plastome of P. decorata contains 105 genes, while Chimaphila japonia contains 102 genes (Table 2). Both contain all 37 photosystem genes, 30 tRNA genes, and 4 rRNA genes. The majority of the variation in plastome genes comes from photorespiration genes (ndh complex). The entire ndh gene complex suffered gene loss and pseudogenization. In addition to ndhJ, which has shown diminished hybridization signals for all green Ericaceae, i.e., for both autotrophic and mixotrophic members of this family (Braukmann and Stefanović, 2012), we found that ndhF has been completely lost in both plants, while ndhI has also been lost in C. japonica. The remaining genes of the ndh complex in both plastomes are pseudogenes. This finding differs from hybridization signal data reported previously (Braukmann and Stefanović, 2012). The gene rpoA RNA polymerase (rpo) has also been lost from both plastomes. Furthermore, both plastomes lack a functional ycf2 gene, which has become a pseudogene with only about 100 bp left. Three essential genes were not detected: clpP, accD and ycf1. Previous studies reported that the hybridization signal for clpP was diminished in most Ericaceae (Braukmann and Stefanović, 2012), so we hypothesized that these genes might actually be lost rather than not detected. Ribosome protein genes also appeared to be absent in both P. decorata (rp123 and rps15) and C. japonica (rps18). The absence of hybridization signal in Pyroleae for rpl23 is unique within Ericaceae and distinguishes this tribe from the rest of the family (Braukmann and Stefanović, 2012). Our results were consistent with both predictions and previous research on plastomes of semiparasitic plants (Jansen et al., 2007; Wicke et al., 2013).
| Photosystem | Pd | Cj | Photorespiration | Pd | Cj | tRNA | Pd | Cj |
| atpA | + | + | ndhA | ψ | ψ | trnA-ugc | + | + |
| atpB | + | + | ndhB | ψ | ψ | trnC-gca | + | + |
| atpE | + | + | ndhC | ψ | ψ | trnD-guc | + | + |
| atpF | + | + | ndhD | ψ | ψ | trnE-uuc | + | + |
| atpH | + | + | ndhE | ψ | ψ | trnF-gaa | + | + |
| atpI | + | + | ndhF | - | - | trnfM-cau | + | + |
| ccsA | + | + | ndhG | ψ | ψ | trnG-gcc | + | + |
| cemA | + | + | ndhH | ψ | ψ | trnG-ucc | + | + |
| petA | + | + | ndhI | ψ | - | trnH-gug | + | + |
| petB | + | + | ndhJ | ψ | - | trnI-cau | + | + |
| petD | + | + | ndhK | ψ | ψ | trnI-gau | + | + |
| petG | + | + | ribisome | Pd | Cj | trnK-uuu | + | + |
| petL | + | + | infA | + | + | trnL-caa | + | + |
| petN | + | + | rpl2 | + | + | trnL-uaa | + | + |
| psaA | + | + | rpl14 | + | + | trnL-uag | + | + |
| psaB | + | + | rpl16 | + | + | trnM-cau | + | + |
| psaC | + | + | rpl20 | + | + | trnN-guu | + | + |
| psaI | + | + | rpl22 | + | + | trnP-ugg | + | + |
| psaJ | + | + | rpl23 | - | - | trnQ-uug | + | + |
| psbA | + | + | rpl32 | + | + | trnR-acg | + | + |
| psbB | + | + | rpl33 | + | + | trnR-ucu | + | + |
| psbC | + | + | rpl36 | + | + | trnS-gcu | + | + |
| psbD | + | + | rps2 | + | + | trnS-gga | + | + |
| psbE | + | + | rps3 | + | + | trnS-uga | + | + |
| psbF | + | + | rps4 | + | + | trnT-ggu | + | + |
| psbH | + | + | rps7 | + | + | trnT-ugu | + | + |
| psbI | + | + | rps8 | + | + | trnV-gac | + | + |
| psbJ | + | + | rps11 | + | + | trnV-uac | + | + |
| psbK | + | + | rps12 | + | + | trnW-cca | + | + |
| psbL | + | + | rps14 | + | + | trnY-gua | + | + |
| psbM | + | + | rps15 | N | + | rRNA | Pd | Cj |
| psbN | + | + | rps16 | + | + | rrn16 | + | + |
| psbT | + | + | rps18 | + | N | rrn23 | + | + |
| psbZ | + | + | rps19 | + | + | rrn4.5 | + | + |
| rbcL | + | + | other | Pd | Cj | rrn5 | + | + |
| ycf3 | + | + | clpP | N | N | rpo | Pd | Cj |
| ycf4 | + | + | accD | N | N | matK | + | + |
| ycf1 | N | N | rpoA | - | - | |||
| ycf2 | ψ | ψ | rpoB | + | + | |||
| ycf15 | + | + | rpoC1 | + | + | |||
| rpoC2 | + | + | ||||||
| Note: Pd, P. decorata; Cj, C. japonia; ψ, pseudogene; þ, present; -, missing; N, undetected. | ||||||||
Strength of selection is commonly measured by calculating the ratio of the rates of nonsynonymous substitution over synonymous substitutions, dN/dS (Muse and Gaut, 1997). To determine whether the cpDNA of plants at different heterotrophic levels experienced differing selective pressures, we calculated the dN/dS values to measure the strength of selection for each species (Figs. 1-5). The estimated d, dN, dS and dN/dS rates of the protein-coding chloroplast genes shared between C. japonica, P. decorata and their autotrophic relatives (R. simsii) are shown in Fig. 1. Overall, the three Ericaceae species, C. japonica, P. decorata and R. simsii, have higher d, dN, dS and dN/dS rates than N. tabacum. Within Ericaceae, the MX C. japonica and P. decorata have higher d, dN, dS rates but lower dN/dS ratios than autotrophic (AH) R. simsii, which suggests that MX plants collectively accumulate more dS and dN than AH plants.
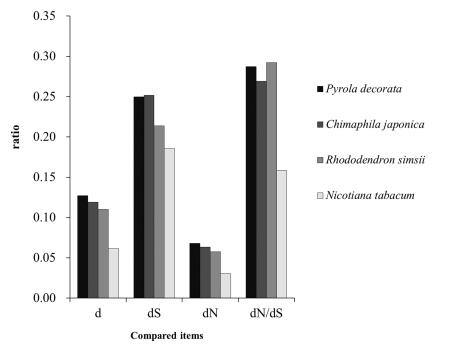
|
| Fig. 1 Deletions (d), synonymous deletions (dS), nonsynonymous deletions (dN), and the ratio of nonsynonymous to synonymous deletions (dN/dS) for 80 genes in four species related to Panax ginseng (NCBI Reference Sequence: NC_006290.1, GI: 52220789). |
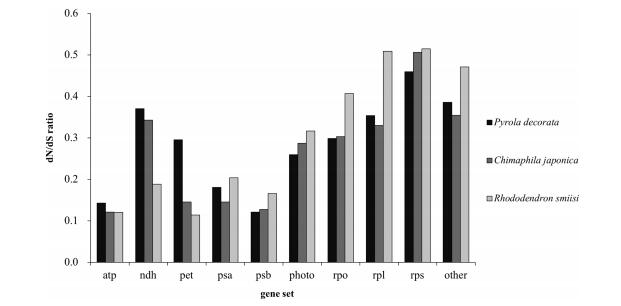
|
| Fig. 2 Comparisons of dN/dS ratio of ten gene sets between mixotrophic plants (P. decorata and C. japonica) and autotrophic plants (R. simsii) in Ericaceae. |
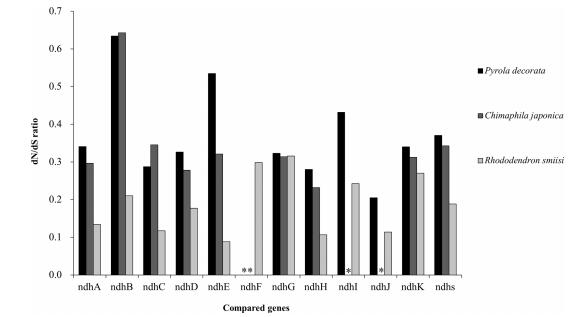
|
| Fig. 3 Comparisons of dN/dS ratio of the ndh gene set between mixotrophic plants (P. decorata and C. japonica) and autotrophic plants (R. simsii) in Ericaceae. *indicates missing data. |
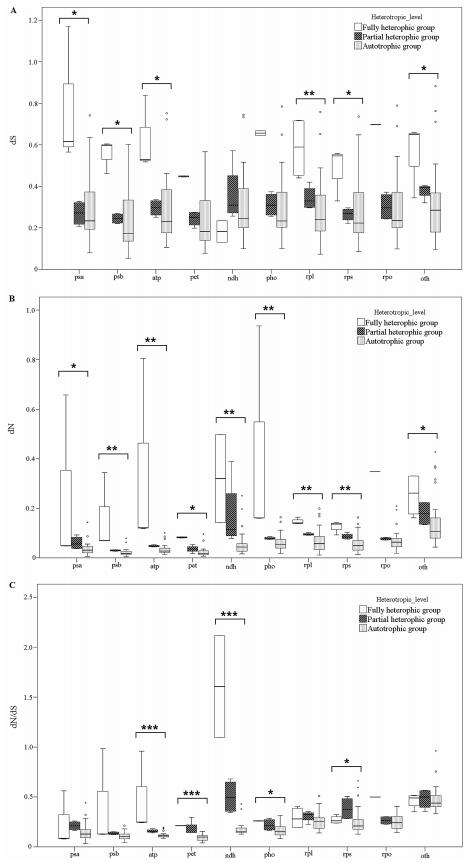
|
| Fig. 4 Boxplots of the value of dS (A), dN (B) and dN/dS (C) for ten groups of genes. Wilcoxon rank sum tests were used to show that dN values for Group Ⅱ were extremely significantly higher than for other angiosperms (P < 0.001), whereas values of dS were significantly different (P = 0.024). The horizontal line is the mean value of dN or dS. For each gene group, the box represents values between quartiles, dotted lines extend to minimum and maximum values, outliers are shown as circles, and the thick, black lines show median values. Small dots show values for gene groups that are statistically different than the values for same gene group in other angiosperms. Boxes include 50% of the distributions. Note: *P < 0.05, **P = 0.05-0.001, ***P < 0.001. |
|
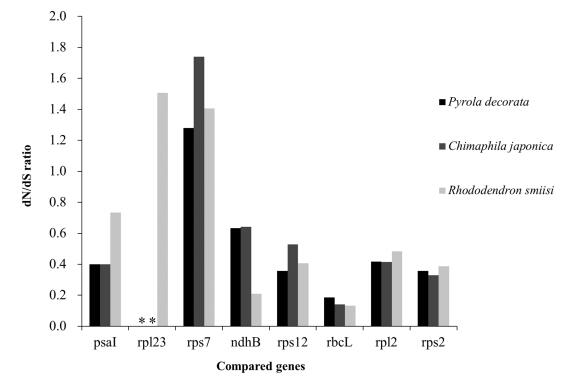
| Fig. 5 Comparisons of genes with dN/dS>1 between mixotrophic plants (P. decorata and C. japonica) and autotrophic plants (R. simsii) in Ericaceae. *indicates missing data. |
The variations in nonsynonymous substitution rates may reflect different selective constraints on plastid gene products in different evolutionary lineages. Using the two newly obtained plastid genome sequences, we tested whether substantial changes have occurred in the selective pressure on genes. The 79 chloroplast protein-coding genes were classified in ten groups according to their function (excluding ycf15, function presently unknown) (Supplementary Table 2). The calculated corrected distances of nonsynonymous nucleotide substitution per nonsynonymous sites (dN) and synonymous substitution per synonymous sites (dS) for C. japonica and P. decorata relative to a common-used outgroup of Panax demonstrates an interesting trend toward relaxed selection in the genome of the fully autotrophic Rododendron (Fig. 2). Of 79 protein-coding genes shared between the two taxa, 56 genes (72.7%) have higher dN/dS value in Rododendron than in Nicotiana (Supplementary Table 3). Furthermore, 12 out of 13 functionally lost genes in C. japonica and P. decorata had higher dN/dS in Rododendron, indicating these genes may have already been under relaxed selection prior to the evolution of heterotrophy in Ericaceae. These findings are consistent with previous studies on Convolvulaceae (McNeal et al., 2007). Analysis of the combined set of ndh genes revealed the ratio of nonsynonymous substitution rates to synonymous substitution rates. We also tested whether ratios of overall selection between classes of genes remaining in two MX plants were similar to autotrophic taxa. These tests showed how patterns of synonymous and nonsynonymous substitution vary between sampled Ericaceae taxa relative to Panax or the various classes of genes in the plastome (Fig. 3). While there were minor changes in synonymous rates between different gene classes, relative ratio tests of synonymous rates showed no significant differences. However, nonsynonymous rate values for psa and psb genes were significantly different from both atp, rpl and rps genes, and there were lower nonsynonymous rates for pet and psa and psb genes in all pairwise comparisons performed (Fig. 2, Supplementary Table 3). Photosynthetic genes showed low nonsynonymous substitution rates, and the dN/dS ratios were higher for photosynthetic genes relative to other gene groups. This suggests that photosynthetic genes have been under strong purifying selection during the evolution of heterotrophic plants. Of the photosynthetic genes, the ndh and pho genes tended to have high dN/dS ratios. Mean dN was higher in Group Ⅰ and Group Ⅱ than in Group Ⅲ, and Wilcoxon rank sum tests showed that the values of dN are significantly different in Group Ⅰ and Ⅱ relative to Group Ⅲ (P < 0.001; Fig. 4); also, the values of dS were significantly different relative to dN and dN/dS (P = 0.024; Fig. 4). Rates for each gene group were compared to reveal patterns of substitutions (Fig. 4). In general, values of dN were higher in Group Ⅱ in comparison to Group Ⅲ, and comparisons revealed that ribosomal protein and RNA polymerase genes were the most significantly different relative to genes involved in photosynthesis. Values of dS exhibited similar yet weaker patterns of variation. Large and small subunit ribosomal protein genes (rpl and rps, respectively) and psb genes were the most significantly different (both P < 0.001). Exceptionally, psbI and psaC had the highest values of dN and dS for photosystem genes (Supplementary Table 3). Nucleotide substitutions were compared and revealed a strong lineage-specific pattern of increases in dN and dS for different heterotrophic levels. However, these results also showed that values of dN were greater overall for Group Ⅱ relative to other groups.
4. Discussions 4.1. Gene loss and pseudogenization of mixotrophic plastomesPlastid gene loss is a general trend in heterotrophic taxa and depends largely on selective pressure on photosynthetic function. As shown in Table 2, none of the genes involved directly in the photosynthetic pathway have been lost in either MX Ericaceae in our study (Table 2), which indicates that plastid-encoded genes for the photosynthetic apparatus are highly conserved in land plant plastomes and evolved under strong selection for maintaining genes directly involved in photosynthesis in mixotrophic taxa (except ndh genes). Gene loss or pseudogenization have only been reported in heterotrophic plants (Wickett et al., 2008). Although MX plants have reduced photosynthetic activity, photosyntheticrelated genes are retained as a whole. This is similar to hemiparasitic Cuscuta, in which genes in the photosynthetic pathways have generally been retained, except for the loss of psaI (Funk et al., 2007; McNeal et al., 2007). It has been previously hypothesized that rbcL potentially has a function unrelated to photosynthesis (Bungard, 2004). The retention of a rbcL open reading frame has been observed in other fully heterotrophic taxa and further investigation is required to elucidate its role outside of photosynthesis (Krause, 2008). Values of dN/dS for photosystem Ⅰ (psa genes group) and photosystem Ⅱ genes (psb genes group) were not significantly different among the three groups studied here; but for group atp, pet and ndh, it was extremely significantly different (P < 0.001). This indicates that photosystem genes have been undergoing stronger selective pressure.
4.1.1. ndh complexThe ndh complex may be involved in chlororespiration and electron recycling (Blazier et al., 2011; Claude and Palmer, 1990) and is presumed to consist of the first genes lost in the transition to heterotrophy. They have been pseudogenized or entirely lost several times during land plant evolution (Martín and Sabater, 2010; McNeal et al., 2007). The loss of the ndh complex is correlated with a decreased reliance on photosynthesis and it is thought to be dispensable in mild environments when maintaining photosynthetic rigor is no longer essential for survival (Martín and Sabater, 2010). This is particularly true for mixotrophic plants living under forest canopies in which the ability to exploit neighboring hosts improves the ability to survive low light conditions (Selosse and Roy, 2009). If the ndh complex is dispensable under these conditions, then we can predict parallel loss of this complex in other lineages of mixotrophic plants with an otherwise preserved photosynthetic apparatus analogous to that in MX species. Within mixotrophic Pyroleae, all the ndh genes have been pseudogenized. The tendency to lose all ndh genes provides an opportunity to investigate the loss of these genes from the plastome (Fig. 3). Throughout land plants, the presence of mycorrhizae and ndh loss appear to be only imperfectly correlated (Braukmann and Stefanović, 2012). Our results here suggest there is strong evidence that ndh gene loss in MX plants is inevitable.
4.1.2. Plastid-encoded polymeraseThe plastid-encoded RNA polymerase (PEP) is a multi-subunit enzyme and is encoded by four plastid genes: rpoA, rpoB, rpoC1, and rpoC2. A loss of the PEP subunits encoded by the plastid rpo genes would therefore deprive plastids of the possibility of transcribing genetic information, making these genes essential for life (Wicke et al., 2011). We found that three of four rpo genes were retained in the plastomes of C. japonica and P. decorata. The absence of plastid-encoded rpoA genes in two MX plants may suggest a shift from PEP to nuclear-encoded plastid RNA polymerase (NEP) for the remaining transcriptional units (Krause, 2008). Similar transitions have been observed in Epifagus Nutt. and Cuscuta and these transitions are presumed to precede loss of photosynthetic capacity (Delannoy et al., 2011; McNeal et al., 2007; Wolfe et al., 1992). To date, the only exceptions are the plastomes of Custuca gronovii and C. obtusiflora, which were the first and only plastomes with a confirmed loss of the entire rpo gene set in a plastome where photosynthesis genes are not only presented (Funk et al., 2007), but, moreover, actively expressed (Berg et al., 2003). This indicates that rpoA gene loss may be the beginning of all rpo gene loss in Ericaceae MX taxa.
4.1.3. Large and small ribosomal protein (rpl and rps) genesThe two MX Ericaceae plants also exhibited some losses of large and small ribosomal protein (rpl and rps) genes, which is more extensive than those observed in Rhizanthella R.S. Rogers, Cuscuta and Epifagus (Delannoy et al., 2011; McNeal et al., 2007; Wolfe et al., 1992). Most genes coding for ribosomal subunit proteins have been transferred to the nuclear genome. However, land plant plastomes commonly encode twelve proteins for the small ribosomal subunits (rps genes) and nine large ribosomal subunit proteins (rpl genes). Loss of rps and rpl genes from plastomes is rare, but has been detected in rosids (Jansen et al., 2007, 2011; Magee et al., 2010) and a variety of non-photosynthetic or minimally photosynthetic angiosperm. The loss of rpl23 in P. decorata and C. japonica was confirmed, but whether rps15 and rps18 were lost or simply undetected is unclear. Loss of rps and rpl genes from plastomes does not affect the expression of ribosomal protein, so these losses indicate a greater reliance on nuclear encoded polymerases and ribosomal proteins to translate the remaining plastid genes (Braukmann and Stefanović, 2012).
4.1.4. ycf1, clpP and accD genes were undetectedOneinteresting result in this study is that ycf1, clpP and accD were undetected in both P. decorata and C. japonica. These genes have functions outside of photosynthesis and were expected to be retained in heterotrophic plants, as previously hypothesized (Barbrook et al., 2006; Bungard, 2004). Chloroplast-encoded clpP has been reported to be involved in multiple processes of chloroplast development, including a housekeeping function that is indispensable for cell survival (Shikanai et al., 2001). Nevertheless, it is known that clpP and accD can be functionally transferred to the nucleus and these loci have been lost multiple times from the plastids of flowering plants (Jansen et al., 2007). Parsimony analyses reconstructing unambiguous changes in gene content among plants revealed that the gene ycf1 was gained in a common ancestor of several green algae and land plants (Maul et al., 2002). Therefore, our detection of a pseudogene copy of ycf2 may provide evidence that those four divergent genes have been transferred to the nucleus in MX Ericaceae. Functions of ycf1 and ycf2 have been hypothesized to involve a number of processes: ATPase-related activities, chaperone-function, cell division, and structural remodeling and/or linkage of plastid chromosomes to protein and/or membrane structures (Wicke et al., 2011). Complete loss of both ycf1 and ycf2 from the plastomes of some, but not all, derived monocot lineages have been reported previously (Guisinger et al., 2010; Maier et al., 1995; Yang et al., 2010). It is therefore possible that these genes may now be less essential in MX Ericaceae plastomes.
4.2. Selective constraint on plastid genes in mixotrophyNonsynonymous substitutions tend to have larger influences on gene function than synonymous substitutions, leading to autotrophic plant group experiencing stronger evolutionary constraints on their cpDNA than heterotrophic plant group. Nonsynonymous mutations are purified through selection to maintain an efficient photosynthetic system. The dN/dS ratios varied among the different genes, implying that different genes accumulated different amounts of deleterious mutations (Fig. 4). Autotrophic plants have lower dN/dS ratios in their cpDNA than heterotrophic species. Thus, the cpDNA of autotrophic plants has experienced stronger purifying selection to maintain efficient photosynthetic function. In heterotrophic plants, the demand for photosynthetic products can be derived from symbiotic, parasitic or other organisms. Because of this heterotrophic individuals that with lower metabolic efficiency are more likely to survive and reproduce in a population than autotrophic individuals under similar circumstances. Taken together, differing constraints on photosynthetic function, as well as the influence of effective population size, explain differences in selective patterns in cpDNA of plants with different photosynthetic capacities.
To assess the selection constraint on plastid genes in P. decorata and C. japonica, we analyzed synonymous and nonsynonymous substitutions in protein-coding genes. Rhododendron, an autotrophic Ericaceae plant, was also included in this analysis. The examination of dN/dS ratio for individual genes that are shared between them demonstrated that most genes in both MX species are evolving under purifying selection (Fig. 2). There were two genes with dN/dS ratios greater than 1: Rhododendron rp123 and C. japonica rps7. The sequence of rpl23 is lost in P. decorata and C. japonica, while rps7 differs by a single substitution (nonsynonymous) (Fig. 5). rpl23 is a pseudogene in several photosynthetic plants (Logacheva et al., 2008). High dN/dS in this gene may indicate the relaxation of selection constraint as an initial stage of pseudogenization. However, in rps14 and rpl33, dN/dS have been found to be increased for short genes (Haddrill et al., 2007), and therefore may not be biologically relevant.
To gain a general view of substitution rates in P. decorata and C. japonica, we calculated substitution values for all genes combined in one sequence. This demonstrated that dN/dS ratios are very similar between two photosynthetic species and between photosynthetic and nonphotosynthetic ones. In contrast, the number of both synonymous and nonsynonymous substitutions differed greatly. The apparent effect of higher substitution rate is often observed in sunshade plants when compared with sunlight plants (Mathews et al., 2003). This implies that this effect is related to heterotrophy. It also suggests that plastid genes in MX Ericaceae plants have higher mutation rates but retain their function. Rhododendron has a much higher dN/dS ratio than P. decorata and C. japonica, indicating that autotrophic Ericaceae plant plastomes are undergoing a degrading stage that may be caused by their relationship with mycorriza (Kerley and Read, 1995, 1997; Stribley and Read, 1980). All classes of genes appear to have evolved under strong negative selection with dN/dS ratios much lower than 1, with photosystem and pet genes remaining the most highly conserved, while the AH plant Rhododendron has a slightly higher dN/dS ratio. Our results suggest that during the evolutionary history of Pyroleae, there was a tendency to increase heterotrophy in the MX group, which is consistent with carbon nutrition research (Matsuda et al., 2012).
5. ConclusionsBy sequencing the plastomes of two MX Ericaceae plants, we have been able to gain a better understanding of the plastome of mixotrophs as dependence on photosynthesis for carbohydrate production decreases. Plastomes of the two mixotrophic plants, P. decorata and C. japonica, are generally very similar to that of nonparasitic plants, although have a greater degree of ndh gene pseudogenization. Genes directly involved in photosynthesis and under strong selection are retained in the two mixotrophs, but with greatly increased overall nucleotide substitution. By studying the similarities and the differences between the two MX and AH plants, we have gained insights into which changes might occur in the plastid genome during the transition from autotrophy to heterotrophy and identified future research directions. Because of unclosed plastome sequences based on present data assembly, further research is also required to gain a clear understanding of the evolution of autotrophs and mixotrophs in Ericaceae.
6. AcknowledgmentsWe gratefully acknowledge four anonymous reviewers for useful comments on the manuscript.
7. Appendix A. Supplementary dataSupplementary data related to this article can be found at http://dx.doi.org/10.1016/j.pld.2016.11.005.
| Barbrook A.C., Howe C.J., Purton S., 2006. Why are plastid genomes retained in non-photosynthetic organisms?. Trends Plant Sci, 11, 101-108. DOI:10.1016/j.tplants.2005.12.004 | ||
| Barkman T.J., McNeal J.R., Lim S.-H., et al., 2007. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol. Biol, 7, 248. DOI:10.1186/1471-2148-7-248 | ||
| Barrett C.F., Freudenstein J.V., Li J., et al., 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. msu252 Mol. Biol. Evol, 31(12), 3095-3112. DOI:10.1093/molbev/msu252 | ||
| Berg S., Krupinska K., Krause K., 2003. Plastids of three Cuscuta species differing in plastid coding capacity have a common parasite-specific RNA composition. Planta, 218, 135-142. DOI:10.1007/s00425-003-1082-8 | ||
| Bidartondo M.I., 2005. The evolutionary ecology of myco-heterotrophy. New Phytol, 167, 335-352. DOI:10.1111/j.1469-8137.2005.01429.x | ||
| Blazier J.C., Guisinger M.M., Jansen R.K., 2011. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol. Biol, 76, 263-272. DOI:10.1007/s11103-011-9753-5 | ||
| Braukmann T., Stefanović S., 2012. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol. Biol, 79, 5-20. DOI:10.1007/s11103-012-9884-3 | ||
| Bungard R.A., 2004. Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. Bioessays, 26, 235-247. DOI:10.1002/(ISSN)1521-1878 | ||
| Cameron D.D., Coats A.M., Seel W.E., 2006. Differential resistance among host and non-host species underlies the variable success of the hemi-parasitic plant Rhinanthus minor. Ann. Bot, 98, 1289-1299. DOI:10.1093/aob/mcl218 | ||
| Claude W.d., Palmer J.D., 1990. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature, 348, 337-339. DOI:10.1038/348337a0 | ||
| Delannoy E., Fujii S., des Francs-Small C.C., et al., 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol, 28, 2077-2086. DOI:10.1093/molbev/msr028 | ||
| Drouin G., Daoud H., Xia J., 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenetics Evol, 49, 827-831. DOI:10.1016/j.ympev.2008.09.009 | ||
| Funk H.T., Berg S., Krupinska K., et al., 2007. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol, 7, 45. DOI:10.1186/1471-2229-7-45 | ||
| Gao L., Su Y.J., Wang T., 2010. Plastid genome sequencing, comparative genomics, and phylogenomics: current status and prospects. J. Syst. Evol, 48, 77-93. DOI:10.1111/jse.2010.48.issue-2 | ||
| Gaut B., Yang L., Takuno S., et al., 2011. The patterns and causes of variation in plant nucleotide substitution rates. Annu. Rev. Ecol. Evol. Syst, 42, 245-266. DOI:10.1146/annurev-ecolsys-102710-145119 | ||
| Gruzdev E.V., Mardanov A.V., Beletsky A.V., et al., 2016. The complete chloroplast genome of parasitic flowering plant Monotropa hypopitys: extensive gene losses and size reduction. Mitochondrial DNA Part B, 1, 1-2. DOI:10.1080/23802359.2016.1156364 | ||
| Guisinger M.M., Chumley T.W., Kuehl J.V., et al., 2010. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J. Mol. Evol, 70, 149-166. DOI:10.1007/s00239-009-9317-3 | ||
| Haddrill P.R., Halligan D.L., Tomaras D., et al., 2007. Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol, 8, R18. DOI:10.1186/gb-2007-8-2-r18 | ||
| Jansen R.K., Cai Z.Q., Raubeson L.A., et al., 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genomescale evolutionary patterns. Proc. Natl. Acad. Sci, 104, 19369-19374. DOI:10.1073/pnas.0709121104 | ||
| Jansen R.K., Saski C., Lee S.-B., et al., 2011. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol, 28, 835-847. DOI:10.1093/molbev/msq261 | ||
| Julou T., Burghardt B., Gebauer G., et al., 2005. Mixotrophy in orchids: insights from a comparative study of green individuals and nonphotosynthetic individuals of Cephalanthera damasonium. New Phytol, 166, 639-653. DOI:10.1111/j.1469-8137.2005.01364.x | ||
| Kerley S.J., Read D.J., 1995. The biology of mycorrhiza in the Ericaceae. New Phytol, 131, 369-375. DOI:10.1111/nph.1995.131.issue-3 | ||
| Kerley S.J., Read D.J., 1997. The biology of mycorrhiza in the Ericaceae. New Phytol, 136, 691-701. DOI:10.1046/j.1469-8137.1997.00778.x | ||
| Kim K.J., Lee H.L., 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res, 11, 247-261. DOI:10.1093/dnares/11.4.247 | ||
| Krause K., 2008. From chloroplasts to "ryptic" plastids: evolution of plastid genomes in parasitic plants. Curr. Genet, 54, 111-121. DOI:10.1007/s00294-008-0208-8 | ||
| Kron K.A., 1997. Phylogenetic relationships of Rhododendroideae (Ericaceae). Am. J. Bot, 84, 973. DOI:10.2307/2446288 | ||
| Kron K.A., Judd W.S., Stevens P.F., et al., 2002. Phylogenetic classification of Ericaceae: molecular and morphological evidence. Botanical Rev, 68, 335-423. DOI:10.1663/0006-8101(2002)068[0335:PCOEMA]2.0.CO;2 | ||
| Kunnimalaiyaan M., Nielsen B.L., 1997. Fine mapping of replication origins (oriA and oriB) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res, 25, 3681-3686. DOI:10.1093/nar/25.18.3681 | ||
| Lemaire B., Huysmans S., Smets E., et al., 2011. Rate accelerations in nuclear 18S rDNA of mycoheterotrophic and parasitic angiosperms. J. Plant Res, 124, 561-576. DOI:10.1007/s10265-010-0395-5 | ||
| Li R.Q., Yu C., Li Y.R., et al., 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics, 25, 1966-1967. DOI:10.1093/bioinformatics/btp336 | ||
| Li X., Zhang T.-C., Qiao Q., et al., 2013. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS One, 8, e58747. DOI:10.1371/journal.pone.0058747 | ||
| Logacheva M.D., Samigullin T.H., Dhingra A., et al., 2008. Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale-a wild ancestor of cultivated buckwheat. BMC Plant Biol, 8, 59. DOI:10.1186/1471-2229-8-59 | ||
| Logacheva M.D., Schelkunov M.I., Penin A.A., 2011. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol. Evol, 3, 1296-1303. DOI:10.1093/gbe/evr102 | ||
| Magee A.M., Aspinall S., Rice D.W., et al., 2010. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res, 20, 1700-1710. DOI:10.1101/gr.111955.110 | ||
| Maier R.M., Neckermann K., Igloi G.L., et al., 1995. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol, 251, 614-628. DOI:10.1006/jmbi.1995.0460 | ||
| Martín M., Sabater B., 2010. Plastid ndh genes in plant evolution. Plant Physiol. Biochem, 48, 636-645. DOI:10.1016/j.plaphy.2010.04.009 | ||
| Mathews S., Burleigh J.G., Donoghue M.J., 2003. Adaptive evolution in the photosensory domain of phytochrome A in early angiosperms. Mol. Biol. Evol, 20, 1087-1097. DOI:10.1093/molbev/msg123 | ||
| Matsuda Y., Shimizu S., Mori M., et al., 2012. Seasonal and environmental changes of mycorrhizal associations and heterotrophy levels in mixotrophic Pyrola japonica (Ericaceae) growing under different light environments. Am. J. Bot, 99, 1177-1188. DOI:10.3732/ajb.1100546 | ||
| Maul J.E., Lilly J.W., Cui L., et al., 2002. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a Sea of Repeats. Plant Cell Online, 14, 2659-2679. DOI:10.1105/tpc.006155 | ||
| McNeal J.R., Kuehl J.V., Boore J.L., et al., 2007. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol, 7, 57. DOI:10.1186/1471-2229-7-57 | ||
| Molina J., Hazzouri K.M., Nickrent D., et al., 2014. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol. Biol. Evol, 31, 793-803. DOI:10.1093/molbev/msu051 | ||
| Moore M.J., Soltis P.S., Bell C.D., et al., 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci, 107, 4623-4628. DOI:10.1073/pnas.0907801107 | ||
| Motomura H., Selosse M.A., Martos F., et al., 2010. Mycoheterotrophy evolved from mixotrophic ancestors: evidence in Cymbidium (Orchidaceae). Ann. Bot, 106, 573-581. DOI:10.1093/aob/mcq156 | ||
| Muse S.V., Gaut B.S., 1997. Comparing patterns of nucleotide substitution rates among chloroplast loci using the relative ratio test. Genetics, 146, 393-399. | ||
| Nickrent, D. L. , Duff, R. J. , Colwell, A. E. , et al. , 1998. Molecular phylogenetic and evolutionary studies of parasitic plants. In: Soltis, Douglas E. , Doyle, J. J. (Eds. ), Molecular Systematics of Plants Ⅱ. Springer, New York, pp. 211-241. | ||
| Selosse M.A., Roy M., 2009. Green plants that feed on fungi: facts and questions about mixotrophy. Trends Plant Sci, 14, 64-70. | ||
| Shikanai T., Shimizu K., Ueda K., et al., 2001. The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol, 42, 264-273. DOI:10.1093/pcp/pce031 | ||
| Stefanović S., Olmstead R.G., 2004. Testing the phylogenetic position of a parasitic plant (Cuscuta, Convolvulaceae, Asteridae): Bayesian inference and the parametric bootstrap on data drawn from three genomes. Syst. Biol, 53, 384-399. DOI:10.1080/10635150490445896 | ||
| Stribley D.P., Read D.J., 1980. The biology of mycorrhiza in the Ericaceae. New Phytol, 86, 365-371. DOI:10.1111/nph.1980.86.issue-4 | ||
| Tamura K., Peterson D., Peterson N., et al., 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol, 28, 2731-2739. DOI:10.1093/molbev/msr121 | ||
| Tedersoo L., Pellet P., Koljalg U., et al., 2007. Parallel evolutionary paths to mycoheterotrophy in understorey Ericaceae and Orchidaceae: ecological evidence for mixotrophy in Pyroleae. Oecologia, 151, 206-217. DOI:10.1007/s00442-006-0581-2 | ||
| Triboush S.O., Danilenko N.G., Davydenko O.G., 1998. A method for isolation of chloroplast DNA and mitochondrial DNA from sunflower. Plant Mol. Biol. Report, 16, 183. DOI:10.1023/A:1007487806583 | ||
| Wicke S., Müller K.F., de Pamphilis C.W., et al., 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell, 25, 3711-3725. DOI:10.1105/tpc.113.113373 | ||
| Wicke S., Schneeweiss G.M., Müller K.F., et al., 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol, 76, 273-297. DOI:10.1007/s11103-011-9762-4 | ||
| Wickett N.J., Zhang Y., Hansen S.K., et al., 2008. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol. Biol. Evol, 25, 393-401. DOI:10.1093/molbev/msm267 | ||
| Wolfe K.H., Morden C.W., Palmer J.D., 1992. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci, 89, 10648-10652. DOI:10.1073/pnas.89.22.10648 | ||
| Wyman S.K., Jansen R.K., Boore J.L., 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics, 20, 3252-3255. DOI:10.1093/bioinformatics/bth352 | ||
| Yang M., Zhang X., Liu G., et al., 2010. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS One, 5, e12762. DOI:10.1371/journal.pone.0012762 | ||
| Zimmer K., Hynson N.A., Gebauer G., et al., 2007. Wide geographical and ecological distribution of nitrogen and carbon gains from fungi in pyroloids and monotropoids (Ericaceae) and in orchids. New Phytol, 175, 166-175. DOI:10.1111/nph.2007.175.issue-1 |



