The species composition of plant assemblages is largely determined by a long history of biogeographic and evolutionary processes (Webb et al., 2002) . Analyses of the composition and assembly of plant assemblages that only focus on present day and local-scale ecological interactions are therefore incomplete and can not successfully explain the primary reasons why plant species composition varies across regions (Swenson, 2011, 2013) . By contrast, integrating phylogenetic information into ecology provides a promising way to explore the ecological, biogeographic, and evolutionary processes that drive plant assemblies at multiple spatial scales (Webb, 2000; Graham and Fine, 2008) . For example, at a global scale, phylogenies permit a re-evaluation of the relative roles of plate tectonics and long-distance dispersal in assemblages of regional and continental biota (Crisp et al., 2004) ; at a local scale, phylogenies permit an examination of phylogenetic relatedness of species and provide insights into the assembly of biological communities (Webb et al., 2002) .
The analysis of phylogenetic community structure attempts to reveal the relative importance of community assembly processes, with a primary focus on habitat filtering and competitive exclusion (Cavender-Bares et al., 2009) . In the most common framework, the presence of phylogenetic clustering indicates the predominance of habitat filtering in structuring the community, while the presence of phylogenetic overdispersion or evenness indicates that competitive exclusion and niche differentiation are playing a structural role in a given community (Webb et al., 2002; Cavender-Bares et al., 2004; Mayfield and Levine, 2010) . However, these interpretations depend on a potentially unsafe assumption that closely related species tend to share similar traits and occupy similar ecological niches than more distantly related species (Darwin, 1859;Webb, 2000;Webb et al., 2002) . This is often termed 'phylogenetic signal' (Losos, 2008) .
Many empirical studies have documented similarities in observed ecological traits among closely related species for a variety of taxa (e.g., Burns and Strauss, 2011; Violle et al., 2011; Münkemüller et al., 2012) . Where ecological traits show a phylogenetic signal, meaning that closely related species display more similar traits than distantly related species, then phylogenetic relatedness among species has been used as a proxy for ecological similarity of co-occurring species to infer mechanisms of community assembly (Webb et al., 2002; Kamilar and Cooper, 2013) .
However, in a review of studies examining phylogenetic signal of ecological traits, Losos (2008) called attention to several instances where no relationship between evolutionary relatedness and ecological similarity was found. For example, Silvertown et al. (2006) found no phylogenetic signal in the hydrological niches of co-occurring meadow plants, while Cavender-Bares et al. (2004) found phylogenetic signal in some traits but not others in Floridian oak communities. The results of these and other studies reviewed by Losos (2008) caution against using phylogenetic relatedness as a proxy for ecological trait similarity in the context of community assembly when those traits show a phylogenetic signal (Cooper et al., 2010; Baldeck et al., 2013; Yang et al., 2014a) .
Most studies that test phylogenetic signal in ecological traits have focused on a local scale (e.g., Schreeg et al., 2010; Pei et al., 2011; Yang et al., 2014b) and only a few studies have been carried out on a regional scale (e.g., Qian and Zhang, 2014; Du et al., 2015) . To better make useful inferences regarding the evolutionary and biogeographic imprint on present day species co-occurrence, we need to examine phylogenetic signal of ecological traits on many regional scales.
The areal-types or distribution patterns of plants are an important trait for regional plant assemblages because they reflect adaptations of plants to specific climate, environmental conditions, and evolutionary history (Wu et al., 2006, 2010) . However, whether there is significant phylogenetic signal in areal-types across a phylogeny that includes all families and genera of plants in a certain region has not been examined.
China is a hotspot for plant diversity. It has the richest northern temperate flora in the world, with at least 30, 000 species of vascular plants, over 50% of them being endemic (Huang et al., 2013) . The formation of rich plant assemblages has been attributed to variety of habitats, unbroken connections from tropical to subtropical, temperate and boreal forest, and a stable paleoclimate influenced less by the Pleistocene glaciations than other temperate areas such as much of North America (Qian and Ricklefs, 1999, 2000) . Based on an analysis of distribution patterns of families and genera of seed plants in China, Wu established a scheme of classification of areal-types for families and genera of seed plants in China (Wu, 1965, 1991; Wu et al., 2003, 2006) , in which 15 arealtypes were recognized (Table 1) .
| Order | Distribution patterns | Number of families | Number of genera |
| 1 | Widespread | 48 | 93 |
| 2 | Pantropical | 91 | 313 |
| 3 | Disjunct between tropical Asia and tropical America | 15 | 41 |
| 4 | Old world tropics | 9 | 170 |
| 5 | Tropical Asia and tropical Australasia | 9 | 206 |
| 6 | Tropical Asia and tropical Africa | 6 | 116 |
| 7 | Tropical Asia | 11 | 548 |
| 8 | Northern temperate | 39 | 288 |
| 9 | Disjunct between eastern Asia and North America | 13 | 115 |
| 10 | Old world temperate | 1 | 173 |
| 11 | Temperate Asia | 0 | 54 |
| 12 | Mediterranean and western to central Asia | 5 | 103 |
| 13 | Central Asia | 2 | 108 |
| 14 | Eastern Asia | 6 | 285 |
| 15 | Endemic to China | 2 | 213 |
In this study, the relationship between the evolutionary relatedness of co-occurring taxa and the similarity in their areal-types in China was examined. I ask a central question: are areal-types of families and genera of seed plants non-randomly distributed across the Chinese tree of life? I expect that areal-types may have strong phylogenetic signal across the phylogeny that includes all families and genera of seed plants in China. If this is supported, then closely related taxa have more similar areal-types than more distant relatives, and phylogenetic relatedness may be used as a proxy for ecological similarity. Alternatively, if closely related taxa have more dissimilar areal-types than more distant relatives, this would indicate that areal-types are highly evolutionarily labile, or convergent.
1 Materials and methods 1.1 Data sourcesThe family list and genus list of seed plants in China were compiled utilizing the Flora of China published in 25 volumes by Wu and his colleagues from 1994 to 2013 (Wu et al., 1994-2013) . The flora represents a comprehensive knowledge of plant taxonomy and phytogeography in China. Families and genera that are not native to China were excluded. I assigned each genus to a family, following the scope of seed plant families in Zanne et al. (2014) . A total of 2826 genera and 257 families were included in this study.
The areal-types of families and genera of seed plants were compiled based on the scheme proposed by Wu et al. (2006) (Table 1) .
According to Angiosperm Phylogeny Group's system (APG IV, 2016) , families and genera of seed plants were put in five taxonomic groups: gymnosperms, magnoliids, monocots, superrosids, and superasterids. Eighteen families and 117 genera, which do not belong to any of these groups, were put in the "unclassified" group.
1.2 Phylogeny constructionI constructed a family-level phylogenetic tree and a genus-level phylogenetic tree by grafting the families and genera present in China, respectively, onto a backbone phylogenetic hypothesis using the function S.PhyloMaker (Qian and Jin, 2015) in R 2.15.3 software (R Development Core Team, 2013) with the 'phytools' package (Revell, 2012) . The backbone of the supertree was Zanne et al. (2014) phylogeny, which is the largest and most up-to-date timecalibrated species-level phylogeny of seed plants and uses seven gene regions in addition to fossil data. Given the scarcity of comprehensive time-calibrated phylogenies within families, I followed previous studies to treat genera as polytomies within families (e.g., Hardy et al., 2012; Li et al., 2015a, 2015b) .
1.3 Statistical analysesI used two statistical approaches to determine whether different areal-types of families and genera of seed plants are randomly distributed across the family-level phylogeny and genus-level phylogeny, respectively. First, I conducted a Chi-square analysis (Zar, 1984) on the contingency table of areal-types and six plant groups in the family-level and genus-level, respectively. Second, areal-types of families and genera of seed plants were treated as categorical variables, and a sankoff parsimony score and a fitch parsimony score were separately calculated based on their distribution on the family- or genus-level phylogeny with equal transition probabilities between areal-types (Maddison and Slatkin, 1991) . The observed parsimony score was compared to a null distribution of parsimony scores derived by permuting areal-types across the tips of the phylogeny 999 times from which a p value could be calculated. I then took p value < 0.05 as an indication that closely related taxa tended to have similar areal-types. Analyses were performed in R 2.15.3 software (R Development Core Team, 2013) using the 'picante' (Kembel et al., 2010) and 'phangorn' (Schliep, 2011) packages.
To show the non-random distribution of areal-types across the family-level phylogeny and genus-level phylogeny of seed plants in China, fifteen areal-types were divided into three groups (tropical elements, temperate elements, and widespread) based on a published monograph (Wu et al., 2010) , in which tropical elements include areal-type 2 to areal-type 7 and temperate elements include areal-type 8 to areal-type 15 in Table 1.
2 Results 2.1 Areal-types and phylogenyA description for the areal-types of families and genera of Chinese seed plants and the numbers of families or genera in each areal-type are shown in Table 1.
The resulting family- and genus-level phylogenies included all the 257 families and 2826 genera of seed plants in China (Fig. 1) . Of 257 families, eight are gymnosperms and 249 are angiosperms; of 2826 genera, 35 are gymnosperms and 2791 are angiosperms.
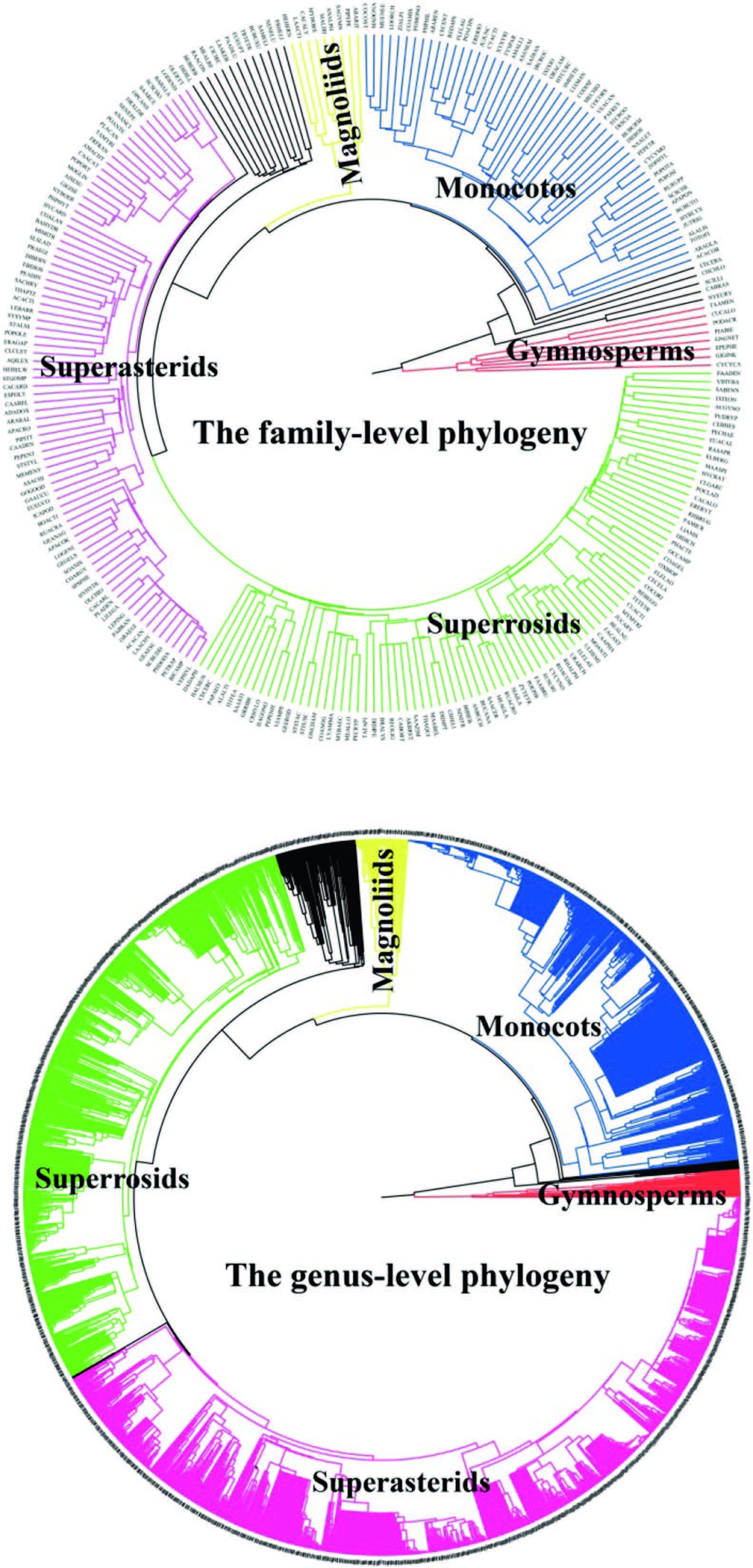
|
| Fig. 1 Phylogenies for all the 257 families and 2826 genera of seed plants in China. Families and genera in the same group (gymnosperms, magnoliids, monocots, superrosids, and superasterids) are shown in the same color; unclassified families and genera are shown in black color. |
Proportions of areal-types were significantly non-randomly distributed among taxonomic groups (χ2=116.1, d.f.=65, p < 0.001 in family-level phylogeny; χ2=333.3, d.f.=70, p < 0.001 in genus-level phylogeny) . When the areal-types of families and genera of seed plants were included in an analysis assessing phylogenetic signal, the p values of the sankoff parsimony score and the fitch parsimony score were less than 0.05 (p=0.003 for both cases in family-level phylogeny; p=0.001 for both cases in genuslevel phylogeny) . Thus, these results are consistent with that of Chisquare analysis, and all these results indicate that the areal-types of families and genera of seed plants exhibited significant phylogenetic signal across the Chinese tree of life (Fig. 2) .
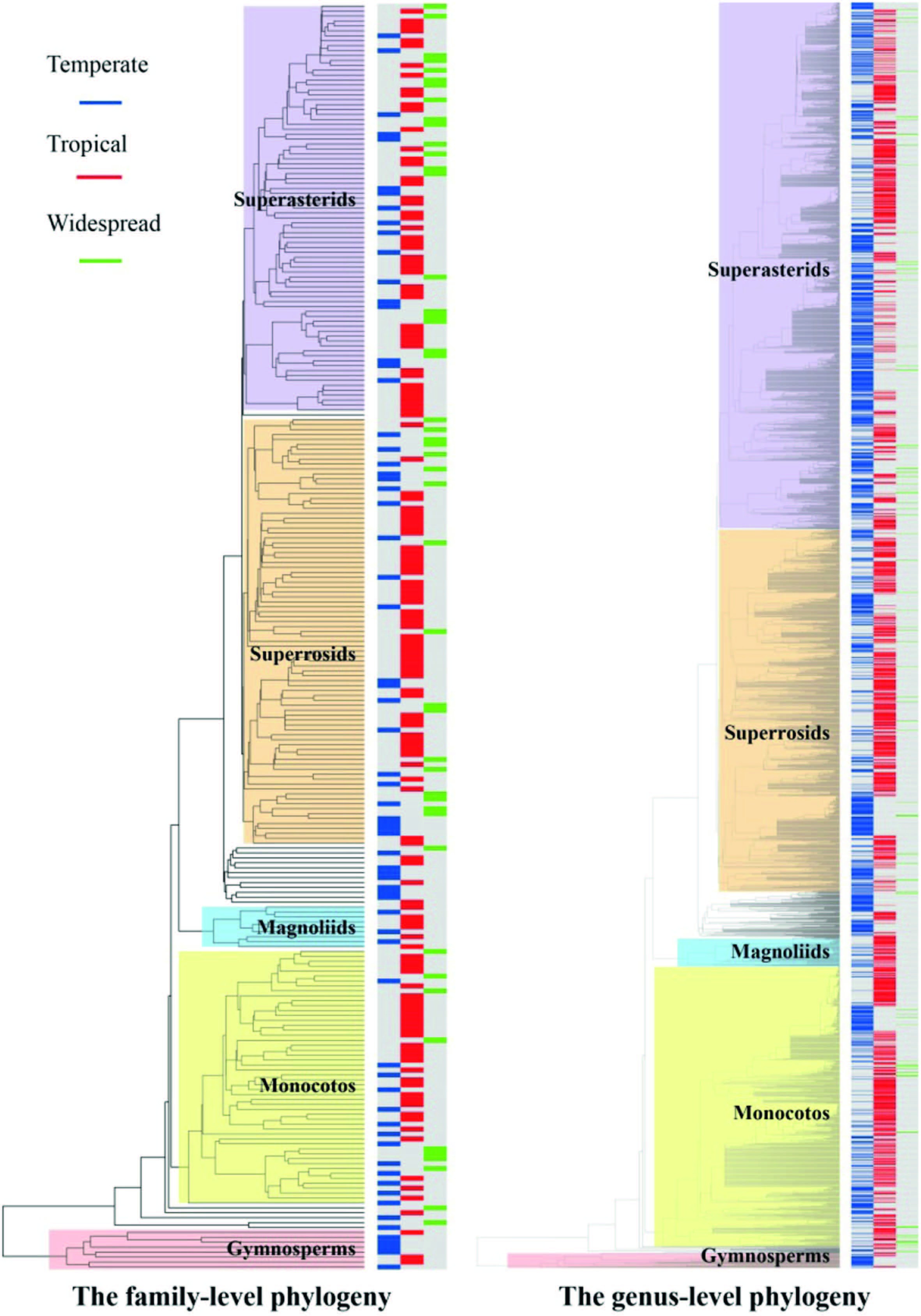
|
| Fig. 2 Phylogenetic signal of areal-types across the family-level phylogeny (left) and genus-level phylogeny (right) of seed plants in China. Areal-types are indicated by three colors in line symbols on branch tips (blue represent temperate elements, red represent tropical elements, and green represent widespread). Families and genera in the same group (gymnosperms, magnoliids, monocots, superrosids, and superasterids) are highlighted with the same colored rectangles. |
In this study, I presented a family-level phylogeny and a genuslevel phylogeny that included all families and genera of extant seed plants in China, respectively. Although the phylogenies generated in my study were based on the phylogeny of Zanne et al. (2014) , they reflect current knowledge about the delineations of, and memberships among families, and provide time-calibrated branch lengths for Chinese seed plants. The phylogenies generated in this study should prove useful for ecological and biogeographic studies in China.
Ideally, a well-resolved phylogeny with time-calibration including all species of seed plants in a certain region will help uncover the evolutionary and biogeographic imprint that potentially explains the assembly processes shaping regional species compositions. However, it is currently not possible to generate a molecular phylogenetic tree for an entire region down to the species level, due to a lack of sequence data for all species. Thus, botanists and plant ecologists, particularly those who work with large data sets covering broad spatial scales, have to rely on phylogenies generated at higher taxonomic ranks such as family- or genus-level phylogeny. If taxa in study samples are widely spread across major clades such as orders and families, rather than restricted to a few clustered major clades in a phylogeny, ecological patterns based on a family- or genus-level phylogeny may not differ substantially from those based on a species-level phylogeny. This is especially true if traits that evolved at deep divisions of major clades play a more important role than those evolved at shallow divisions (e.g., within genera) in driving ecological and biogeographical patterns (Qian and Zhang, 2014, 2016) . As shown in the present study, the areal-types of families and genera of seed plants are significantly, non-randomly distributed and phylogenetic signal of areal-types is significant across the family- and the genus-level phylogeny of seed plants in China.
Across the Chinese tree of life, closely related taxa have more similar areal-types than more distant relatives, which indicates that closely related taxa have similar biogeographic and evolutionary histories. For example, at the family level, Adoxaceae and Caprifoliaceae are only two families in the order Dipsacales, which is a monophyletic clade. Both families are common shrubs (rarely herb) in many broad-leaved forests across the China and their areal-type is northern temperate. The close relationship between them is supported by the shared presence of opposite leaves, tricellular pollen grains, and cellular endosperm without haustoria (Donoghue et al., 2001) . However, Adoxaceae can be easily distinguished from Caprifoliaceae by its actinomorphic flowers, stigmas lobed, and pollen smooth. In contrast, Caprifoliaceae possesses more or less zygomorphic flowers, capitate stigmas, and pollen spinulose (Zhang et al., 2003) . The origin of both families was probably in the northern hemisphere (Beaulieu et al., 2013) . Their diversification exhibited a pronounced correlation with movement into new geographic areas, particularly the dispersal of lineages into new mountainous regions relatively recently, occurring within perhaps ca. 10 mya (Bell and Donoghue, 2005; Moore and Donoghue, 2007) . At the genus level, the family Dipterocarpaceae includes five genera (Dipterocarpus, Hopea, Parashorea, Shorea, Vatica) in China, which are dominant large trees in tropical rain forests from India and Sri Lanka toWest Malesia (Alexander, 1989) . The five genera are trees that may be recognized by their often tworanked and coriaceous leaves with strong and parallel secondary veins, monochasial inflorescences with the flowers have a conspicuously contorted corolla, and distinctive single-seeded nuts surrounding by the unequal sepals (Tsumura et al., 2011) . Within the Dipterocarpaceae, only member of the genus Dipterocarpus possess the trait that calyx in fruit with a distinct tube. Of the remaining genera of the family, all except for Vatica (reticulate tertiary leaf veins) possess scalariform tertiary leaf veins. The genera with scalariform tertiary leaf veins may be clearly differentiated: Hopea has distinct stylopoidum in the ovary, whereas Parashorea and Shorea have no stylopodium in the ovary. Parashorea can be distinguished from Shorea by its prominently lenticellate bark. On the contrary, Shorea possesses scarcely lenticellate bark (Ashton, 1982; Yulita et al., 2005) . The family Dipterocarpaceae has originated in India and later dispersal to South East Asia- Malesia after contact of the Indian and Asian plates in the early Tertiary (Dutta et al., 2011; Shukla et al., 2012) .
The scheme of classification of areal-types or distribution patterns of families and genera of seed plants has been widely used in analyzing national and regional floras of China at various levels and proved helpful in understanding biogeographical issues, such as floristic divisions, endemism and disjunctive distributions (e.g., Wu, 1979, 1983; Li and Li, 1997; Wu et al., 2005; Li et al., 2007) . However, previous studies did not examine phylogenetic signal of areal-types in the context of phylogeny at different spatial scales. Integrating phylogenetic information, my study has successfully found significant biogeographical patterns with respect to phylogenies used, confirming that biogeographical and evolutionary processes played a role in assemblages of Chinese flora.
Acknowledgments: This study was supported by the National Natural Science Foundation of China (grant no. 31370243, 31570212), Natural Science Foundation of Yunnan (grant no. 2014FB169), and the Talent Project of Yunnan (grant no. 2015HB092). I am grateful to Professor Zhekun Zhou for invitation to contribute to this special issue.| Alexander, I.J., 1989. Mycorrhizas in tropical forests. In: Proctor, J. (Ed.), Mineral Nutrients in Tropical Forest and Savanna Ecosystems. Blackwell, Oxford, pp. 169-188. | ||
| APG IV, 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc, 181 -120. | ||
| Ashton, P.S., 1982. Dipterocarpaceae. ser. 1, vol. 9(2). In: van Steenis, C.G.G.J. (Ed.), Flora MalesianaMartinus Nijhoff, The Hague, pp. 237-552. | ||
| Baldeck, C.A., Kembel, S.W., Harms, K.E., et al., 2013. A taxonomic comparison of local habitat niches of tropical trees. Oecologia, 173, 1491 -1498. DOI:10.1007/s00442-013-2709-5 | ||
| Beaulieu, J.M., Tank, D.C., Donoghue, M.J., 2013. A Southern Hemisphere origin for campanulid angiosperms, with traces of the break-up of Gondwana. BMC Evol. Biol, 13, 80 . DOI:10.1186/1471-2148-13-80 | ||
| Bell, C.D., Donoghue, M.J., 2005. Dating the Dipsacales: comparing models, genes, and evolutionary implications. Am. J. Bot, 92, 284 -296. DOI:10.3732/ajb.92.2.284 | ||
| Burns, J.H., Strauss, S.Y., 2011. More closely related species are more ecologically similar in an experimental test. Proc. Natl. Acad. Sci. U.S.A, 108, 5302 -5307. DOI:10.1073/pnas.1013003108 | ||
| Cavender-Bares, J., Ackerly, D.D., Ba um, D ., et al., 2004. Phylogenetic overdispersion in Floridian oak communities. Am. Nat, 163, 823 -843. DOI:10.1086/386375 | ||
| Cavender-Bares, J., Kozak, K.H., Fine, P.V., et al., 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett, 12, 693 -715. DOI:10.1111/ele.2009.12.issue-7 | ||
| Cooper, N., Jetz, W., Freckleton, R.P., 2010. Phylogenetic comparative approaches for studying niche conservatism. J. Evol. Biol, 23, 2529 -2539. DOI:10.1111/jeb.2010.23.issue-12 | ||
| Crisp, M.C., Cook, L., Steane, D., 2004. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities. Philos.Trans. R. Soc. Lond. B. Biol. Sci, 359, 1551 -1571. DOI:10.1098/rstb.2004.1528 | ||
| Darwin, C., 1859. On the Origin of Species. John Murray, London. | ||
| Donoghue, M.J., Eriksson, T., Reeves, P.A., et al., 2001. Phylogeny and phylogenetic taxonomy of Dipsacales, with special reference to Sinadoxa and Tetradoxa (Adoxaceae). Harv. Pap. Bot, 6, 459 -479. | ||
| Du, Y., Mao, L., Queenborough, S.A., et al., 2015. Phylogenetic constraints and trait correlates of flowering phenology in the angiosperm flora of China. Glob. Ecol. Biogeogr, 24, 928 -938. DOI:10.1111/geb.12303 | ||
| Dutta, S., Tripathi, S.M., Mallick, M ., et al., 2011. Eocene out-of-India dispersal of Asian dipterocarps. Rev. Palaeobot. Palynol, 63 -68. | ||
| Graham, C.H., Fine, P.V.A., 2008. Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecol. Lett, 11, 1265 -1277. DOI:10.1111/ele.2008.11.issue-12 | ||
| Hardy, O.J., Couteron, P., Munoz, F ., et al., 2012. Phylogenetic turnover in tropical tree communities: impact of environmental filtering, biogeography and mes- oclimatic niche conservatism. Glob. Ecol. Biogeogr, 21, 1007 -1016. DOI:10.1111/geb.2012.21.issue-10 | ||
| Huang, H.W., Oldfield, S., Qian, H., 2013. Global significance of plant diversity in China. In: Hong, D.Y., Blackmore, S. (Eds.), Plants of China, a Companion to the Flora of China. Science Press, Beijing, pp. 7-34. | ||
| Kamilar, J.M., Cooper, N., 2013. Phylogenetic signal in primate behaviour, ecology and life history. Lond. B. Biol. Sci, 368, 20120341 . DOI:10.1098/rstb.2012.0341 | ||
| Kembel, S.W., Cowan, P.D., Helmus, M.R., et al., 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics, 26, 1463 -1464. DOI:10.1093/bioinformatics/btq166 | ||
| Li, R., Dao, Z.L., Ji, Y.H., et al., 2007. A floristic study on the seed plants of the northern Gaoligong mountains in western Yunnan, China. Acta Bot Yunnanica, 29, 601 -615. | ||
| Li, R., Kraft, N.J.B., Ya ng, J ., et al., 2015a. A phylogenetically informed delineation of floristic regions within a biodiversity hotspot in Yunnan. China. Sci. Rep, 5, 9396 . DOI:10.1038/srep09396 | ||
| Li, R., Kraft, N.J.B., Yu, H.Y., et al., 2015b. Seed plant phylogenetic diversity and species richness in conservation planning within a global biodiversity hotspot in eastern Asia. Conserv. Biol, 29, 1552 -1562. DOI:10.1111/cobi.12586 | ||
| Li, X.W., L i, J ., 1997. The Tanaka-Kaiyong line- an important floristic line for the study of the flora of East Asia. Ann. Mo. Bot. Gard, 84, 888 -892. DOI:10.2307/2992033 | ||
| Losos, J.B., 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett, 11, 995 -1003. DOI:10.1111/ele.2008.11.issue-10 | ||
| Maddison, W.P., Slatkin, M ., 1991. Null models for the number of evolutionary steps in a character on a phylogenetic tree. Evolution, 45, 1184 -1197. DOI:10.2307/2409726 | ||
| Mayfield, M.M., Levine, J.M., 2010. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett, 1085, 1085 -1093. | ||
| Moore, B.R., Donoghue, M.J., 2007. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations. Am. Nat, 170, S28 -S55. DOI:10.1086/519460 | ||
| Münkemüller, T., Lavergne, S., Bzeznik, B ., et al., 2012. How to measure and test phylogenetic signal. Methods Ecol. Evol, 3, 743 -756. DOI:10.1111/mee3.2012.3.issue-4 | ||
| Pei, N., Lian, J.Y., Erickson, D.L., et al., 2011. Exploring tree-habitat associations in a Chinese subtropical forest plot using a molecular phylogeny generated from DNA barcode loci. PLoS One, 6, e21273 . DOI:10.1371/journal.pone.0021273 | ||
| Qian, H., Jin, Y., 2015. An updated megaphylogeny of plants, a tool for generating plant phylogenies, and an analysis of phylogenetic community structure. J. Plant Ecol . | ||
| Qian, H., Ricklefs, R.E., 1999. A comparison of the taxonomic richness of vascular plants in China and the United States. Am. Nat, 154, 160 -181. DOI:10.1086/303230 | ||
| Qian, H., Ricklefs, R.E., 2000. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature, 407, 180 -182. DOI:10.1038/35025052 | ||
| Qian, H., Zhang, J., 2014. Using an updated time-calibrated family-level phylogeny of seed plants to test for non-random patterns of life forms across the phylogeny. J. Syst. Evol, 52, 423 -430. DOI:10.1111/jse.v52.4 | ||
| Qian, H., Zhang, J., 2016. Are phylogenies derived from family-level supertrees robust for studies on macroecological patterns along environmental gradients?. J.Syst. Evol, 54, 29 -39. DOI:10.1111/jse.v54.1 | ||
| R Development Core Team, 2013. R: a Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. | ||
| Revell, L.J., 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol, 217 -223. | ||
| Schliep, K.P., 2011. Phangorn: phylogenetic analysis in R. Bioinformatics, 27, 592 -593. DOI:10.1093/bioinformatics/btq706 | ||
| Schreeg, L.A., Kress, W.J., Erickson, D.L., et al., 2010. Phylogenetic analysis oflocal-scale tree soil associations in a lowland moist tropical forest. PLoS One, 5, e13685 . DOI:10.1371/journal.pone.0013685 | ||
| Shukla, A., Guleria, J.S., Mehrotra, R.C., 2012. A fruit wing of Shorea Rocb.from the Early Miocene sediments of Kachchh, Gujarat and its bearing of palaeoclimatic interpretation. J. Earth Syst. Sci, 121, 195 -201. DOI:10.1007/s12040-012-0142-5 | ||
| Silvertown, J ., McConway, K., Gowing, D., et al., 2006. Absence of phylogenetic signal in the niche structure of meadow plant communities. Proc. R. Soc. Lond., B273, 39 -44. | ||
| Swenson, N.G., 2011. The role of evolutionary processes in producing biodiversity patterns, and the interrelationships between taxonomic, functional and phylogenetic biodiversity. Am. J. Bot, 472 -480. | ||
| Swenson, N.G., 2013. The assembly of tropical tree communities- the advances and shortcomings of phylogenetic and functional trait analyses. Ecography, 36, 264 -276. DOI:10.1111/j.1600-0587.2012.00121.x | ||
| Tsumura, Y., Kado, T., Yoshida, K., et al., 2011. Molecular database for classifying Shorea species (Dipterocarpaceae) and techniques for checking the legitimacy of timber and wood products. J. Plant Res, 124, 35 -48. DOI:10.1007/s10265-010-0348-z | ||
| Violle, C., Nemergut, D.R., P u, Z ., et al., 2011. Phylogenetic limiting similarity and competitive exclusion. Ecol. Lett, 14, 782 -787. DOI:10.1111/ele.2011.14.issue-8 | ||
| Webb, C.O., 2000. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat, 156, 145 -155. DOI:10.1086/303378 | ||
| Webb, C.O., Ackerly, D.D., McPeek, M.A., et al., 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Evol. Syst, 33, 475 -505. DOI:10.1146/annurev.ecolsys.33.010802.150448 | ||
| Wu, Z.Y., 1965. On the tropical affinities of Chinese flora. Chin. Sci. Bull, 1, 25 -33. | ||
| Wu, Z.Y., 1979. The regionalization of Chinese flora. Acta Bot. Yunnanica, 1, 1 -22. | ||
| Wu, Z.Y., 1983. On the significance of Pacific intercontinental discontinuity. Ann. Mo. Bot. Gard, 70, 577 -590. DOI:10.2307/2398977 | ||
| Wu, Z.Y., 1991. Areal-types of Chinese genera of seed plants. Acta Bot. Yunnanica 1-139. Suppl. IV. | ||
| Wu, Z.Y., Raven, P.H., Hong, D.Y., 1994-2013. Flora of China. Science Press, Beijing and Missouri Botanical Garden Press, St. Louis. | ||
| Wu, Z.Y., Sun, H., Zhou, Z.K., et al., 2005. Origin and differentiation of endemism in the flora of China. Acta Bot. Yunnanica, 27, 577 -604. | ||
| Wu, Z.Y., Sun, H., Zhou, Z.K., et al., 2010. Floristics of Seed Plants from China. Science Press, Beijing. | ||
| Wu, Z.Y., Zhou, Z.K., Li, D.Z., et al., 2003. The areal-types of the world families of seed plants. Acta Bot. Yunnanica, 25, 245 -257. | ||
| Wu, Z.Y., Zhou, Z.K., Sun, H., et al., 2006. The Areal-types of Seed Plants and Their Origin and Differentiation. Yunnan Science and Technology Press, Kunming. | ||
| Yang, J., Ci, X.Q., Lu, M.M., et al., 2014a. Functional traits of tree species with phylogenetic signal co-vary with environmental niches in two large forest dynamics plots. J. Plant Ecol, 7, 115 -125. DOI:10.1093/jpe/rtt070 | ||
| Yang, J., Zhang, G.C., Ci, X.Q., et al., 2014b. Functional and phylogenetic assembly in a Chinese tropical tree community across size classes, spatial scales and habitats. Funct. Ecol, 28, 520 -529. DOI:10.1111/fec.2014.28.issue-2 | ||
| Yulita, K.S., Bayer, R.J., West, J.G., 2005. Molecular phylogenetic study of Hopea and Shorea (Dipterocarpaceae): evidence from the trnL-trnFand internal transcribed spacer regions. Plant Spec. Biol, 20, 167 -182. DOI:10.1111/psb.2005.20.issue-3 | ||
| Zanne, A.E., Tank, D.C., Cornwell, W.K., et al., 2014. Three keys to the radiation of angiosperms into freezing environments. Nature, 506, 89 -92. | ||
| Zar, J.H., 1984. Biostatistical Analysis, second ed. Prentice Hall, Englewood Cliffs. | ||
| Zhang, W.H., Chen, Z.D., Li, J.H., et al., 2003. Phylogeny of the Dipsacales s.l. based on chloroplast trnL-F and ndhF sequences.. Mol. Phyl. Evol, 26, 176 -189. DOI:10.1016/S1055-7903(02)00303-2 |


