b. Genetics Institute, University of Florida, Gainesville, FL, USA;
c. Department of Biology, University of Florida, Gainesville, FL, USA
Recent developments in phylogenetics coupled with emerging cyberinfrastructure and new data sources provide unparalleled opportunities for mobilizing and integrating massive amounts of biological data, driving the discovery of complex patterns and new hypotheses for further study. Over the past few decades, biologists have witnessed stunning progress toward the resolution of Darwin's evocative vision of the “Great Tree of Life”(Darwin, 1859; Collins et al., 2013). This holy grail of evolutionary biology promises new insights into the evolutionary process that were not previously possible(e.g. Darwin, 1859; Stebbins, 1950; Dobzhansky, 1973). However, extracting biological knowledge from the large and complex datasets that will be gathered to facilitate this progress will require highly integrated and powerful tools - tools that can leverage the developing cyberinfrastructure for evolutionary biology to enable new and innovative research.
The biological community nowbenefits from major investments in cyberinfrastructure(CI), analytical platforms, and analyses on a grand scale. For example, the analysis of Phylogenetic Diversity(PD)on large phylogenetic trees has become a crucial tool in conservation(e.g., Faith, 1992, 2007, 2008; Faith et al., 2010). Numerous studies now implement methods for calculating PD and the topic and approaches are well reviewed in Mishler et al.(2014). Many investigators use the program Biodiverse Package(Laffan et al., 2010; http://purl.org/biodiverse).
The Open Tree of Life(blog.opentreeoflife.org; Collins et al., 2013; Hinchliff et al., 2015)provides phylogenetic data and a tree for all of Earth's 2.3 million named species and is becoming an increasingly valuable tool for evolutionary biology, as modifications are yielding branch lengths and a timetree. Data aggregators are amassing spatial data and information on species distributions at an amazing pace - after decades of near abandonment, many natural history collections are being revitalized through digitization and mobilization of data online while citizen scientists are exploring the planet and sharing their observations. The Global Biodiversity Information Facility(GBIF; gbif.org)currently serves nearly 625 million occurrences, most of them unvouchered observations but validation tools are under development. National and regional aggregators of natural history specimen records serve key information that bears on species distributions and forms the nexus to which other biological data - traits, DNA, etc. - can be linked. iDigBio(idigbio.org; Matsunaga et al., 2013; Page et al., 2015)is the U.S. national coordinating center for digitization of biodiversity collections and currently serves over 70 million specimen records and associated media and metadata.
Geospatial analyses using these specimen locality data may be conducted using custom software but large-scale analytical platforms(e.g., Map of Life, MOL.org; Lifemapper, lifemapper.org)enable integrated analyses. Connections among data providers(Open Tree of Life, iDigBio)and analytical platforms(Lifemapper)using modular workflows such as those found in Arbor(Harmon et al., 2013)are facilitating synthetic perspectives on historical, current, and future species distributions. Linkage of CI, data, and tools in this way will enable novel, data-driven discovery in biodiversity science and the genealogy of life and will be explored further later in this paper. This integration will be of broad value to systematists, evolutionary biologists, and ecologists and will be of great utility. BiotaPhy (www.biotaphy.org) is an ongoing project creating the linkages required for this integration, enabling novel data collection and analysis pipelines blending the existing resources provided by Open Tree of Life, iDigBio, and Lifemapper.
These linkages will provide all researchers the opportunity to synthesize rich data sets rapidly and to use these data to perform detailed hypothesis tests addressing diverse evolutionary questions. Here we overview some of the linkages and opportunities in the “big data” world of biodiversity analysis and the possibilities these research developments will enable.
2 Biodiversity hotspots: Florida, USA and ChinaFlorida is a hyper-diverse region of the U.S.A. and part of the North American Coastal Plain biodiversity hotspot(Noss et al., 2015). In the U.S.A., only California and Hawaii have more hotspots than Florida(Fig. 1). Florida is home to approximately 4300 species of vascular plants(Atlas of Florida Plants; http://florida.plantatlas.usf.edu/) representing temperate to subtropical floristic elements. China exhibits immense biodiversity, with over 29, 000 species of vascular plants and numerous prominent hotspots, including well-known areas in southwest China(http://www.cepf.net/where_we_work/regions/asia_pacific/southwest_china/Pages/default.aspx).
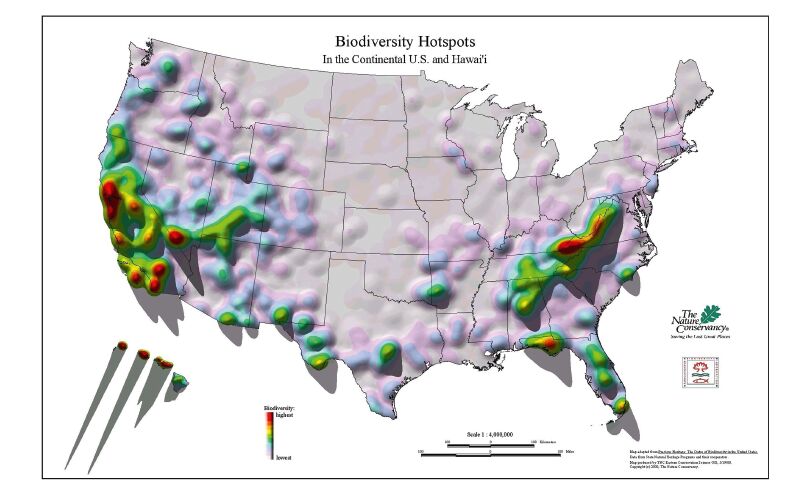
|
| Fig. 1 Biodiversity hotspots in the U.S.A. from Conservation International. Map adapted from Precious Heritage: The status of Biodiversity in the United States. Data from State Natural Heritage Programs and their cooperators. Map produced by TNC Eastern Conservation Science GIS, 5/19/00. © 2000 The Nature Conservancy. Used with permission. |
These areas represent key regions for the application of biodiversity toolkits to implement a better understanding of biodiversity in these pivotal areas, including the response to ongoing climate change and plant response in a phylogenetic context. Our own research is focused on expanding and linking biodiversity toolkits. We explore these applications here and overview our ongoing integrative studies in Florida. Similar integrated studies are well underway in California(Baldwin et al., unpubl.)and Australia(e.g., Gonzalez-Orozco et al., 2014, 2016). These types of big datainformed biodiversity studies may be useful models for application to other areas of the world, including China.
3 Layering the data of life-big data tools for big questionsThe past few years have seen the development of powerful informatic tools for the study of biodiversity that were unimaginable only a decade ago. These developments are not trivial in that biodiversity data on the global scale now being collected and analyzed are inherently complex. The ongoing integration and maturation of biodiversity tools as discussed here is transforming biodiversity science, enabling what we broadly term “next-generation” investigations in systematics, ecology, and evolution(i.e., “biodiversity science”). Dobzhansky(1973)famously stated, “nothing makes sense except in light of evolution.” A modern corollary(stated by many researchers)is “things make a lot more sense in the light of a phylogeny.” As diverse fields move forward with the recognition of the fundamental importance of a phylogenetic underpinning, the need for powerful, new, integrative tools in downstream evolutionary analysis are widely recognized by the biological community.
3.1 Using specimen dataHerbarium and other museum specimens represent a gold mine of datadnot only as a source of DNA and morphology, but the label data of specimens is an invaluable record of present and past distribution. A current goal of biodiversity science is to make data for millions of biological specimens available in electronic format via customized cloud computing. The information includes taxonomy, geographic location, images, vocalizations, and molecular resources. As reviewed in more detail below, these diverse data promote integrative biodiversity research and provide an immense baseline for assessing impacts of climate change, invasive species, and other environmental issues.
3.2 iDigBioiDigBio - Integrated Digitized Biocollections(www.idigbio.org) - serves information associated with vouchered specimens held in neontological and paleontological research collections. State-of-the- art CI using cloud computing technologies(http://portal.idigbio.org) supports the project and makes data broadly available - not only to the research community, but also to government agencies, students, educators, and the general public. iDigBio continues to expand its resources, with the goal of mobilizing the estimated 1 billion specimens collected, studied, and curated in the U.S. over the past fewcenturies. Currently, iDigBio is the conduit for over 72, 000, 000 specimen records and this number is growing rapidly.
The specimen portal offers search capability on many specimen attributes, multiple views of the search result, and visualization of the location of the specimen on a map, as well as the images associated with the specimens. Data are also made available through a RESTful Application Programming Interface(API)that allows others to build tools that can programmatically make use of specimen information. A more comprehensive overview of the technologies and rationale behind the CI choices is given in Matsunaga et al.(2013).
3.3 Other similar aggregatorsAs noted above, GBIF represents a global repository for both specimen-based and observational data on localities. However, large-scale digitization projects are likewise underway, or recently completed, elsewhere in the world. Here we focus on herbaria, but note that digitized resources likewise exist on national and regional scales for other clades of life as well, as well as for other biodiversity information. The Chinese Virtual Herbarium(http://www.cvh.org.cn/), one of the National Science and Technology Infrastructures of China and a collaboration of approximately 50 herbaria in China with collective holdings of 16 million specimens, aggregates and serves specimen records from the flora of China, with nearly 3 million records currently online. The herbarium of the Museum National d'Histoire Naturelle(MNHN)in Paris(P)has recently digitized over 5 million specimens, or 90% of its entire collection of vascular plants, approximately 6 million specimens, and these records are searchable at both the MNHN's site(https://science.mnhn.fr/institution/mnhn/collection/p/item/search) and iDigBio. Australia's Virtual Herbarium(http://avh.chah.org.au/), a consortium of Australian institutions, now serves over 7 million specimen records of plants, algae, and fungi. Collaborations in Brazil have produced the Institutos Nacionals de Ciencia - Tecnologia - Herbario Virtual da Flora - Dos Fungos(http://inct.florabrasil.net/), with over 5 million specimen records currently online. Other national and international efforts include Canadensys, a consortium of Canadian institutions for digitizing specimen and occurrence records especially for plants(which make up the bulk of the 3 million records digitized), insects, and fungi; and the JACQ Virtual Herbarium(http://herbarium.univie.ac.at/database/index.php), which has at its core 5.5 million specimens of the Naturhistorisches Museum Wien(W; http://www.nhmwien.ac.at/en/research/botany) and 1.4 million specimens of the University of Vienna(WU; http://herbarium.univie.ac.at/), as well as more than 25 additional institutions, primarily from central and eastern Europe. Other similar efforts are underway in other parts of the world, ingFinland(LUOMUS, www.luomus.fi/en/botanical-andmycological-collections; digitizing botanical and mycological collections), Museum National d'Histoire Naturelle, Paris, France(SYNTHESYS; http://www.synthesys.info/tafs/fr-taf/museumnational-dhistoire-naturelle/). Digitization of key international herbaria, such as the Royal Botanic Garden Kew and the Royal Botanic Garden Edinburgh, also promises massive data in the near future. Moreover, digitization of regional collections may provide key data for specific geographic areas or vegetation types.
3.4 Big trees-Open treeThe Open Tree of Life(blog.opentreeoflife.org; tree.open treeoflife.org/opentree/argus/opentree7.0@ott93302)provides a phylogenetic window into the history of life on Earth, comprising a record of the genealogical connections among 2.3 million species(Hinchliff et al., 2015). Although much of the tree is poorly resolved, reflecting both conflict among source trees and the use of taxonomy for placing the 80% of the tree that lacks DNA sequence data, this tree is a huge resource for those wanting to employ a tree with complete species sampling. Innovations are improving the usefulness of the tree for downstream analyses, and various tools and toolkits are under development for manipulating and using both the tree and the underlying data sets.
Construction of the draft tree required the collection and curation of thousands of phylogenetic trees from the literature. These data sets, which represent most of the phylogenetic information known about life on Earth, are stored in the OT treestore. Connecting these diverse trees to one another required a standardized taxonomy of life against which all tips in the trees in the treestore were matched. For this purpose, the OT project curated a taxonomy(the Open Tree Taxonomy or OTT), based on the GBIF and NCBI taxonomies, but which ismore inclusive and biologically accurate than either of these alone. Together, these datasets represent an unprecedented resource for evolutionary biology, by providing in one place, for the first time, the combined public phylogenetic and taxonomic information known for all recognized species. The OT treestore, for example, is an enormous resource.
3.5 Ecological niche modelsEcological niche modeling has recently emerged as an extremely powerful tool in the study of biodiversity(Elith and Leathwick, 2009; 2003; Phillips et al., 2006; e.g. https://www.cs.princeton.edu/schapire/maxent/). The approach is now widely used in ecology, evolutionary studies and in conservation. Niche modeling affords the opportunity to predict the geographic distribution of species based on data representing the current known distribution(= realized ecological niche). The approach relies heavily on precise distributional data points coupled with available environmental data. The latter is typically represented by climate data(e.g., temperature, precipitation), and information such as soil type. The availability of such data varies greatly from one part of the world to the next which greatly impacts the power of inference using these methods. Application of these models now plays a pivotal research role in diverse research in conservation biology, ecology and evolution.
3.6 LifemapperLifemapper (http://lifemapper.org/) provides amodular, scalable, computational platform for species distribution and biodiversity modeling(Cavner et al., 2012). It adds spatial and temporal dimensions to the analysis of species ranges and biological diversity, along with a rich set of open source software tools and software engineering expertise. Lifemapper now consists of two modeling subsystems: Species Distribution Modeling(LmSDM)for single species distribution models and Range and Diversity(LmRAD)for analysis of multi-species, continental, and global-scale biodiversity patterns. Lifemapper infrastructure is composed of a central management component, LmDbServer, which manages data and analysis operations with a “data pipeline” written in Python and a PostgreSQL/ PostGIS database; multiple instances of LmCompute, actual or virtual compute clusters at KU, UF, and San Diego Supercomputer Center with Lifemapper and third-party computational software(openModeller(Munoz et al., 2009), Maxent(Phillips et al., 2006)); and LmWebServer, which manages all communications between LmDbServer and LmCompute and client applications.
Lifemapper is an open-source platform designed to be extendable in backend functionality, web service accessibility, and client integration. All of Lifemapper’s data and computational methods are published through standard web service formats. Lifemapper includes a large data store of species information updated continuously from GBIF’s species occurrence web services. Lifemapper computes Species Distribution Models(SDMs)for the most current GBIF terrestrial taxa occurrence data, joined with observed climate data from Worldclim(Hijmans et al., 2005), and predicted future climate data based on International Panel on Climate Change(IPCC)scenarios, as inputs for ecological niche algorithms, resulting in publicly accessible species distribution maps. For formal research, Lifemapper's functions are accessible through web services and in QGIS(www.qgis.org), an open-source GIS platform. QGIS is useful for data integration and visual and statistical exploration of research questions involving analysis of phylogenetic, biogeographical, and character data.
3.7 BiotaPhyThis new project is contributing to a new generation of research to benefit the biodiversity research community, in which new sets of questions at the interface of phylogenetics, ecology, evolutionary biology, biogeography, and biodiversity science can be addressed in a streamlined, effective manner. To facilitate this work, integrated research pipelines are being developed to link data, CI, and analytical tools established by recent NSF investments in biodiversity sciencedthe Open Tree of Life, iDigBio, Lifemapper, and tools for comparative methods in Arbor. For example, Arbor(arborworkflows.com/: Harmon et al., 2013), hopes to provide streamlined, powerful workflows for evolutionary analyses. Although workflows/linkages are in development, the underlying biodiversity programs developed by diverse authors as R packages are already available for use.
This integrative approach permits the linkage of data sets consisting of(1)molecular sequence data, (2)specimens, (3)fossils, (4)geographic coordinates, (5)climate models, and(6)trait data. This type of integration will enable the pursuit of large-scale biodiversity analyses of spatial and temporal variation, trait evolution, ecological interaction and community assembly, and speciation and extinction, across landscapes and through time, with the goal of greatly increasing the efficiency of research in biodiversity-focused disciplines and facilitating synthetic research in a computationally straightforward manner.
4 Enabled research-examples of workflowsAn obvious question is what types of novel “next-generation” biodiversity questions can now be addressed via the linkage of big data resources and available tools? The cross-linking of massive data sets and use of high-powered tools will facilitate the development of synergisticworkflows capable of addressingmajor questions, andwe imagine a number of possible integrativeworkflows(Fig. 2). Some of the more prominent questions that can be addressed include those below(although by no means limited to these options):
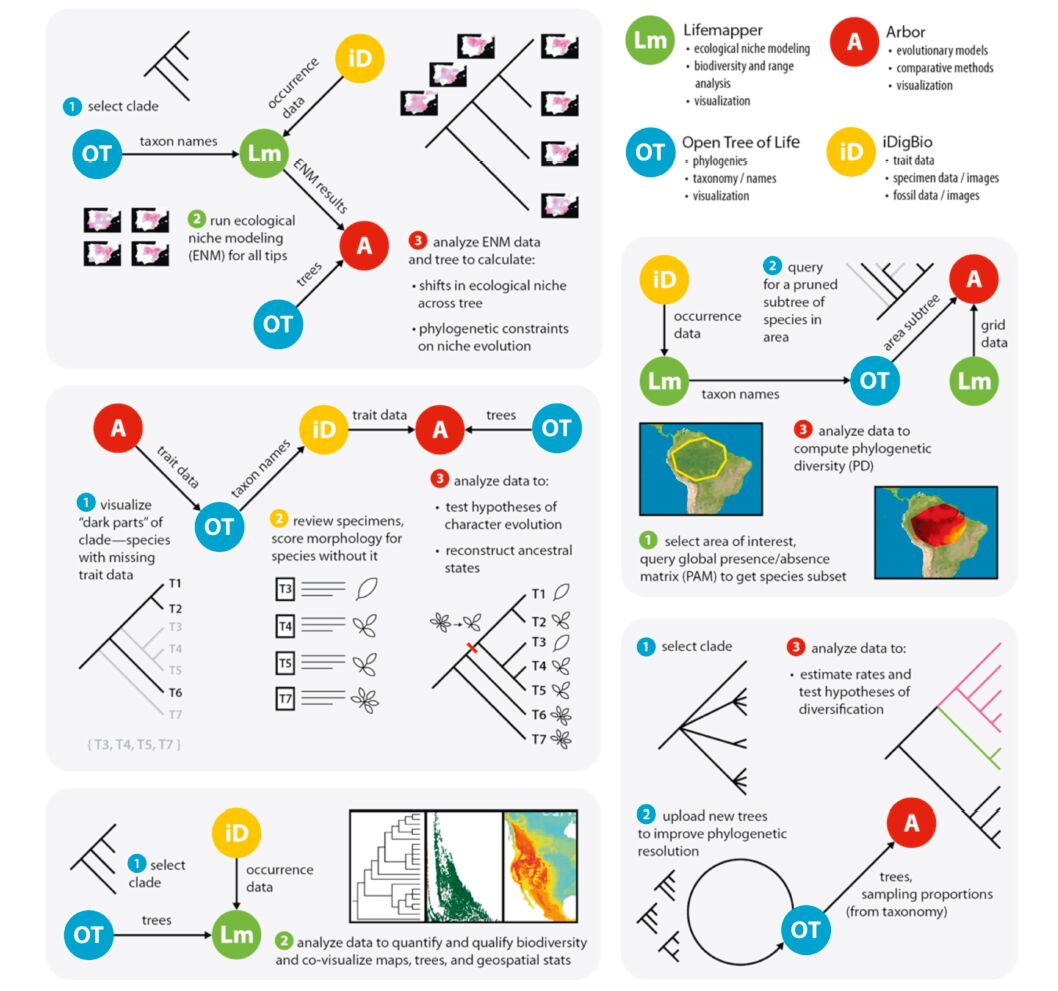
|
| Fig. 2 Some example of workflows that will be enabled by the linkages we propose to create. We emphasize that these are just examples; many more workflows will be possible through unique combinations of resources and tools, with even more becoming possible as the tools and data mature. Resources at top right are listed along with some of their largest contributions. |
1. How are processes such as speciation and extinction associated with niche divergence?
2. Howhave traits and ecological niches changed through time as a function of changes to geographic range and fluctuations in climate and geology?
3. How is the evolution of phenotype correlated with changes in ecological niche at deep time scales?
4. How is the phylogenetic diversity of a given area related to evolution of phenotype in that area?
5. How have diversification processes interacted with climate and geology to shape modern biotas?
6. How can fossil locality data inform our understanding of range and ecological niche evolution?
7. How do traits change over time? Can within-species phenotypic variation with respect to climate and ecology inform our understanding of trait lability and evolution at deep time scales?
5 Floridaddiversity and niche modelsFlorida is home to a highly diverse flora and fauna. The state spans from the southern margin of the temperate flora of the southeastern U.S. to subtropical habitats in the southern part of the state. The state spans three broad ecozones as defined by the U.S. Environmental Protection Agency(www3.epa.gov)and contributes to two of the world’s 35 biodiversity hotspots identified by Conservation International, the Caribbean hotspot in the south and the most recently recognized hotspot, the Southeastern Coastal Plain, in the north. The flora of Florida includes over 4300 species of vascular plants(Atlas of Florida Plants; http://florida.plantatlas.usf.edu/) that span a very broad range ecosystems and terrestrial, coastal, and various aquatic habitats. Importantly, Florida is also home to a very high concentration of federally sensitive, threatened, and endangered species. Florida also represents a critical test case for the role of invasive plants. For example, in south Florida roughly one third of the flora is considered to be naturalized or invasive. The Lake Wales Ridge in central Florida supports numerous endemic species, but is highly fragmented due to citrus farming and urbanization(e.g., retirement communities). Both south Florida and the Lake Wales Ridge illustrate the extensive ecological and conservation concerns for the state in general. In a sense, Florida and similarly biodiverse states(California and Hawaii)represent ground zero in a growing global biodiversity crisis. Our ability to understand the present status of biodiversity and make future projections in these areas represent crucial test cases.
We have been integrating phylogenetic analyses with niche modeling studies to understand vascular plant diversity in Florida. In addition to clarifying present distributions of phylogenetic diversity(Allen et al., in prep.), we are also projecting future distributions of the species comprising the flora of Florida. We have developed phylogenetic trees specifically for the purpose of a broader integration with spatial ecological and biodiversity data. Here we provide a brief overviewof work in progress and projected research.
Two species, Abigaardia ovata(flatspike sedge)and Prunus geniculata(scrub plum), have restricted ranges today but illustrate alternative outcomes when future distributions are projected using typical models of climate change(Fig. 3). Albigaardia ovata, currently found in south Florida, will have suitable habitat restricted to only the Miami area based on climate models for 2050. This projected distribution assumes that there will in fact be areas free of concrete in the Miami area in 2050, with limited effects of sea-level rise. In contrast, P. geniculata, which is adapted to the hot, dry conditions of the Lake Wales Ridge, will thrive under the predicted climate of 2050, and its suitable habitat will cover much of Florida. Similar modeling for 1500 plant species in Florida is allowing us to consider regional distribution patterns and how these may be altered by climate change. Moreover, linkage of these models to a phylogenetic tree of Florida plants reveals whether all parts of the evolutionary tree will be affected equivalently to climate change or whether certain clades may experience greater impact. These analyses of the Florida flora, while extensive, represent only minor spatial and phylogenetic scales from a global perspective. We hope that the workflows we have established through this study will be beneficial as others scale up to larger regional or continental analyses.
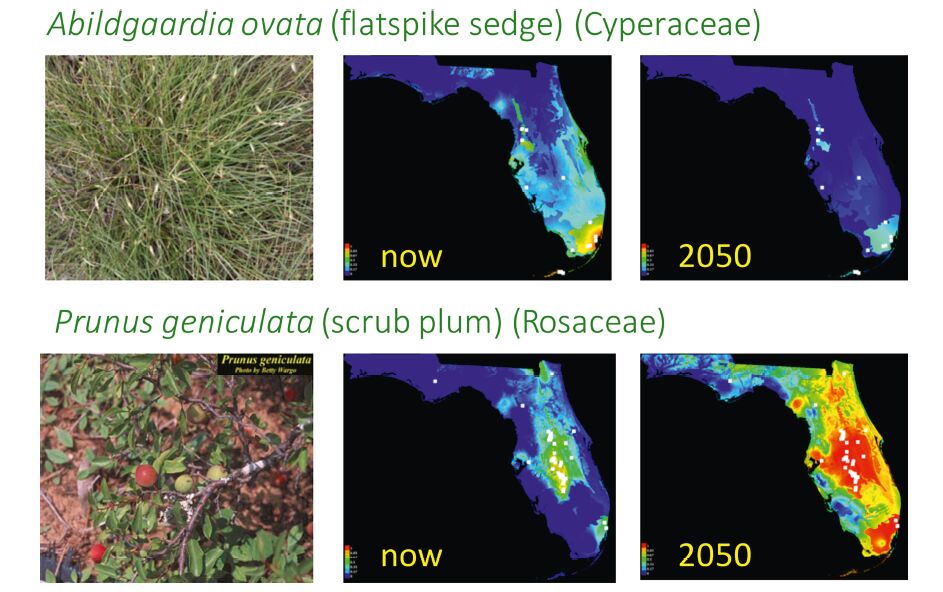
|
| Fig. 3 Florida niche projections for two species, Abigaardia ovata (flatspike sedge) and Prunus geniculata (scrub plum). Using herbarium records and niche models we reconstructed present day potential distributions and projected distributions in 2050. |
The current global challenges that threaten biodiversity are immense and rapidly growing. These biodiversity challenges demand approaches that meld bioinformatics, large-scale phylogeny reconstruction, use of digitized specimen data, and complex post-tree analyses(e.g. nichemodeling, niche diversification, and other ecological analyses). However, tool development for biodiversity science is in its infancy and generally does not yet scale as needed. Moreover, extensive trait information - relevant to evolutionary and ecological study as well as conservation - remains ‘trapped’ in metadata and images of natural history collections, and our ability to extract these data is limited. New training that integrates domain knowledge in biodiversity and data science skills is needed to accelerate research in these areas. Integrative biodiversity science is crucial to the future of global biodiversity. We cannot simply react to continued threats to biodiversity, but via the use of an integrative, multifaceted, big data approach, researchers must now make biodiversity projections to provide crucial data not only for scientists, but also for the public, land managers, policy makers, urban planners, and agriculture.
Acknowledgments: This work was supported in part by US NSF grants EF-1115210, DBI-1547229, DBI-1458640, DEB-1442280, and DEB-1208809.| Allen, J., Germain-Aubrey, C., Barve, N., Neubig, K.M., Majure, L., Whitten, W.M., Abbott, J.R., Laffan, S.W., Mishler, B., Owens, H., Guralnick, R., Soltis, D.E., Soltis P.S. Spatial phylogenetics of the vascular plants of Florida: the effects of tree uncertainty and ultrametricity. Glob. Ecol. Biogeogr., in prep. | ||
| Cavner J.A, Stewart A.M, Grady C.J, Beach J.H, 2012. An innovative web processing services based GIS architecture for global biogeographic analyses of species distributions. OSGeo J, 15, 10,15 -25. | ||
| Collins, T., Kearney, M., Maddison, D., 2013 Mar 7. The ideas lab concept, assembling the tree of life, and AVAToL. PLOS Curr. Tree Life. Edition 1. | ||
| Darwin, C., 1859. The Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. Cambridge Univ. Press, Cambridge, UK. | ||
| Dobzhansky T, 1973. Nothing in biology makes sense except in the light of evolution. Amer. Biol. Teach, 35, 125 -129. | ||
| Elith J, Leathwick J.R, 2009. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst, 40, 677 -697. | ||
| Faith D.P, 1992. Conservation evaluation and phylogenetic diversity. Biol. Cons. 61, 61, 1 -10. | ||
| Faith D.P, 2007. Phylogeny and conservation. Syst. Biol. 56, 56, 690 -694. | ||
| Faith, D.P., 2008. Phylogenetic diversity and conservation. In: Carroll, S.P., Fox, C.W. (Eds.), Conservation Biology: Evolution in Action. Oxford University Press, Oxford, UK, pp. 99-115. | ||
| Faith D.P, Magallon S, Hendry A.P, Conti E, Yahara T, Donoghue M.J, 2010. Ecosystem services: an evolutionary perspective on the links between biodiversity and human well-being. Curr. Op. Env. Sust. 2, 2, 66 -74. | ||
| Gonzalez-Orozco, C.E., Ebach, M.C., Laffan, S., Thornhill, A.H., Knerr, N.J., Schmidt- Lebuhn, A.N., Cargill, C.C., Clements, M., Nagalingum, N.S., Mishler, B.D., Miller, J.T., 2014. Quantifying phytogeographical regions of Australia using geospatial turnover in species composition. Plos One 9 e92558. | ||
| Gonzalez-Orozco C.E, Pollock L.J, Thornhill A.H, Mishler B.D, Knerr N, Laan S.W, Miller J.T, Rosauer D.F, Faith D.P, Nipperess D.A, Kujala H, Linke S, Butt N, Kulheim C, Crisp M.D, Gruber B, 2016. Phylogenetic approaches reveal biodiversity threats under climate change. Nat. Clim. Change 6, 6, 1110 -1114. | ||
| Harmon, L.J., Baumes, J., Hughes, C., Soberon, J., Specht, C.D., Turner, W., Thacker, R.W., 2013. Arbor: comparative analysis workflows for the tree of life. Plos Curr. 5 ecurrents.tol.099161de5eabdee073fd3d21a44518dc. | ||
| Hijmans R.J, Cameron S.E, Parra J.L, Jones P.G, Jarvis A, 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 25, 1965 -1978. | ||
| Hinchliff C.E, Smith S.A, Allman J.F, Burleigh J.G, Chaudhary R, Coghill L.M, Crandall K.A, Deng J, Drew B.T, Gazis R, Gude K, Hibbett D.S, Katz L.A, Laughinghouse H.D, McTavish E.J, Midford P.E, Owen C.L, Reed R.H, Rees J.A, Soltis D.E, Williams T, Cranston K.A, 2015. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc. Natl. Acad. Sci. U.S.A, 112, 12764 -12769. | ||
| Laffan S.W, Lubarsky E, Rosauer D.F, 2010. Biodiverse, a tool for the spatial analysis of biological and related diversity. Ecography 33, 33, 643 -647. | ||
| Matsunaga, A., Thompson, A., Figueiredo, R.J., Germain-Aubrey, C.C., Collins, M., Beaman, R.S., MacFadden, B.J., Riccardi, G., Soltis, P.S., Page, L.M., Fortes, J.A.B., 2013. A Computational- and Storage-Cloud for Integration of Biodiversity Collections. In: Proceedings of the 2013 IEEE 9th International Conference on e- Science, Beijing, China, pp. 78-87. http://dx.doi.org/10.1109/eScience.2013.48. | ||
| Mishler B.D, Knerr N, Gonzalez-Orozco C.E, Thornhill A.H, Laffan S.W, Miller J.T, 2014. Phylogenetic measures of biodiversity and neo- and paleo- endemism in Australian Acacia. Nat. Comm. 5, 5, 5473 . | ||
| Munoz, M.E.S., Giovanni, R., Siqueira, M.F., Sutton, T., Brewer, P., Pereira, R.S., Canhos, D.A.L., Canhos, V.P., 2009. openModeller: a generic approach to species’ potential distribution modelling. GeoInformatica. http://dx.doi.org/10.1007/s10707-009-0090-7. | ||
| Noss R.F, Platt W.J, Sorrie B.A, Weakley A.S, Means D.B, Costanza J, Peet R.K, 2015. How global biodiversity hotspots may go unrecognized: lessons from the North American Coastal plain. Divers. Distrib, 21, 236 -244. | ||
| Page L.M, MacFadden B.J, Fortes J.A, Soltis P.S, Riccardi G, 2015. Digitization of biodiversity collections reveals biggest data on biodiversity. BioScience, 65, 841 -842. | ||
| Peterson A.T, 2003. Predicting the geography of species' invasions via ecological niche modeling. Quart. Rev. Biol. 78, 78, 419 -433. | ||
| Phillips S.J, Anderson R.P, Schapire R.E, 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 190, 231 -259. | ||
| Stebbins G.L, 1950. Variation and Evolution in Plants. Columbia University Press, |



