b. Yunnan Tobacco Industrial Hi-tech Material CO., LTD, Kunming, 650106, Yunnan, China;
c. Kunming Botanical Garden, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, Yunnan, China
Genetic variation within and among natural populations is important for the species future adaptive changes and evolution (Frankham et al., 2010; Frankham, 2012). Knowledge of genetic variation of endangered species can provide critical information needed to understand the evolutionary history of populations and identify short-or long-term risks for the species (Avise and Hamrick, 1996). Knowledge of extent and structure of genetic variation of endangered species is essential for designing efficient conservation practices (Hedrick and Miller, 1992; Hamrick and Godt, 1996a, b; Woodruff, 2001).
The genus Craigia W. W. Smith & W. E. Evans (Tiliaceae) was widespread throughout the Northern Hemisphere, and abundant across Europe, North America and East Asia during the Tertiary period (Jin et al., 2009; Kvaček et al., 2002), but only two Craigia species existed in modern time: C. yunnanensis W. W. Smith & W. E. Evans (distributed in southern China and northern Vietnam), and Cragia kwangsiensis H. H. Hsue (endemic to China). In the IUCN (International Union for Conservation of Nature) Red List, C. yunnanensis is listed as 'endangered' (http://www.iucnredlist.org/details/32335/0) and C. kwangsiensisis listed as 'critically endangered' (http://www.iucnredlist.org/details/32395/0). C. kwangsiensis known only from type and not found in the wild since 1975 apparently got extinct due to deforestation (Tang et al., 2007). C. yunnanensis is a deciduous canopy tree occupying limestone mountainous forests. As a result of deforestation most of the species habitat was destroyed and the remaining habitat is severely fragmented. Survey and mapping of the remaining populations is the first step of a conservation program to prevent extinction of this species, followed by a carefully designed action plan (Corlett, 2016; Volis, 2016).
Based on all the available information and our extensive survey of the natural habitats of C. yunnanensis, six small, remnant populations were located in Yunnan province (Gao et al., 2010). No material could be found in Guizhou or Guangxi, indicating that the species may already be extinct in these provinces. Its status in Vietnam is currently unknown due to travel restrictions and logistical limitations. The survey also revealed that in order to grow economically important plants such as tea, Amomum tsaoko and Cunninghamia lanceolata, locals chop down C. yunnanensis or change its habitat into farmlands. Facing a very high risk of extinction, C. yunnanensis is now listed as a plant species with extremely small populations of China (PSESP) in the national-level Implementation Plan of Rescuing and Conserving China's PSESP (2010-2015). A conservation strategy focusing on PSESP and aimed at rescuing the most endangered plants was approved by the Chinese government in 2009 (Ma et al., 2013; Ren et al., 2012). Conservation of C. yunnanensis based on this strategy must include seed collection, seedling propagation, ex-situ conservation at the Kunming Botanical Garden (KBG) and reintroductions.
Knowledge of extent and structure of genetic variation in C. yunnanensis may have important implications for management of the species germplasm collection, in-situ/ex-situ conservation and later reintroduction (Hoban and Schlarbaum, 2014; Li et al., 2008; Volis, 2015; Yang et al., 2015). This analysis is a preliminary step for selecting breeding material and establishing conservation strategies for C. yunnanensis. Here, we use AFLP (amplified fragment length polymorphism) to study extent and structure of genetic variation in C. yunnanensis. AFLP is commonly used in population genetic studies of rare and endangered species (Bensch and ÅKesson, 2005; Juan et al., 2004; Nybom, 2004). The results of this study can be useful for setting an appropriate conservation strategy for C. yunnanensis.
2 Materials and methods 2.1 Study species and samplingC. yunnanensis (Tiliaceae) is a deciduous canopy tree. It is a predominantly outcrossing with partial self-compatibility species. The major pollinator is a blowfly (Chrysomyia megacephala). The flowering time is from middle August to late September. Fruit of C. yunnanensis is samara containing about 6 seeds (around 34 mg per seed). Upon maturation in early December, the fruits are dispersed by wind and gravity. On the bare soil, the seeds could germinate easily. The experiments (Gao et al., 2010) showed that for some populations, germination capacity in canopy gaps was significantly higher than that in closed canopy, indicating that canopy removal might promote regeneration from seed. Small seedlings and resprouts are abundant but very few of them reach the sapling stage.
Sampling was done in six populations representing two disjunct regions of Yunnan province, China (Fig. 1). Four populations, W-FD (Fadou county), W-LH (Lianhuatang town), W-ML (Malipo county) and W-MG (Maguan county), are from Wenshan prefecture (southeastern Yunnan); two populations, D-JD (Jiangdong town) and DHG (Huguo town) are from Dehong prefecture (south-western Yunnan). Wenshan and Dehong are more than 600 km apart. Locality information of the six wild populations is provided in Table 1. The number of reproductive individuals in each population was counted and in total 105 reproductive individuals were sampled (Table 1). There were only 11 reproductive individuals in the W-MG population, and 10 of them were sampled for the AFLP analysis.

|
| Fig. 1 The geographic locations of six studied C. yunnanensis populations. Numbers correspond to the population codes in Table 1. |
| Population code | Locations | Sample size | Latitude | Longitude | Altitude | Population sizea |
| W-FD (1) | Fadou county | 20 | 23°22' | 104°46' | 1461 | 89 |
| W-LH (2) | Lianhuatang town | 20 | 23°09' | 104°51' | 1460 | 151 |
| W-ML (3) | Malipo county | 20 | 23°11' | 104°43' | 1449 | 125 |
| W-MG (4) | Maguan county | 10 | 23°03' | 104°15' | 1410 | 11 |
| D-JD (5) | Jiangdong town | 15 | 24°31' | 98°22' | 1694 | 106 |
| D-HG (6) | Huguo town | 20 | 24°33' | 98°03' | 1696 | 173 |
| a Population size, the number of reproductive individuals detected in each population. | ||||||
Total genomic DNA was extracted using a modified CTAB method (Doyle, 1987). AFLP analysis was performed following Vos et al. (1995) with minor modifications. 200 ng samples of genomic DNA were digested with EcoRI and MseI (New England Biolabs), followed by ligation of appropriate adapters. Pre-selective PCR amplifications were performed using the primer pair EcoRI + 1 and MseI + 1. For selective PCR amplification, three EcoRI + 3/ MseI + 3 primer combinations (E-ACR/M-CAG, E-ACA/M-CAG and EAAG/M-CTC) were chosen among 64 selective primer pairs available from PerkineElmer AFLP Selective Amplification Start-Up Module. PCR products were separated on a 6% poly acrylamide gel (acrylamide:bisacrylamide=19:1) and stained using the silver nitrate method.
2.3 Data analysisAFLP bands between 100 and 500 bp in the poly acrylamide gels were scored manually as present (1) or absent (0), and transformed into a binary data matrix. The following genetic diversity parameters were calculated using POPGENE version 1.31 (Yeh et al., 1999): the percentage of polymorphic loci (PPL), expected heterozygosity (HE) and Shannon's information index (IS=-Pilog2Pi) (Lewontin, 1972). The relationship between population size (the number of reproductive individuals detected in each population, Table 1) and genetic diversity (estimated as PPL, HE and IS) was analyzed via Pearson's product moment correlation using SPSS statistical package version 15.0 (SPSS, Inc., Chicago, IL, USA).
Nei's genetic distance between populations (Nei, 1978), the coefficient of genetic differentiation (GST) and gene flow (Nm) were also estimated in POPGENE and a dendrogram was constructed from Nei's genetic distance with the unweighted pair-group method of averages (UPGMA) using the TFPGA software version 1.3 (Miller, 1997). Using the same software, a Mantel test was performed to calculate the correlation between the Nei's genetic distance and geographical distances. Analysis of molecular variation (AMOVA) within/among populations and between geographic regions was calculated using Arlequin 3.1 (Excoffier et al., 2005). The Bayesian clustering program STRUCTURE (Pritchard et al., 2000) was used to estimate proportion of ancestry in each of the clusters identified. The analysis used the admixture ancestry model with independent allelic frequencies. A burn-in period of 10, 000 and a run length of the Monte Carlo Markov Chain (MCMC) of 10, 000 iterations were performed with 10 replicates for each K ranging from one to seven. The STRUCTURE Harvester (Earl and vonHoldt, 2012) was conducted to determine the most likely number of ge netic groups (K) according to the ΔK and Mean LnP (K) (Evanno et al., 2005).
3 Results 3.1 Genetic diversityFrom three AFLP primer combinations, a total of 185 bands ranging from 100 to 500 bp were produced across the 105 indi vidual samples from six populations, of which 128 were poly morphic (69.19%). Genetic diversity parameters are presented in Table 2. AFLP analysis showed that the percentage of polymorphic loci (PPL) ranged from 16.22% in population W-MG to 35.14% in population W-LH. The expected heterozygosity (HE) and Shannon's information index of diversity (IS) of the species level were 0.221 and 0.333, respectively. Across the six populations, W-MG had the lowest diversity (PPL=16.22%, HE=0.066, IS=0.095), and the WLH had the highest (PPL=35.14%, HE=0.133, IS=0.195).
| Region | Population | N | PPL (%) | HE | IS | GST | Nm |
| Wenshan | W-FD | 20 | 30.81 | 0.111 | 0.163 | ||
| W-LH | 20 | 35.14 | 0.133 | 0.195 | |||
| W-ML | 20 | 29.19 | 0.1 | 0.149 | |||
| W-MG | 10 | 16.22 | 0.066 | 0.095 | |||
| Pooled | 70 | 52.43 | 0.161 | 0.245 | 0.367 | 0.863 | |
| Dehong | D-JD | 15 | 21.08 | 0.069 | 0.103 | ||
| D-HG | 20 | 24.86 | 0.089 | 0.132 | |||
| Pooled | 35 | 34.05 | 0.096 | 0.149 | 0.166 | 2.506 | |
| Population average | 18 | 26.22 | 0.095 | 0.14 | |||
| Species-level value | 105 | 69.19 | 0.221 | 0.333 | 0.575 | 0.369 | |
The regional diversity estimates for Wenshan were higher than that for Dehong (PPL=52.43%, HE=0.161, IS=0.245, vs. PPL=34.05%, HE=0.096, IS=0.149, respectively). There was no significant relationship between population size and any estimate of genetic diversity.
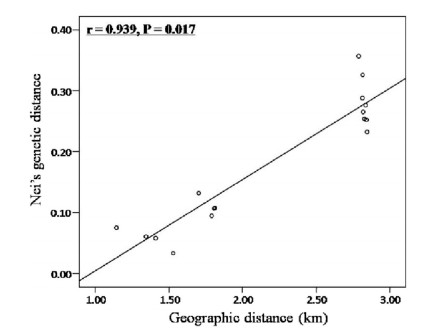
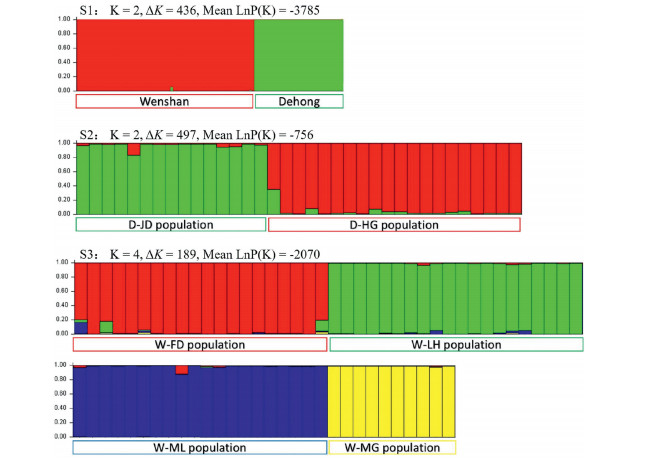
3.2 Population structureAMOVA revealed slightly higher genetic variation within pop ulations (25.36%) than that of among populations (17.05%), while most of the variation was between the two regions of Wenshan and Dehong (57.59%). Similarly, within populations, among populations and among regions GST was 0.16, 0.37 and 0.57, respectively. The cluster dendrogram produced using Nei's genetic distance values revealed two distinct clusters corresponding to the two regions (Fig. 2). The Mantel tests detected a significant relationship be tween genetic and geographic population distances (r=0.939, p=0.017) (Fig. 3). Bayesian clustering for six populations was consistent with the results of UPGMA analyses and revealed virtually no admixture not only between the two regions, but also among the populations within regions (Fig. 4). The greatest infor mative representation of genetic structure was achieved with K=2 (ΔK=436 and Mean LnP (K)=-3785).

|
| Fig. 2 UPGMA dendrogam based on Nei's unbiased genetic distance among six populations of C. yunnanensis. |
|
| Fig. 3 Correlation between geographical distance and Nei's genetic distance revealed by the Mantel test. |
|
| Fig. 4 Model-based ancestry for each individual assuming admixture: S1, six populations from two regions with K=2; S2, two populations from Dehong region with K=2; S3, four populations from Wenshan region with K=4. |
Our derived estimate of HE=0.221 is close to the average values presented in a review of available AFLP studies by Nybom (2004) for plants with long life spans (Hpop=0.25), mixed breeding sys tems (Hpop=0.18) and dispersal of seeds by gravity (Hpop=0.19). A study using AFLP data to investigate the genetic diversity in Tilia amurensis (Tiliaceae), a national second-class protected plant in China, and also a woody deciduous species, showed HE=0.21 and IS=0.34 (Wang et al., 2014), which differed little from our estimates for C. yunnanensis (HE=0.221 and IS=0.333). These comparisons suggest that C. yunnanensis exhibit moderate genetic diversity at the species level but low genetic diversity at the pop ulation level (population average: HE=0.095 and IS=0.140) and in the two separate regions (Wenshan region: HE=0.161 and IS=0.245; Dehong region: HE=0.096 and IS=0.149).
The breeding system of C. yunnanensis is predominant out crossing with partial self-compatibility, and the artificial geito nogamous pollination resulted in a fruit set of 33.33% (Gao et al., 2012). The main effective pollinator is a blowfly (C. megacephala). The blowflies generally make long visits visiting several flowers on the same tree (Gao et al., 2012), and fly only short distances (less than 100 m in a human-modified landscape, Rader et al., 2011). Though the fruit of C. yunnanensis has narrow wings allowing its dispersal by wind, the dispersal distance, by our observation, is not longer than 50 m. Combined limited pollen/ seed dispersal ability and isolation of individuals, small sized populations of C. yunnanensis can suffer from Alee effect, i.e. low pollinator visitation rate and increased selfing, inbreeding and genetic drift, which will result in decrease of genetic variation (Barrett and Kohn, 1991; Young et al., 1996). However, the correlation between decreased population size and lower genetic diversity was weak, suggesting that the decline of population size is likely to have a recent anthropogenic cause. The possible reason is that the longevity of woody perennials may have buffered against the deleterious genetic consequences of the recent population size reductions (Kang et al., 2005; Llorens et al., 2004).
4.2 Population genetic structureVarious evolutionary processes, such as geographic isolation, limited seed/pollen dispersal and habitat fragmentation, can have important impact on population genetic structure (Loveless and Hamrick, 1984; Slatkin, 1985; Foré et al., 1992; Ge et al., 1998).
Craigia occurred in the past throughout the Northern Hemisphere from the early Palaeogene (Kvaček, 2004). During the Eocene global warming, Craigia rapidly expanded its range across North America, Spitzbergen, East and Central Asia, and it had reached its modern distribution center in South China no later than in the early late Eocene (Jin et al., 2009). Today, as a relic of the genus, C. yunnanensis occurs only in southern China (Yunnan province) and Northern Vietnam (Tonkin) in broad-leaved evergreen and deciduous mixed forests and seasonally wet forests on limestone (Kvaček et al., 2002).
Although our study revealed strong genetic differentiation among populations within regions, a differentiation between the Wenshan and Dehong regions was even stronger. The estimates of GST at the species level were higher than the average values for the species with similar characteristics (Nybom, 2004). The distance between Wenshan and Dehong regions is around 600 km, which hardly allows gene flow between these regions. Taking into consideration that C. yunnanensis is a relic species that survived only in refuges, it seems that the two regions became isolated long time ago and their genetic identity was maintained over time.
In rare and endangered plants, a high degree of amongpopulation genetic differentiation is a commonly observed pattern (Gitzendanner and Soltis, 2000; Hamrick and Godt, 1996a, b). Genetic admixture even among geographically close populations was found to be close to zero. Both, distance-limited pollen flow and short-distance seed dispersal appear to contribute to the strong genetic differentiations among the populations (see above). From our observation, when undisturbed, one C. yunnanensis tree can produce thousands of seeds per fruiting season, but most of the seedlings and resprouts are found around the fruiting trees.
4.3 Conservation implicationsIn-situ conservation hardly has an alternative as a way to conserve endangered plants. Of the six populations studied, population W-ML is located in the buffer area of Laoshan provincial nature reserve and the habitat is relatively intact. This nature reserve was established in 2002, and two ethnic villages are located near this population. Although chopping down trees are strictly prohibited and cutting or coppicing are not observed, human disturbance through growing A. tsaoko under the C. yunnanensis trees is common. Also the bark girdling on one individual of C. yunnanensis was observed. Many seedlings were found growing under Amomum but they get eradicated by locals during weeding. Situation with the other five populations is even more troublesome because they are all located near human settlements in the privateowned forests or beside the roads and farmlands. In all these populations the regrowth sprouts from stumps were found indicating logging and cutting. In W-LH and D-JD populations located in private-owned forests, felling C. yunnanensis trees (among other species) to make space for growing tee and timber was observed. The local people told that when the trees of C. yunnanensis hinder the growth of economic plants, they chop them down. All the above makes the protection of the existing populations very problematic. We can suggest the following. First, all the reproductive adults in these populations must be designated as strictly protected national heritage trees, and used as mother trees for seed collection. Second, if the current interference between the local human activities and regeneration in populations of C. yunnanensis can not be resolved in favor of C. yunnanensis, ready germination of seeds and development into seedlings under Amomum should be used as an opportunity of propagating material for relocation actions. These wildlings can be dug out and transplanted into suitable protected habitats (Pritchard et al., 2014; Volis, 2016).
The population genetic characteristics of C. yunnanensis can be summarized as low within-population genetic diversity and high differentiation among populations. Considering that only six populations and fewer than seven hundred individuals still exist in China, all extant populations must be ex-situ conserved. In 2015, we collected seeds from six populations and produced 700 seedlings in Kunming Botanical Garden (KBG). Other collecting missions followed by establishing permanent seed and adult plant collections in KBG and other botanical gardens, as well as designing a plan of C. yunnanensis relocation is underway.
To conclude, this study indicated large differentiation between the populations of Dehong and Wenshan, thus germplasm from the two regions should not be mixed and used separately. Crosses between these two regions must be prevented because of the high risk of outbreeding depressions (Lynch, 1991; Fischer and Matthies, 1997; Ruane et al., 2015). For this purpose, planting of trees from both regions in the same ex-situ site is undesirable (as already accepted by the stakeholders of the two ex-situ conservation sites in Dehong and Maguan). The plants maintained in KBG will be carefully separated by regional origin, kept in different greenhouses, and might be used in further experiments testing for outbreeding depressions. To increase intra-population genetic diversity, introduction of individuals from other populations from the same region is recommended.
Acknowledgments: This study was funded by the National Science Foundation of China (NSFCU1302262), and the Department of Forestry of Yunnan Province, project name "Rescue and conservation of PSESP species Craigia yunnanensis". It was also supported by the Science and Technology Research Program of Kunming Institute of Botany, the Chinese Academy of Sciences (KIB2016005).| Avise J.C., Hamrick J.L., 1996. Conservation Genetics: Case Histories from Nature. Chapman and Hall, New York. | ||
| Barrett, S.C.H., Kohn, J.R., 1991. Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk, D.A., Holsinger, K.E. (Eds.), Genetics and Conservation of Rare Plants. Oxford University Press, USA, pp. 3-30. | ||
| Bensch S., ÅKesson M., 2005. Ten years of AFLP in ecology and evolution: why so few animals? Mol. Ecol, 14, 2899 -2914. | ||
| Corlett R.T., 2016. Plant diversity in a changing world: status, trends, and conservation needs. Plant Divers, 1016 . | ||
| Doyle J.J., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull, 19, 11 -15. | ||
| Earl D.A., vonHoldt B.M., 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour, 4, 359 . DOI:10.1007/s12686-011-9548-7 | ||
| Excoffier L., Laval G., Schneider S., 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform, 1, 47 -50. | ||
| Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol, 14, 2611 -2620. | ||
| Fischer M., Matthies D., 1997. Mating structure and inbreeding and outbreeding depression in the rare plant Gentianella germanica (Gentianaceae). Am. J. Bot, 84, 1685 -1685. DOI:10.2307/2446466 | ||
| Frankham R., Ballou J.D., Briscoe D.A., 2010. Introduction to Conservation Genetics. Cambridge University Press, Cambridge. | ||
| Frankham R., 2012. How closely does genetic diversity in finite populations conform to predictions of neutral theory? Large deficits in regions of low recombination. Heredity, 108, 167 -178. DOI:10.1038/hdy.2011.66 | ||
| Foré S.A., Hickey R.J., Vankat J.L., Guttman S.I., Schaefer R.L., 1992. Genetic structure after forest fragmentation: a landscape ecology perspective on Acer saccharum. Can. J. Bot, 70, 1659 -1668. DOI:10.1139/b92-205 | ||
| Gao Z.R., Zhang C.Q., Han Z.Q., Li Z., Wei J., Shi H.L., 2012. Pollination biology and breeding system of Craigia yunnanensis in fragmented habitat. Chin. J. Ecol, 31, 2217 -2224. | ||
| Gao Z.R., Zhang C.Q., Milne R.I., 2010. Size-class structure and variation in seed and seedling traits in relation to population size of an endangered species Craigia yunnanensis (Tiliaceae). Aust. J. Bot, 58, 214 -223. DOI:10.1071/BT09190 | ||
| Ge S., Hong D.Y., Wang H.Q., Liu Z.Y., Zhang C.M., 1998. Population genetic structure and conservation of an endangered conifer, Cathaya argyrophylla (Pinaceae). Int. J. Plant Sci, 159, 351 -357. DOI:10.1086/297556 | ||
| Gitzendanner M.A., Soltis P.S., 2000. Patterns of genetic variation in rare and widespread plant congeners. Am. J. Bot, 87, 783 -792. DOI:10.2307/2656886 | ||
| Hamrick, J.L., Godt, M.J.W., 1996a. Conservation genetics of endemic plant species. In: Avise, J.S., Hamrick, J.L. (Eds.), Conservation Genetics: Case Histories from Nature. Chapman and Hall, New York, pp. 281-304. | ||
| Hamrick, J.L., Godt, M., 1996b. Effects of life history traits on genetic diversity in plant species. Philos. Trans. Roy. Soc. B 351, 1291-1298. | ||
| Hedrick P.W., Miller P.S., 1992. Conservation genetics: techniques and fundamentals. Ecol. Appl, 2, 30 -46. DOI:10.2307/1941887 | ||
| Hoban S., Schlarbaum S., 2014. Optimal sampling of seeds from plant populations for ex-situ conservation of genetic biodiversity, considering realistic population structure. Biol. Conserv, 177, 90 -99. DOI:10.1016/j.biocon.2014.06.014 | ||
| Jin J., Kodrul T.M., Liao W., Wang X., 2009. A new species of Craigia from the Eocene Changchang formation of Hainan Island, China. Rev. Palaeobot. Palynol, 155, 80 -82. DOI:10.1016/j.revpalbo.2009.02.003 | ||
| Juan A., Crespo M.B., Cowan R.S., Lexer C., Fay M.F., 2004. Patterns of variability and gene flow in Medicago citrina, an endangered endemic of islands in the western Mediterranean, as revealed by amplified fragment length polymorphism (AFLP). Mol. Ecol, 13, 2679 -2690. DOI:10.1111/mec.2004.13.issue-9 | ||
| Kang M., Ye Q., Huang H., 2005. Genetic consequence of restricted habitat and population decline in endangered Isoetes sinensis (Isoetaceae). Ann. Bot, 96, 1265 -1274. DOI:10.1093/aob/mci277 | ||
| Kvaček Z., Manchester S.R., Zetter R., Pingen M., 2002. Fruits and seeds of Craigia bronnii (Malvaceae-Tilioideae) and associated flower buds from the late Miocene Inden Formation, Lower Rhine Basin, Germany. Rev. Palaeobot. Palynol, 119, 311 -324. DOI:10.1016/S0034-6667(01)00135-X | ||
| Kvaček Z., 2004. Early Miocene records of Craigia (Malvaceae sl) in the most basin North Bohemia-whole plant approach. J. Czech Geol. Soc, 49, 161 -171. | ||
| Lewontin R.C., 1972. The apportionment of human diversity. Evol. Biol, 6, 381 -398. | ||
| Li X.X., Ding X.Y., Chu B.H., Zhou Q., Ding G., Gu S., 2008. Genetic diversity analysis and conservation of the endangered Chinese endemic herb Dendrobium officinale Kimura et Migo (Orchidaceae) based on AFLP. Genetica, 133, 159 -166. DOI:10.1007/s10709-007-9196-8 | ||
| Llorens T.M., Ayre D.J., Whelan R.J., 2004. Evidence for ancient genetic subdivision among recently fragmented populations of the endangered shrub Grevillea caleyi (Proteaceae). Heredity, 92, 519 -526. DOI:10.1038/sj.hdy.6800444 | ||
| Loveless M.D., Hamrick J.L., 1984. Ecological determinants of genetic structure in plant populations. Annu. Rev. Ecol. Syst, 15, 65 -95. DOI:10.1146/annurev.es.15.110184.000433 | ||
| Lynch M., 1991. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution, 45, 622 -629. DOI:10.2307/2409915 | ||
| Ma Y.P., Chen G., Grumbine R.E., Dao Z.L., Sun W.B., Guo H.J., 2013. Conserving plant species with extremely small populations (PSESP) in China. Biodivers. Conserv, 22, 803 -809. DOI:10.1007/s10531-013-0434-3 | ||
| Miller, M.P., 1997. Tools for Population Genetic Analyses (TFPGA) 1.3: a Windows Program for the Analysis of Allozyme and Molecular Population Genetic Data. Computer software distributed by author, vol. 4, p. 157. | ||
| Nei M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89, 583 -590. | ||
| Nybom H., 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol, 13, 1143 -1155. DOI:10.1111/mec.2004.13.issue-5 | ||
| Pritchard J.K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155, 945 -959. | ||
| Pritchard H.W., et al., 2014. Innovative approaches to the preservation of forest trees. For. Ecol. Manag, 333, 88 -98. DOI:10.1016/j.foreco.2014.08.012 | ||
| Rader R., Edwards W., Westcott D.A., Cunningham S.A., Howlett B.G., 2011. Pollen transport differs among bees and flies in a human-modified landscape. Divers. Distrib, 17, 519 -529. DOI:10.1111/j.1472-4642.2011.00757.x | ||
| Ren H., et al., 2012. Wild plant species with extremely small populations require conservation and reintroduction in China. AMBIO, 41, 913 -917. DOI:10.1007/s13280-012-0284-3 | ||
| Ruane L.G., Dickens M.E., Wall M.E., 2015. Fitness consequences of short-and long distance pollinations in Phlox hirsuta, an endangered species. Am. J. Bot, 102, 1659 -1665. DOI:10.3732/ajb.1500270 | ||
| Slatkin M., 1985. Gene flow in natural populations. Annu. Rev. Ecol. Syst, 16, 393 -430. DOI:10.1146/annurev.es.16.110185.002141 | ||
| Tang Y., Gilbert M.G., Dorr L.J., 2007. Flora of China. vol. 12, Science Press, Beijing. | ||
| Volis S., 2015. Species-targeted plant conservation: time for conceptual integration. Isr. J. Plant Sci . | ||
| Volis S., 2016. How to conserve threatened Chinese plant species with extremely small populations?. Plant Divers . | ||
| Vos P., et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res, 23, 4407 -4414. DOI:10.1093/nar/23.21.4407 | ||
| Wang D.S., Xin H., Xing S.Y., Li J.H., Liu X.J., Wu Q.K., 2014. Analysis of Tilia amurensis Ruprecht genetic diversity from Shandong province by AFLP markers. J. Fujian Agric. For. Univ. Nat. Sci. Ed, 43, 44 -48. | ||
| Woodruff, D.S., 2001. Populations, species and conservation genetics. In: Levin, S.A. (Ed.), Encyclopedia of Biodiversity. Academic Press, London. | ||
| Yang J., Zhao L.L., Yang J.B., Sun W.B., 2015. Genetic diversity and conservation evaluation of a critically endangered endemic maple, Acer yangbiense, analyzed using microsatellite markers. Biochem. Syst. Ecol, 60, 193 -198. DOI:10.1016/j.bse.2015.04.027 | ||
| Yeh, F.C., Yang, R.C., Boyle, T., 1999. PopGene Version 1.31: Microsoft Window-based Freeware for Population Genetic Analysis. University of Alberta and Tim Boyle, Centre for International Forestry Research. | ||
| Young A., Boyle T., Brown T., 1996. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol, 11, 413 -418. DOI:10.1016/0169-5347(96)10045-8 |



