b. Key Laboratory of Biodiversity and Biogeography, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, 650201, China
Genome size is the amount of DNA in a non-replicated, basic, gametic chromosome set (Soltis et al., 2003). It has been estimated in several thousand plant species since 1950(Hanson et al., 2001), including at least 152 gymnosperms species (Leith et al., 2001) and 7542 angiosperms species (Bennett and Leitch, 2011). Genome size variation in angiosperms is especially impressive, ranging from 0.06 pg (Genlisea tuberosa Rivadavia) to 152.23 pg (Paris japonica (Franch. & Sav.) Franch.)(Pellicer et al., 2010; Fleischmann et al., 2014). Such large-scale analyses have been enabled in part by using flow cytometry (FCM), a highthroughput method of estimating DNA content from isolated nuclei that are stained with a DNA-selective fluorochrome (Bainard et al., 2011). The popularity of FCM lies in its numerous advantages:(1) easy and convenient sample preparation;(2) high accuracy that permits the detection of minute variations in nuclear DNA amount;(3) rapid detection of mixed samples or endopolyploidy;(4) its non-destructive nature, which permits the comprehensive investigation of rare and endangered species or seedlings in a very early ontogenetic stage; and (5) low operating costs (Dolezel, 1991).
The tribe Ophiopogoneae (Asparagaceae) has three genera: Ophiopogon Ker Gawl., Liriope Lour., and Peliosanthes Andr., which are mainly distributed in tropical, subtropical, and temperate regions of East and Southeast Asia. DNA content has been reported for only a few species (Bennett, 1972; Bharathan et al., 1994; Zonneveld et al., 2005; Lattier and Ranney, 2014). Best practices for FCM generally recommend that DNA content is measured using fresh tissue. Lack of fresh material is cited as one of the main reasons for the dearth of studies on plants from remote locations. Therefore, analyzing dehydrated tissue might be an attractive alternative. Tissue desiccation, using either herbarium presses or silica gel, is a rapid and undemanding approach that has been used traditionally for sample preservation in field botany (Suda and Travnicek, 2006a). Voglmayr (2000)was the first who estimated genome size in herbarium voucher specimens of mosses. The number of FCM studies that successfully used desiccated plant material has increased considerably, e.g., in Juncus biglumis (Schonswetter et al., 2007), Lychnis spp.(Popp et al., 2008), and Senecio carniolicus (Suda et al., 2007). Bainard et al.(2011)showed that rapidly dried tissue with silica gel can be efficiently used in genome size studies. Bai et al.(2012)also successfully used rapidly desiccated tissue for estimating genome size in 37 taxa.
Here, we compare the use of fresh and desiccated tissue for estimating genome size in the Ophiopogoneae and five model plant species (Arabidopsis thaliana (L.) Heynh., Zea mays L., Oryza sativa L., Glycine max (L.) Merr., and Lycopersicon esculentum Mill.).
2 Materials and methods 2.1 Plant materialWe sampled 22 accessions representing 16 species of Ophiopogon, Liriope, and Peliosanthes in the Ophiopogoneae, and collected five model plant species from different families spanning a nearly 16-fold range of genome sizes (from 0.32 pg/2C to 5.43 pg/2C). The genome sizes of the model species according to the literature are: A. thaliana 0.32 pg (Bennett et al., 2003), O. sativa spp. japonica 0.86 pg (Goff et al., 2002), L. esculentum 2.05 pg (Bennett and Smith, 1976), G. max 2.25 pg (Greilhuber and Obermayer, 1997), Z. mays (B73)5.43 pg. Living plants were cultivated in a greenhouse of Kunming Institute of Botany (KUN) in Yunnan, China. Leaf samples were taken from three individual plants per accession. Fresh leaf material was stored at 4 ℃ for no more than 3 d until it could be processed. The same leaf material was dried in silica gel. Desiccated tissues were stored in a hermetically sealed box in a deep freezer (at -80 ℃, for no more than 6 months). Voucher specimens were deposited at KUN.
2.2 Determining 2C-valuesAbout 0.5 cm2 leaf material was chopped up using a new razor blade in a Petri dish containing 1500-2000 μL of Galbraith nuclear solution buffer (45 mmol/L MgCl2, 30 mmol/L C6H5O7Na3•2H2O, 20 mmol/L MOPS, 0.1%(v/v) Triton X-100, pH 7.0(Galbraith et al., 1983)). The nuclear suspension was then filtered through disposable filters (30 μm) to remove the cell debris, and then stained with 150 μL propidium iodide (PI)(50 μg/mL; included RNAse (500 μg/mL)) for 10 min. Samples were analyzed on a CyFlow Space (Partec, Münster, Germany) flow cytometer equipped with a blue laser operating at 488 nm. For all samples, at least 5000 nuclei were measured. FlowMax 2.82 analyzed the resulting histograms. Z. mays (B73)(2C = 5.43 pg) was chosen as an internal standard. The 2C-value of a sample was calculated as follows:(mean of sample peak/mean of standard peak)* 2C-value (pg) of the standard species (Table 1). These values per sample were used to obtain the accession averages.
| Taxa | Herbarium voucher number | Locality in China (Province, City) | 2C-Value ±SE (fresh tissue) | 2C-value ±SE (desiccated tissue) | Paired t-test |
| Study species | |||||
| O. bodinieri Lévl. | 20081615 | Yunnan, KIB | 11.22±0.11 | 11.73±0.58 | t = 0.987, P = N.S. |
| O. bockianus Diels | HGWZ506 | Hunan, Yongshun | 18.40±0.42 | 16.68±0.68 | t = 1.993, P = N.S. |
| nie2350 | Guangxi, Longzhou | 17.49±0.29 | 17.39±0.90 | t = 0.140, P = N.S. | |
| O. chingii Wang et Tang | nie3739 | Yunnan, Malipo | 11.98±0.29 | 12.26±0.18 | t = -0.216, P = N.S. |
| O. chingii var. glaucifolious | nie2325 | Guangxi, Fangcheng | 15.16±2.57 | 14.50±0.85 | t = 0.153, P = N.S. |
| F. T. Wang & L. K. Dai | |||||
| O. dracaenoides (Baker) | HGWZ557 | Yunnan, Mengla | 13.31±1.07 | 14.53±0.34 | t = -0.581, P = N.S. |
| Hook. f. | |||||
| O. mairei Levl. | B673 | Hunan, Zhangjiajie | 12.31±0.11 | 12.91±0.67 | t = -1.060, P = N.S. |
| O. marmortus Pierre ex | HGWZ625 | Yunnan, Puer | 11.22±0.08 | 12.05±0.19 | t = -3.609, P = N.S. |
| Rodrig. | |||||
| O. peliosanthoides Wang | HGWZ570 | Yunnan, Ninglang | 8.84±0.10 | 8.90±0.20 | t = -0.203, P = N.S. |
| et Tang | |||||
| O. pingbienensis Wang | nie3535 | Yunnan, Lijiang | 13.97±0.11 | 15.18±0.44 | t = -3.476, P = N.S. |
| et Dai | |||||
| O. platyphyllus Merr. et | HGWZ655 | Hainan, Baoting | 18.50±0.14 | 19.45±1.61 | t = -0.647, P = N.S. |
| Chun | |||||
| nie2340 | Guangxi, Longzhou | 14.73±0.15 | 14.29±0.81 | t = 0.616, P = N.S. | |
| O. revolutus Wang et Dai | HGWZ556 | Yunnan, Mengla | 12.58±0.16 | 12.76±1.24 | t = -0.144, P = N.S. |
| HGWZ636 | Yunnan, Puer | 9.57±0.10 | 9.77±0.28 | t = -0.560, P = N.S. | |
| HGWZ598 | Yunnan, Jinghong | 11.33±0.17 | 11.82±0.28 | t = -1.611, P = N.S. | |
| O. szechuanensis Wang et | HGWZ593 | Yunnan, Jinghong | 11.07±0.13 | 10.21±0.33 | t = 3.983, P = N.S. |
| Dai | |||||
| O. umbraticola Hance | HGWZ00791 | Yunnan, KIB | 13.38±0.19 | 13.70±0.23 | t = -1.273, P = N.S. |
| L. spicata (Thunb.) Lour. | nie3830 | Yunnan, Maguan | 11.90±0.10 | 12.81±0.40 | t = -3.268, P = N.S. |
| P. ophiopogoniodes F. T. | HGWZ536 | Yunnan, Pingbian | 21.75±0.10 | 23.73±0.84 | t = -2.404, P = N.S. |
| Wang & Tang | |||||
| P. sinica Wang et Tang | HGWZ587 | Yunnan, Puer | 29.36±1.05 | 30.38±0.60 | t = -0.624, P = N.S. |
| nie2364 | Guangxi, Longzhou | 25.75±0.56 | 26.27±1.12 | t = -0.279, P = N.S. | |
| P. yunnanensis Wang et Tang | nie3724 | Yunnan, Malipo | 24.55±0.89 | 22.72±0.27 | t = 1.250, P = N.S. |
| Model species | |||||
| Arabidopsis thaliana (L.) | 0.32±0.01 | 0.34±0.01 | t = -3.463, P = N.S. | ||
| Heynh. | |||||
| Oryza sativa L. spp. japonica | 0.98±0.03 | 1.05±0.01 | t = -2.945, P = N.S. | ||
| Lycopersicon esculentum Mill. | 1.98±0.04 | 1.89±0.08 | t = 1.096, P = N.S. | ||
| Glycine max (L.) Merr. | 2.30±0.08 | 2.27±0.05 | t = 0.319, P = N.S. | ||
| Zea mays L. | 4.97±0.25 | 4.65±0.08 | t = 1.364, P = N.S. |
Pearson correlation was used to analyze the relationships between genome sizes estimated from fresh and desiccated tissue using the whole data set, and Paired t-test was applied to FCM data obtained using dessicated vs. fresh tissues for each accession separately; SPSS 16.0(SPSS Inc., Chicago, IL, USA) was used to perform the statistical analyses.
3 ResultsThe 2C-value for the fresh and desicated tissue samples of 16 study species and five model species are presented in Table 1.
A highly significant correlation between values obtained measuring fresh and desiccated tissues was observed combining data for all studied accessions (r = 0.986, P < 0.001; Fig. 1). For every accession, the 2C-values of fresh and desiccated tissue also showed no significant difference (Table 1).
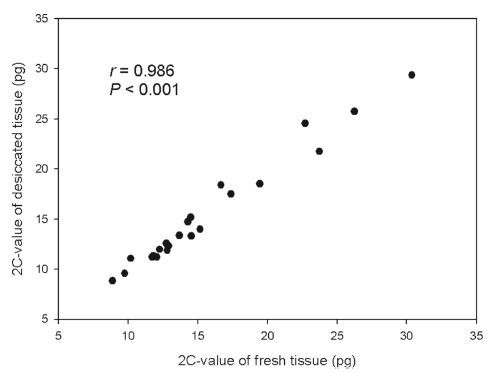
|
| Fig. 1 Comparison showing 2C-value for fresh and desiccated tissue from 16 species (22 accessions) in Ophiopogoneae. |
Similarly, a significant correlation was observed for the five model plant species (r = 0.959, P < 0.001; Fig. 2) with no difference between the 2C-values of fresh and desiccated tissue (Table 1).
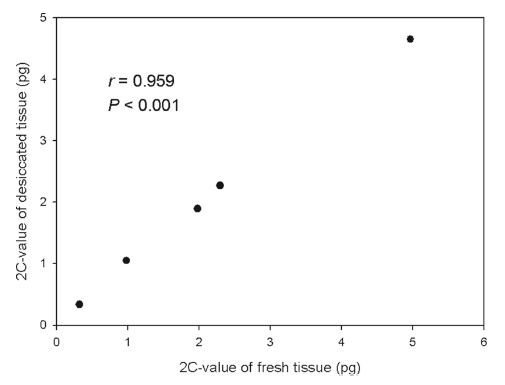
|
| Fig. 2 Comparison showing 2C-value for fresh and desiccated tissue from 5 model plant species. |
Fresh, young leaf tissues are the usual source material for FCM analysis (Suda and Travnicek, 2006a). However, need in fresh material has been repeatedly stressed as a crucial limitation for FCM application in botany (especially for expeditions to remote areas when rapid dispatch or maintenance of fresh samples is difficult)(Suda and Travnicek, 2006b). Desiccated tissue has been proved to be suitable for FCM in some but not all cases (Kolar et al., 2012). It was firstly used in mosses (Voglmayr, 2000), and the desiccated moss tissue contained intact nuclei after drying. Two-year-old silica gel-dried samples of J. biglumis L.(Juncaceae) used in FCM showed variation in nuclear DNA content within instrumental error (Schonswetter et al., 2007). Air desiccation had no negative effects on nuclei integrity and subsequent DAPI fluorescence in Vaccinium subg. Oxycoccus (Suda and Travnicek, 2006b). In general, variation in genome size detected by FCM using desiccated tissue falls within an acceptable range of error (Bai et al., 2012), but some authors cast doubt on wide applicability of dessicated tissues. Bainard et al.(2011)thought that tissue treatment caused larger differences than other methodological factors, and Suda and Travnicek (2006a, b) proposed that desiccated tissue in FCM analysis should be restricted to determine ploidy, but not the applied to absolute genome size quantification. The previous studies comparing usage of fresh vs. desiccated tissue for estimation of genome sizes produced mixed results. Little et al.(2007), Suda et al.(2007), and Balao et al.(2009)reported the genome size difference between fresh and desiccated tissues for Selaginella, S. carniolicus, and Dianthus broteri to be less than 1.2%. Bainard et al.(2011)showed that desiccated tissue can alleviate the constraint of fresh tissue; about 75% plant species were able to yield adequate histograms after 1 year storage at room temperature, although the remaining 25% produced no fluorescence signal.
In the present study, all studied species produced high quality histograms with highly significant correlation of the 2C-values obtained from fresh versus desiccated tissues. There was comparably low level of variation in 2C-values obtained with fresh and dessicated tissues with no significant difference between the later for any studied species. Thus, the observed intraspecific variation in genome size, which is a common phenomenon (Moscone et al., 2003), should be attributed to instrumental errors (Greilhuber and Obermayer, 1997; Greilhuber, 1998, 2005; Wang et al., 2015).
The 2C-values of the tested Ophiopogoneae species were similar to those reported earlier in the literature, but with some noticeable differences. For example, for L. spicata the detected 1Cx-value was 5.95 pg while it was earlier reported to be 3.99 pg (Lattier and Ranney, 2014). A reason for the observed discrepancy can be that Lattier and Ranney (2014)used Pisum sativum L. 'Ctirad' (2C = 8.76 pg) as an internal standard, while we used Z. mays (B73)(2C = 5.43 pg). Usage of different reference standards may result in different estimates for the same material (Dolezel et al., 1998), with other potential causes being instrumental and methodological differences.
To conclude, our results show suitability of the dessicated tissues for reliable estimation of genome size if the issue after desiccation remains green without brown or yellow spots and is stored at ×80 ℃ for no more than 6 months. All the samples stored in this way should give FCM results of acceptable quality with good histogram resolution and little loss of fluorescence intensity.
Acknowledgement
The study was supported by grants from the General Project of Natural Science Research in Anhui Province (AQKJ2015B018), and Major projects of the National Natural Science Foundation of China (31590823).
| Bai C, Alverson W.S, Follansbee A, et al., 2012. New reports of nuclear DNA content for 407 vascular plant taxa from the United States. Ann. Bot., 110(8), 1623 -1629. DOI:10.1093/aob/mcs222 | ||
| Bainard J.D, Husband B.C, Baldwin S.J, et al., 2011. The effects of rapid desiccation on estimates of plant genome size. Chromosome Res., 19(6), 825 -842. DOI:10.1007/s10577-011-9232-5 | ||
| Balao F, Casimiro-Soriguer R, Talavera M, et al., 2009. Distribution and diversity of cytotypes in Dianthus broteri as evidenced by genome size variations. Ann. Bot. Lond., 105, 965 -973. | ||
| Bennett M.D, 1972. Nuclear DNA content and minimum generation time in Her-baceous plants. P. Roy. Soc. B-Biol. Sci., 181(1063), 109 -135. DOI:10.1098/rspb.1972.0042 | ||
| Bennett M.D, Leitch I.J, 2011. Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann. Bot., 107(3), 467 -590. DOI:10.1093/aob/mcq258 | ||
| Bennett M.D, Leitch I.J, Price H.J, et al., 2003. Comparisons with Caenorhabditis (~100 Mb) and Drosophila (~175 Mb) using flow cytometry show genome size in Arabidopsis to be ~157 Mb and thus ~25% larger than the Arabidopsis genome initiative estimate of ~125 Mb. Ann. Bot., 91(5), 547 -557. DOI:10.1093/aob/mcg057 | ||
| Bennett M.D, Smith J, 1976. Nuclear DNA amounts in angiosperms. Philos. T. R. Soc. B, 274(933), 227 -274. DOI:10.1098/rstb.1976.0044 | ||
| Bharathan G, Lambert G, Galbraith D.W, 1994. Nuclear DNA content of mono-cotyledons and related taxa. Amer. J. Bot., 81(3), 381 -386. DOI:10.2307/2445466 | ||
| Dolezel J, 1991. Flow cytometric analysis of nuclear DNA content in higher plants. Phytochem. Anal., 2, 143 -154. DOI:10.1002/(ISSN)1099-1565 | ||
| Dolezel J, Greilhuber J, Lucretti S, Meister A, Lysak M.A, Nardi L, Obermayer R, 1998. Plant genome size estimating by flow cytometry: inter-laboratory comparison. Ann. Bot. Suppl. A, 82, 17 -26. | ||
| Fleischmann A, Michael T.P, Rivadavia F, et al., 2014. Evolution of genome size and chromosome number in the carnivorous plant genus Genlisea (Lentibu- lariaceae), with a new estimate of the minimum genome size in angiosperms. Ann. Bot., 114, 1651 -1663. DOI:10.1093/aob/mcu189 | ||
| Galbraith D, Harkins K, Maddox J, et al., 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science, 220(4601), 1049 -1051. DOI:10.1126/science.220.4601.1049 | ||
| Goff S.A, Ricke D, Lan T.H, et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296(5565), 92 -100. | ||
| Greilhuber J, 1998. Intraspecific variation in genome size: a critical reassessment. Ann. Bot., 82, 27 -35. DOI:10.1006/anbo.1998.0725 | ||
| Greilhuber J, 2005. Intraspecific variation in genome size in Angiosperms: iden-tifying its existence. Ann. Bot, 95, 91 -98. DOI:10.1093/aob/mci004 | ||
| Greilhuber J, Obermayer R, 1997. Genome size and maturity group in Glycine max (soybean). Heredity, 78(5), 547 -551. DOI:10.1038/hdy.1997.85 | ||
| Hanson L, Mcmahon K, Johnson M, et al., 2001. First nuclear DNA C-values for 25 angiosperm families. Ann. Bot., 87, 251 -258. DOI:10.1006/anbo.2000.1325 | ||
| Kolar F, Lucanova M, Tesitel, J , et al., 2012. Glycerol-treated nuclear suspensionsan efficient preservation method for flow cytometric analysis of plant samples. Chromosome Res., 20(2), 303 -315. DOI:10.1007/s10577-012-9277-0 | ||
| Lattier J.D, Ranney T.G, 2014. Identification, nomenclature, genome sizes, and ploidy levels of Liriope and Ophiopogon Taxa. HortScience, 49(2), 145 -151. | ||
| Leith I, Hanson L, Winfield M, et al., 2001. Nuclear DNA C-values complete fa-milial representation in gymnosperms. Ann. Bot., 88, 843 -849. DOI:10.1006/anbo.2001.1521 | ||
| Little D.P, Moran R.C, Brenner E.D, et al., 2007. Nuclear genome size in Selaginella. Genome 50, 50, 351 -356. DOI:10.1139/G06-138 | ||
| Moscone E.A, Baranyi M, Ebert I, et al., 2003. Analysis of nuclear DNA content in Capsicum (Solanaceae) by flow cytometry and Feulgen densitometry. Ann. Bot., 92, 21 -29. DOI:10.1093/aob/mcg105 | ||
| Pellicer J, Fay M, Leitch I, 2010. The largest eukaryotic genome of them all. Bot. J. Linn. Soc., 164, 10 -15. DOI:10.1111/boj.2010.164.issue-1 | ||
| Popp M, Gizaw A, Nemomissa S, et al., 2008. Colonization and diversification in the African 'sky islands' by Eurasian lychnis L. (Caryophyllaceae). J. Biogeogr., 35, 1016 -1029. | ||
| Schonswetter P, Suda J, Popp M, et al., 2007. Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Mol. Phylogenet. Evol., 42(1), 92 -103. DOI:10.1016/j.ympev.2006.06.016 | ||
| Soltis D.E, Soltis P.S, Leitch I.J, et al., 2003. Evolution of genome size in the an- giosperms. Amer. J. Bot., 90(11), 1596 -1603. DOI:10.3732/ajb.90.11.1596 | ||
| Suda, J., Travnicek, P., 2006a. Estimation of relative nuclear DNA Content in dehy-drated plant tissues by Flow Cytometry. In: Robinson, J.P., et al. (Eds.), Current Protocols in Cytometry. Wiley, New York. | ||
| Suda J, Travnicek P, 2006b. Reliable DNA ploidy determination in dehydrated tissues of vascular plants by DAPI flow cytometry-new prospects for plant research. Cytom. Part A, 69A(4), 273 -280. DOI:10.1002/(ISSN)1552-4930 | ||
| Suda J, Weiss-Schneeweiss H, Tribsch A, et al., 2007. Complex distribution pat-terns of di-, tetra-, and hexaploid cytotypes in the European high mountain plant Senecio carniolicus (Asteraceae). Amer. J. Bot., 94(8), 1391 -1401. DOI:10.3732/ajb.94.8.1391 | ||
| Voglmayr H, 2000. Nuclear DNA amounts in mosses (Musci). Ann. Bot., 85, 531 -546. DOI:10.1006/anbo.1999.1103 | ||
| Wang J, Liu J, Kang M, 2015. Quantitative testing of the methodology for genome size estimation in plants using flow cytometry: a case study of the Primulina genus. Front. Plant Sci, 6, 354 . | ||
| Zonneveld B.J.M, Leitch I.J, Bennett M.D, 2005. First nuclear DNA amounts in more than 300 angiosperms. Ann. Bot., 96(2), 229 -244. DOI:10.1093/aob/mci170 |



