b. University of Chinese Academy of Sciences, Beijing 100049, China;
c. Missouri Botanical Garden, P.O. Box 299, St. Louis, MO, 63166-0299, USA
The Brassicaceae (Cruciferae) comprises 51 tribes, 340 genera, and 3840 species distributed worldwide except Antarctica (Al- Shehbaz and German unpublished preliminary compilation). The family is economically and scientifically important, and it contains many species of ornamentals (e.g., Orychophragmus Bunge), crops (e.g., Brassica L.), and model organisms [e.g., Arabidopsis thaliana (L.) Heynh.]. It is also well known as a taxonomically difficult family, as most morphological characters used for generic delimitation have undergone extensive convergent evolution, and many traditionally defined genera and tribes were found to be artificially delimited (Al-Shehbaz, 2012). Fortunately, molecular phylogenetic studies during the past 20 years have greatly improved our understanding of the phylogenetic relationships within Brassicaceae. Indeed, a number of genera, including, for example, Solms-laubachia Muschl.(Yue et al., 2008), Eutrema R.Br.(Warwick et al., 2006), and Arabidopsis (DC.) Hyenh.(O′Kane and Al-Shehbaz, 2003) and tribes such as Eutremeae (Warwick et al., 2006) and Euclidieae (Warwick et al., 2007) were redefined morphologically based on the utilization of molecular sequence data.
The first Brassicaceae-wide molecular phylogeny was carried out by Beilstein et al.(2006)using the chloroplast ndhF sequences of 113 species from 101 genera. Three major lineages (Lineages I-III) within the core Brassicaceae were identified, and using these results Al-Shehbaz et al.(2006)established the first phylogenetic tribal classification of the family, in which 25 tribes were recognized. The three-lineage backbone phylogeny and 25 tribes were later confirmed by nuclear phytochromeA (Beilstein et al., 2008), as well as nuclear ITS (Bailey et al., 2006; Warwick et al., 2010), nad4 intron1(Franzke et al., 2009), and combined molecular data sets (Couvreur et al., 2010; Koch et al., 2007). The molecularly wellsupported major monophyletic clades in the family have been recognized as tribes. To date, 51 tribes have been recognized, of which 13 are unigeneric (Al-Shehbaz, 2012; Al-Shehbaz et al., 2014; German and Friesen, 2014).
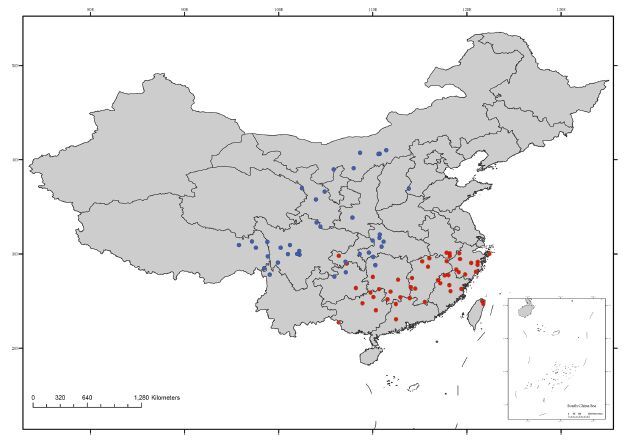
The unigeneric tribe Yinshanieae was recognized by Warwick et al.(2010), and in their family-level phylogeny based on ITS sequences from 96 genera, two Yinshania Y.C.Ma & Y.Z.Zhao taxa, Y. acutangula (O.E.Schulz) Y.H.Zhang and Y. acutangula ssp. wilsonii (O.E.Schulz) Al-Shehbaz et al., formed a strongly supported clade occupying a relatively solitary position used to represent this new tribe. As currently delimited, the Yinshanieae contains the single genus Yinshania (Warwick et al., 2010; Al-Shehbaz, 2012). However, the taxonomy on Yinshania has long been in dispute, and its generic boundary was mixed up with those of Hilliella (O.E.Schulz) Y.H.Zhang & H.W.Li, Cochleariella Y.H.Zhang & Vogt, and Cochlearia L. The taxonomic revision by Al-Shehbaz et al.(1998)united the three Chinese genera into Yinshania, which consequently included 13 species and 4 subspecies (Fig. 1). By contrast, Zhang (2003) concluded that Yinshania and Hilliella should be kept as two separate genera. These two genera, however, show dissimilarities in both morphology and geographic distribution (Fig. 2), and therefore the unigeneric identity of Yinshanieae came into dispute and waited to be tested.

|
| Fig. 1 Selected species of Yinshanieae.(A) Y. yixianensis;(B) Y. lichuanensis;(C) Y. rivulorum;(D) Y. hunanensis;(E) Y. fumarioides;(F) and (I) Y. rupicola ssp. shuangpaiensis;(G) and (J) Y. hui;(H) and (K) Y. sinuata;(L) and (N) Y. acutangula ssp. wilsonii;(M) Y. henryi;(O) and (P) Y. zayuensis |
|
| Fig. 2 Distributions of Yinshanieae based on field and herbarium collections. Blue and red dots represent specimens records of Yinshania and Hilliella, respectively |
In this study, we present the most comprehensive species-level phylogeny of Yinshanieae covering 12 out of the 13 recognized species and using two nuclear DNA (ITS and ETS) and four chloroplast DNA ((trnL-F, trnH-psbA, rps16, rpL32-(trnL) markers, with analyses at family and tribal levels. Our goals are to test the identity of Yinshanieae and to clarify the infratribal relationships within the tribe.
2 Materials and methods 2.1 Plant materials and molecular dataPlant materials included 12 species and 2 subspecies of Yinshanieae (Table 1). Dry leaf material of Y. exiensis, Y. rupicola ssp. rupicola, and Y. paradoxa were obtained from herbarium specimens, but material for all other species were collected from the wild in China, and that of Y. rupicola ssp. shuangpaiensis was cultivated in the Kunming Botanical Garden. We were unable to obtain material of Y. furcatopilosa, Y. acutangula ssp. microcarpa, and Y. sinuata ssp. qianwuensis. The taxonomic circumscription of Yinshanieae follows Al-Shehbaz (2012)and Al-Shehbaz et al.(1998).
| Species | Geographical origin (China) | Collection number (Herbarium) | Genbank No. | |||||
| ETS | ITS | rpL32-trnL | rps16 | trnH-psbA | trnL-F | |||
| Y. acutangula ssp. acutangula | Kangding, Sichuan | Boufford et al. 37855(KUN) | KX244360 | KX244386 | KX244410 | KX244434 | KX244458 | KX244483 |
| Luolong, Xizang | Boufford et al. 40929(KUN) | KX244361 | KX244387 | KX244411 | KX244435 | KX244459 | KX244484 | |
| Y. acutangula ssp. wilsonii | Kangding, Sichuan | MCQ063(KUN) | KX244366 | KX244392 | KX244416 | KX244440 | KX244464 | KX244489 |
| Wenxian, Gansu | MCQ107 (KUN) | KX244367 | KX244393 | KX244417 | KX244441 | KX244465 | KX244490 | |
| Y. henryi | Shennongjia, Hubei | zdg6185(KUN) | KX244362 | KX244388 | KX244412 | KX244436 | KX244460 | KX244485 |
| Shennongjia, Hubei | zdg7062(KUN) | KX244364 | KX244390 | KX244414 | KX244438 | KX244462 | KX244487 | |
| Y. zayuensis | Shennongjia, Hubei | zdg6330(KUN) | KX244363 | KX244389 | KX244413 | KX244437 | KX244461 | KX244486 |
| Shennongjia, Hubei | SunHang18133(KUN) | KX244368 | KX244394 | KX244418 | KX244442 | KX244466 | KX244491 | |
| Y. exiensis | Wushan, Chongqing | 1414 (PE) | KX244369 | KX244395 | KX244419 | KX244443 | KX244467 | |
| Y. fumarioides | Jinhua, Zhejiang | Chen.HL 165 (KUN) | KX244356 | KX244381 | KX244406 | KX244430 | KX244454 | KX244478 |
| Y. yixianensis | Yixian, Anhui | H.L.Chen069 (KUN) | KX244347 | KX244372 | KX244398 | KX244422 | KX244446 | KX244470 |
| Y. lichuanensis | Wuning, Jiangxi | H.L.Chen084 (KUN) | KX244349 | KX244374 | KX244400 | KX244424 | KX244448 | KX244472 |
| Y. hunanensis | Lushan, Jiangxi | H.L.Chen081 (KUN) | KX244348 | KX244373 | KX244399 | KX244423 | KX244447 | KX244471 |
| Y. hui | Yanling, Hunan | H.L.Chen105 (KUN) | KX244350 | KX244375 | KX244401 | KX244425 | KX244449 | KX244473 |
| Y. sinuata | Xinning, Hunan | H.L.Chen128 (KUN) | KX244352 | KX244377 | KX244403 | KX244427 | KX244451 | KX244475 |
| Y. rivulorum | Shuangpai, Hunan | H.L.Chen123 (KUN) | KX244351 | KX244376 | KX244402 | KX244426 | KX244450 | KX244474 |
| Y. rupicola ssp. rupicola | Shuangpai, Hunan | 219156 (KUN) | KX244354 | KX244379 | KX244405 | KX244429 | KX244453 | KX244477 |
| Y. rupicola ssp. shuangpaiensis | Cultivated in KBG | No vochuer, Fig. 1 F&I | KX244353 | KX244378 | KX244404 | KX244428 | KX244452 | KX244476 |
| Y. paradoxa | Beibei, Chongqing | He3926(PE) | KX244355 | KX244380 | ||||
| Cardamine flexuosa | Shennongjia, hubei | zdg4044(KUN) | KX244365 | KX244391 | KX244415 | KX244439 | KX244463 | KX244488 |
| Descurainia sophia | Tongren, Qinghai | ZH379(KUN) | KX244370 | KX244396 | KX244420 | KX244444 | KX244468 | KX244492 |
| Eutrema heterophylhum | Banma, Qinghai | ZH551(KUN) | KX244357 | KX244382 | KX244407 | KX244431 | KX244455 | KX244479 |
| Megacarpaea delavayi | Lijiang, Yunnan | YangBChen-221(KUN) | KX244385 | KX244482 | ||||
| Sinalliaria limprichtiana | Lin'an, Zhejiang | H.L.Chen032(KUN) | KX244358 | KX244383 | KX244408 | KX244432 | KX244456 | KX244480 |
| Pegaeophyton scapiflorum | Shangri-La, Yunnan | NY&WQ 14(KUN) | KX244359 | KX244384 | KX244409 | KX244433 | KX244457 | KX244481 |
| Smelowskia tibetica | Yushu, Qinghai | ZH641(KUN) | KX244371 | KX244397 | KX244421 | KX244445 | KX244469 | KX244493 |
| *KBG: Kunming Botanical Garden. | ||||||||
Phylogenetic studies were initially conducted to determine the monophyly of Yinshanieae within the Brassicaceae, and later to establish the phylogenetic relationships within the tribe. For analyses at the family level, 95 ITS and 69 trnL-F sequences were used, representing 48 and 36 tribes, respectively. Based on these familywide analyses, six species (Smelowskia tibetica, Descurainia sophia, Cardamine flexuosa, Sinalliaria limprichtiana, Pegaeophyton scapiflorum, and Eutrema heterophylhum) were selected as outgroups at the tribal-level analyses using two nuclear DNA markers (ITS, ETS) and four chloroplast DNA markers (trnL-F, trnH-psbA, rps16, rpL32- (trnL). Except for these six species and all Yinshania taxa, DNA sequences of all other studied taxa were downloaded from GenBank. Taxa and GenBank accession numbers are listed in Table 1 and Appendix A.
2.2 DNA extraction, PCR amplification, and sequencingTotal genomic DNA was extracted from silica gel-dried leaf materials or herbarium specimens using the Plant Genomic DNA Kit (Tiangen Bioteke, Beijing, China) following the manufacturer's protocol. The ITS region was amplified with the primers ITS-18 as modified by Mummenhoff et al.(1997)and ITS-25R (White et al., 1990); the ETS region was amplified with the primers18S-IGS (Baldwin and Markos, 1998) and Bur-ETS1F (Weeks et al., 2005); the (trnL-F regionwas amplified with the primers c/f (Taberlet et al., 1991); the trnH-psbA region was amplified with the primers trnH/ psbA (Tao et al., 1997); the rps16 region was amplified with the primers rps16F/rps16R (Shaw and Small, 2005); and the rpL32-(trnL region was amplified with the primers (trnL (UAG) and rpL32-F (Shaw et al., 2007). All polymerase chain reactions (PCR) were performed in a 25 μL volume consisting of 1-2 μL sample DNA (approx. 1-10 ng), 2.5 μL 10 × buffer, 1 μL MgCl2(25 μM stock), 2.5 μL dNTPs, 1 μL of 10 μM stock of each primer, and 0.2 μL Taq polymerase, adjusted to 25 μL with ddH2O. The PCR cycling conditions of rpL32-(trnL region were template denaturation at 80 ℃ for 5 min followed by 34 cycles of denaturation at 95 ℃ for 1 min, primer annealing at 50 ℃ for 1 min, followed by a ramp of 0.3 ℃/s to 65 ℃, and primer extension at 65 ℃ for 4 min, followed by a final extension step of 5 min at 65 ℃ (Shaw et al., 2007). The PCR protocol of the remaining regions involved a hot start with 4-5 min at 94 ℃, and 32-35 cycles of amplification (1 min denaturing at 94 ℃, 30-60 s annealing at 48-55 ℃, 60-90 s extension at 72 ℃), and a final elongation step for 7-10 min at 72 ℃. The sequencing primers are the same with amplified primers, the sequencing reactions mixes were analyzed on an ABI 3730 automated sequencer (Applied Biosystems, Foster City, California, USA). The cpDNA (including (trnL-F, trnH-psbA, rps16 and rpL32-(trnL) of Y. paradoxawas not sequenced due to the low-quality specimen material.
2.3 Phylogenetic analysesOriginal chromatograms were evaluated with Sequencher 4.1.4 for base confirmation and contiguous sequences editing, and sequences were aligned and manually adjusted with BioEdit v.5.0.9(Hall, 1998). The aligned sequences were analyzed with maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI).
Parsimony analyses were performed with heuristic searches of 1000 replicates with random stepwise addition using tree bisection reconnection (TBR) branch swapping as implemented in PAUP* 4.0b10(Swofford, 2003). All characters were weighted equally, and gaps were treated as missing data. The bootstrap probabilities (BP) were calculated from 1000 replicates using a heuristic search with simple addition with the TBR and MULPARS options implemented (Felsenstein, 1985).
For ML and BI analyses, jModeltest v2.1.7(Darriba et al., 2012) was used to select the best-fitted model of nucleotide substitution based on the Akaike information criterion (AIC). For family-level analyses, the GTR+I+G model was selected for the ITS and (trnLF datasets. For tribal-level analyses, the GTR+G model was selected for the nDNA (combined ITS and ETS) and cpDNA (combined (trnL-F, trnH-psbA, rps16 and rpL32-(trnL) datasets in Yinshania and Hilliella. The ML analyses were carried out in RA × ML v8.2.4(Stamatakis, 2014) on the CIPRES Science Gateway V 3.3(Miller et al., 2010), using 1000 bootstrap replicates. Due to the debate about the correlation between parameters I and G (Kelchner and Thomas, 2007; Ren et al., 2005) and the GTRGAMMA+I model not being recommended by the developer of RA × ML (Mayrose et al., 2005; Stamatakis, 2006), all ML analyses were run under the GTR+G model. Bayesian inference (BI) based on the Markov chain Monte Carlo methods (Yang and Rannala, 1997) was performed using MrBayes v3.2.5(Ronquist et al., 2012). For family-level analyses, four simultaneous Monte Carlo Markov chains (MCMCs) were run for eight million generations (ITS) and three million generations ((trnL-F), and one tree sampled every 1000 generations. The first 2000 trees (ITS dataset) and 750 trees ((trnL-F dataset)(25% of total trees) were discarded as burnin. The remaining trees were summarized in a 50% majority-rule consensus tree, and the posterior probabilities (PP) were calculated. For tribal-level analyses, datasets of nDNA and cpDNA were analyzed separately and combined, following the same methods described above. The levels of incongruence among data partitions (nDNA and cpDNA) were evaluated by incongruence-length difference (ILD) test (Farris et al., 1994) with 1000 replicates of heuristic search using TBR branch swapping with random sequence additions. The datasets were not incongruent in Yinshania [P = 0.381], while P = 0.02 in Hilliella means incongruent (P < 0.05) between nDNA and cpDNA. Datasets were combined, though there is a slight incongruence in Hilliella. All analyses were conducted using two runs for one million generations, sampling one tree every 100 generations and discarding the first 2500 trees (25% of total trees).
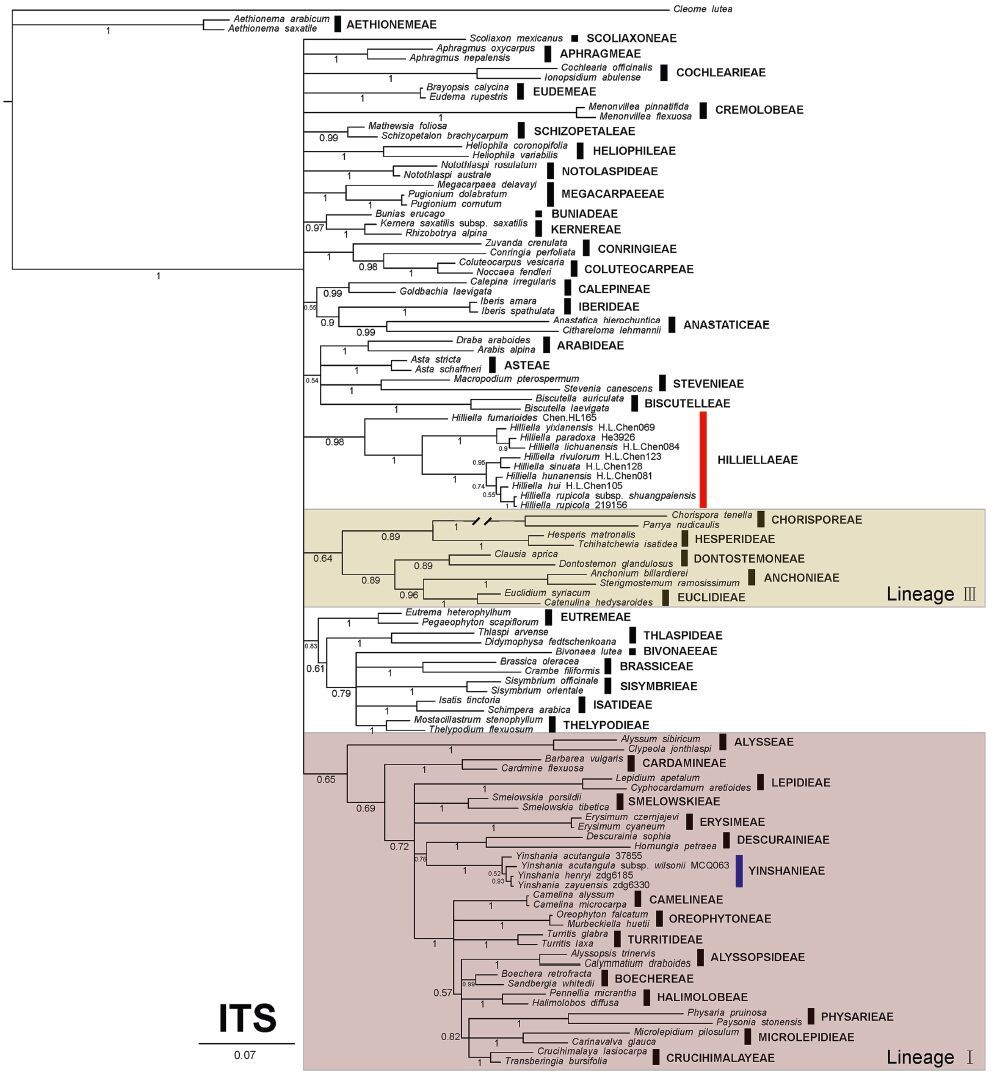
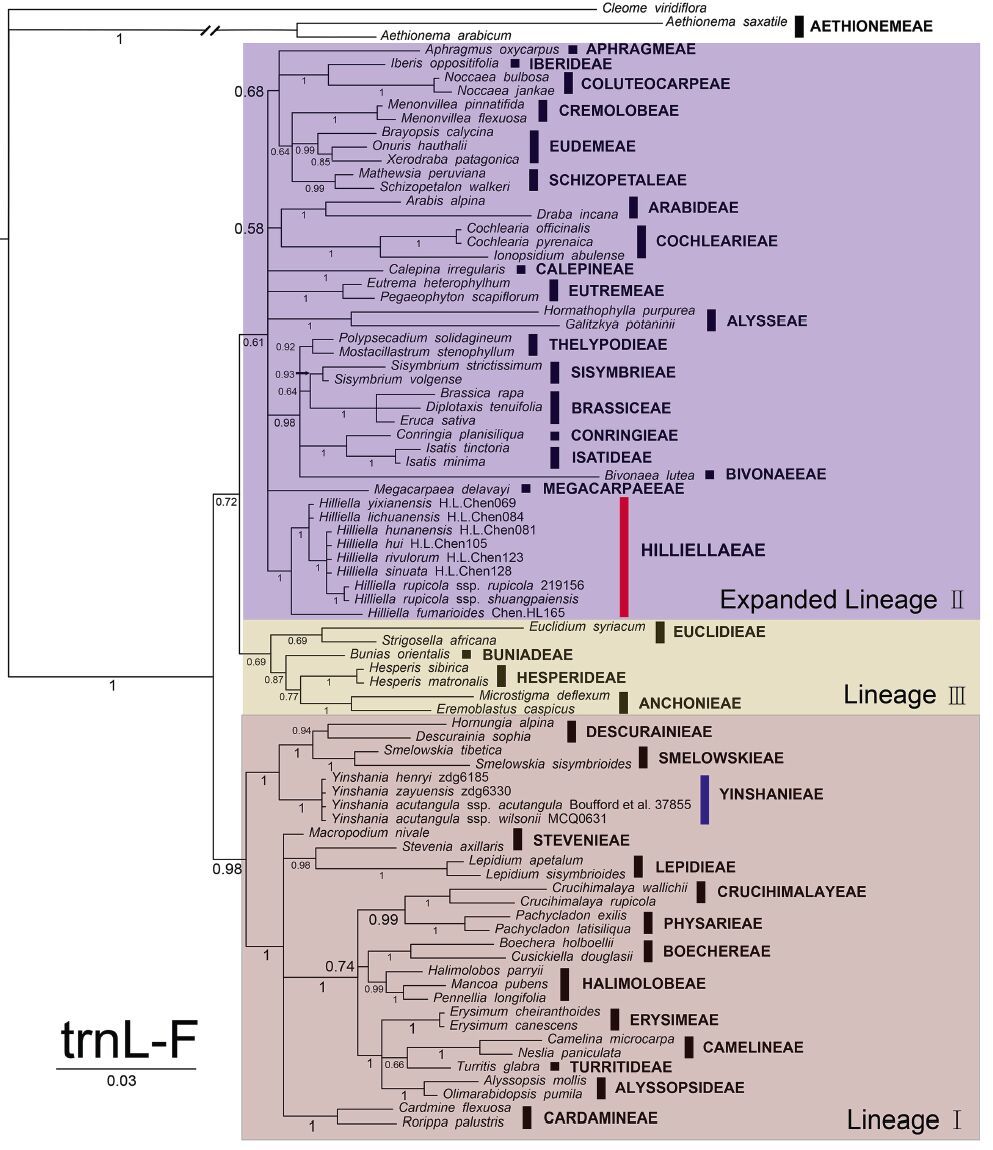
3 Results 3.1 Non-monophyly of YinshanieaeThe aligned ITS matrix included 109 sequences and was 643 bp long with 316(49.1%) parsimonious informative sites. The aligned (trnL-F matrix included 82 sequences and was 1078 bp long with 279(25.9%) parsimonious informative sites. Node labels and descriptions of support within the text include MP bootstrap values, ML bootstrap values and Bayesian posterior probabilities in the following format:(MP/ML/PP). All MP, ML, and BI analyses of both regions suggested Yinshanieae was split into two distantly related clades and, therefore, only the BI topologies are shown (Figs. 3 and 4). Yinshania formed a strongly supported monophyletic clade (ITS, 100/100/1; (trnL-F, 99/100/1) close to the tribes Descurainieae and Smelowskieae (ITS, d/93/0.76; (trnL-F, 84/d/1), while species from the previously recognized Hilliella formed a moderately to strongly supported clade (ITS, 83/82/0.98; (trnL-F, 72/75/1). However, the relationships of Hilliella to the other genera or tribes was not resolved.
|
| Fig. 3 Phylogenetic tree resulting from Bayesian analysis of the ITS sequences of the 108 Brassicaceae species from 82 genera and Cleome lutea as outgroup. The taxa represents 49 currently recognized tribes, and their tribal assignments are given to the right. Posterior probability values are given. Hilliellaeae and Yinshanieae are highlighted in red and blue bars, respectively |
|
| Fig. 4 Phylogenetic tree resulting from Bayesian analysis of the (trnL-F sequences of 81 Brassicaceae species from 58 genera and Cleome viridiflora as outgroup. The taxa represent 37 currently recognized tribes, and their tribal assignments are given to the right. Posterior probability values are given. Hilliellaeae and Yinshanieae are highlighted in red and blue bars, respectively |
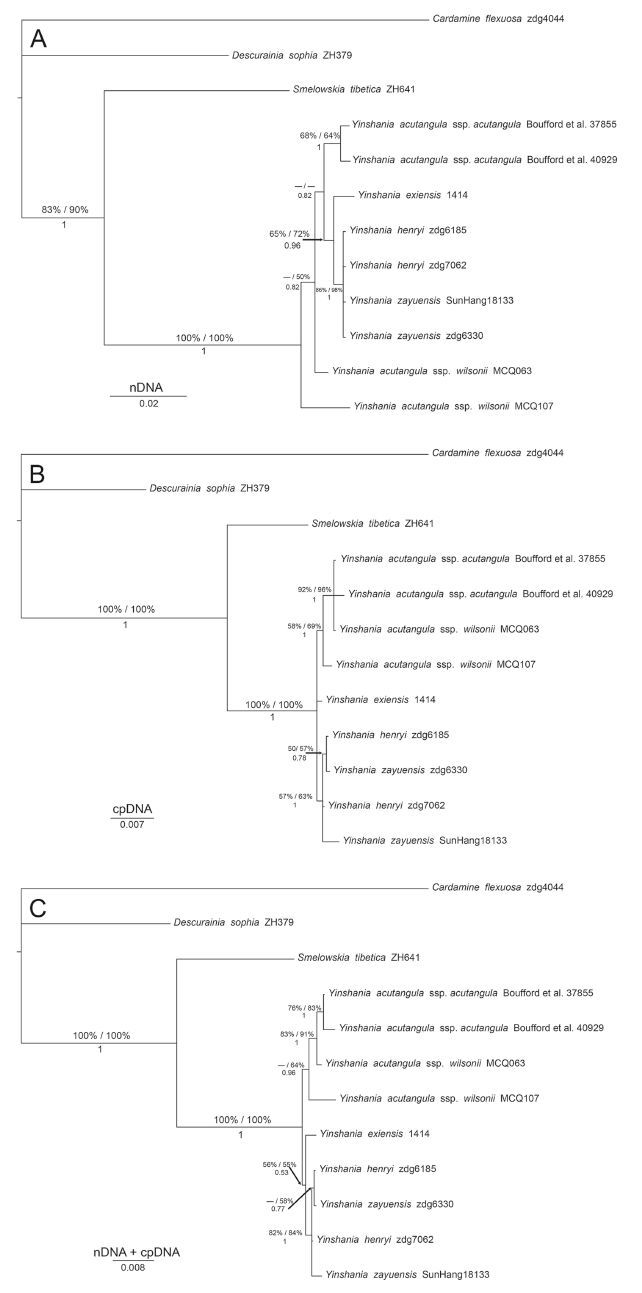
Dataset characteristics and summary statistics for phylogenetic analyses are given in Table 2. The three phylogenetic analyses (MP, ML and BI) of the nDNA (combined ITS and ETS) and cpDNA (combined (trnL-F, trnH-psbA, rps16 and rpL32-(trnL) datasets of Yinshania and Hilliella yielded similar topologies and only the BI topologies are shown (Fig. 5). The systematic position of Y. acutangula ssp. wilsonii showed a conflict between nDNA- and cpDNA-derived phylogenies; the subspecies formed an early branching lineage in nDNA phylogeny (Fig. 5A), while in the cpDNA phylogeny (Fig. 5B) it formed a lineage with Y. acutangula ssp. acutangula. When the nDNA and cpDNA data were combined (Fig. 5C), topology of the tree was mostly congruent with cpDNA results. Y. exiensis, which was treated as a synonym of Y. zayuensis, formed an independent clade (Fig. 5). By contrast, Y. henryiand Y. zayuensiswere nested together (nDNA, 86/98/1; cpDNA, 57/63/1; n+cpDNA, 82/84/1), and Y. henryizdg6185 and Y. zayuensiszdg6330 formed a clade in cpDNA and n+cpDNA phylogeny trees (cpDNA, 50/57/0.78; n+cpDNA, -/58/0.77) as sister to Y. henryizdg7062 and Y. zayuensisSunHang 18133.
|
| Fig. 5 Phylogenetic relationships within Yinshania inferred from Bayesian analysis of:(A) the nDNA (combined ITS and ETS) dataset;(B) the cpDNA (combined (trnL-F, trnH-psbA, rps16 and rpL32-(trnL) dataset;(C) the nDNA + cpDNA dataset, Cardamine flexuosa, Descurainia sophia, and Smelowskia tibetica as outgroups. Values above braches are maximum parsimony/maximum likelihood bootstrap (only show if > 50%), and values below braches are Bayesian posterior probabilities. |
| ITS | ETS | nDNA | trnL-F | trnH-psbA | rps16 | rpL32-trnL | cpDNA | n+cpDNA | ||||||||||
| Y | H | Y | H | Y | H | Y | H | Y | H | Y | H | Y | H | Y | H | Y | H | |
| No. of sequences | 12 | 13 | 12 | 13 | 12 | 13 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 13 |
| Alignment length | 656 | 663 | 418 | 420 | 1074 | 1083 | 915 | 750 | 415 | 362 | 847 | 790 | 945 | 1104 | 3122 | 3006 | 4196 | 4089 |
| No. of parsimony- informative characters | 38 | 107 | 46 | 97 | 84 | 204 | 8 | 19 | 14 | 40 | 8 | 26 | 47 | 60 | 77 | 145 | 161 | 349 |
| Retention index | 0.7970 | 0.8863 | 0.8199 | 0.8326 | 0.7952 | 0.8628 | ||||||||||||
| Consistency index | 0.9050 | 0.8674 | 0.9382 | 0.9208 | 0.9190 | 0.8911 | ||||||||||||
| Best tree length | 287 | 445 | 340 | 505 | 630 | 955 | ||||||||||||
| Model selected by AIC | GTR+G | GTR+G | GTR+G | GTR+G | ||||||||||||||
| *Y: Yinshania, H: Hilliella. | ||||||||||||||||||
Within the Hilliella clade there are three subclades, with H. fumarioidesforming an independent A Clade (Fig. 6). The rest of the genus falls into two strongly supported clades: B Clade (nDNA, 100/ 100/1; cpDNA, 100/100/1; n + cpDNA, 100/100/1) includes H. yixianensis, H. lichuanensis,and H. paradoxa ; C Clade (nDNA, 100/ 100/1; cpDNA, 98/100/1; n + cpDNA, 100/100/1) includes H. hui, H. hunanensis, H. rupicola, H. rivulorum, and H. sinuata. The systematic position of H. hui was in conflict between the nDNA- and cpDNAderived phylogenies (Fig. 6A and B). In the nDNA phylogenetic tree, H. hui was sister to H. hunanensis and H. rupicola (79/75/0.99), whereas in the cpDNA phylogenetic tree, H. hui formed a clade with H. rivulorum and H. sinuata (98/100/1), and H. rivulorum was sister to H. hui and H. sinuata. When the nDNA and cpDNAwere combined (Fig. 6C), topology of the tree was congruent with the cpDNA results.
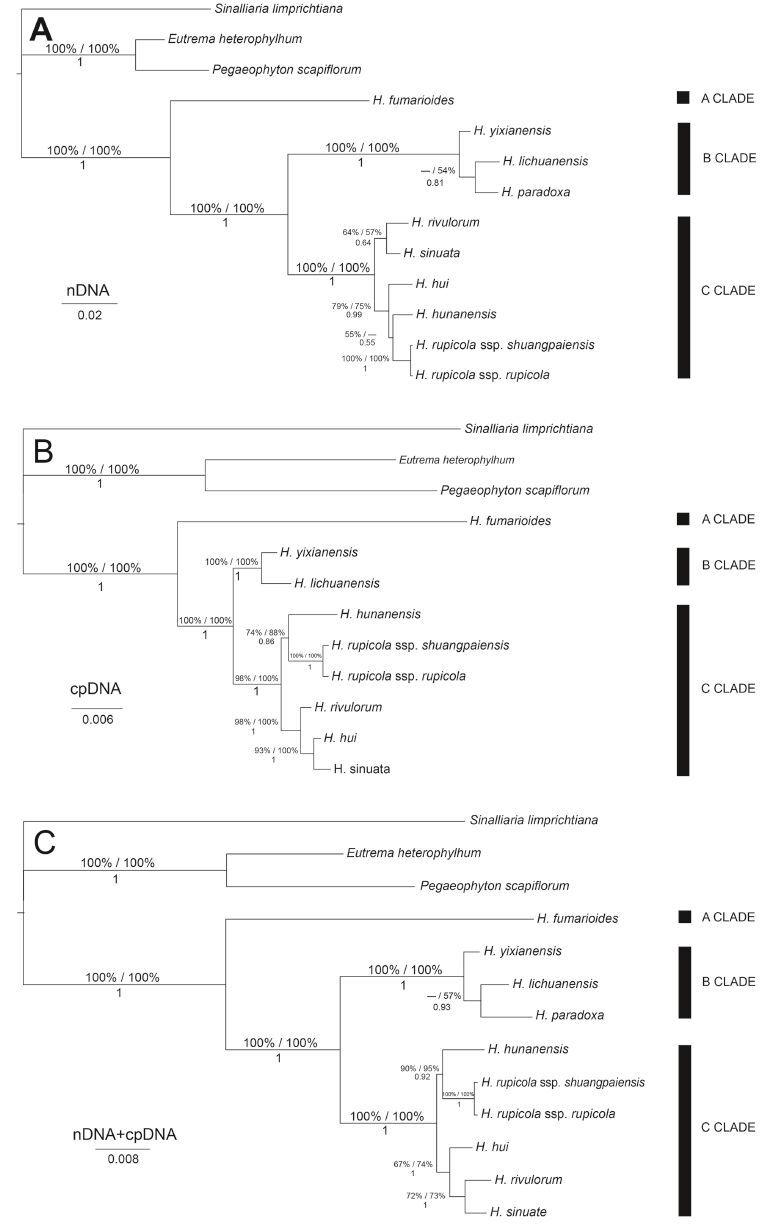
|
| Fig. 6 Phylogenetic relationships within Hilliella inferred from Bayesian analysis of:(A) the nDNA (combined ITS and ETS) dataset;(B) the cpDNA (combined (trnL-F, trnH-psbA, rps16 and rpL32-(trnL) dataset;(C) the nDNA + cpDNA dataset. Sinalliaria limprichtiana, Eutrema heterophylhum, and Pegaeophyton scapiflorum were selected as outgroups, for the sister group of Hilliella is not clear. Values above braches are maximum parsimony/maximum likelihood bootstrap (only show if > 50%), and values below braches are Bayesian posterior probabilities. Three clades (A-C) are given on the right. |
Our analyses indicate that Yinshanieae is not a monophyletic tribe. Both ITS and (trnL-F phylogenetic trees show the species within Yinshanieae split into two distantly related clades (Figs. 3 and 4): Yinshania clade and Hilliella clade. The Yinshania clade (ITS, 100/100/1; (trnL-F, 99/100/1) fell into Lineage I (Beilstein et al., 2006) and as a sister group of tribes Descurainieae and Smelowskieae, whereas the Hilliella clade was separated from Yinshanieae and formed a moderately to strongly supported clade (ITS, 83/82/0.98; (trnL-F, 72/75/1) embedded in the Expanded Lineage II recognized by Franzke et al.(2011).
Koch and Al-Shehbaz (2000)previously reported that the YinshaniaeHilliella clade was weakly supported (<30% in ITS, <50% in (trnL-intron) due to the incongruent position of Y. qianningensis.In the ITS phylogeny the species fell in the Yinshania clade, while in the (trnL-intron phylogeny it fell in the Hilliella clade. The species was treated as a synonym of Y. acutangula ssp. wilsonii by Al-Shehbaz et al.(1998), whereas Hilliella was merged into Yinshania. However, the incongruencies in Koch and Al-Shehbaz (2000)were caused by a different treatment to the gaps in (trnLintron data. When gaps were considered as additional unweighted binary characters, Y. qianningensiswas placed in the Hilliella clade, but when the gaps were considered as missing data, Y. qianningensiswas nested with Yinshania and consistent with nrDNA phylogeny (Zhang, 2003). Morphologically, taxa of these two clades can be easily distinguished by a series of characters shown in Table 3: species of Hilliella have eseptate fruits and tuberculate seeds, while those of Yinshania have septate fruits and reticulate seeds. Furthermore, the leaves of Hilliella are compound with craspedodromous venation, whereas those of Yinshania are predominantly pinnatipartite to pinnatisect and with half craspedodromous venation. Finally, the trichomes of Hilliella are absent or simple, whereas those of Yinshania are simple, forked, and bifurcate (Zhang, 2003; Zhou and Wei, 2001). In addition, cytological data has shown that species of Hilliella are polyploid whereas Yinshania are diploid (Tian, 1990; Zhang, 1995, 1996; Zhang and Ma, 2001).
| Characters | Hilliella | Yinshania |
| Septum | Absent | Complete or fenestrate |
| Seed | Tuberculate | Reticulate |
| Leaf | Compound, with 3 or 3-5 (-9) leaflets sometimes simple in H. sinuata | Predominantly pinnatipartite to pinnatisect |
| Trichomes | Absent or simple | Simple, forked, and bifurcate |
| Venation | Craspedodromous | Half craspedodromous |
| Chromsome | 2n = 42(44) (based on 7 spp.) | 2n = 12(14) (based on 4 spp.) |
| Habitat | Shady moist places | Sunny and dry places |
| Distribution | S and E China, N Vietnam | SW to N China |
Therefore, on the bases of previous morphological and cytological research by Al-Shehbaz et al.(1998)and Zhang (2003), as well as on our present molecular results, Yinshania and Hilliella should be retained as two genera, with the former retained in tribe Yinshanieae, and Hilliella excluded from it.
4.2 Phylogenetic relationships within the redefined genus YinshaniaYinshania was originally established by Ma and Chao (1979)and was placed in tribe Sisymbrieae by An (1987). Our molecular analyses suggest that the redefined Yinshania is a monophyletic genus close to Descurainieae and Smelowskieae, which is congruent with previous studies (German et al., 2009; Warwick et al., 2010). The redefined genus is endemic to SW to N China, and its species grow at relatively high altitudes (800-3300 m). The accepted species number has varied from four to eight depending on differences in species delimitation (Al-Shehbaz et al., 1998; Zhang, 2003).
Although two nuclear and four chloroplast sequences were combined for phylogenetic analyses, the relationships within this genus remained unresolved. The systematic position of Y. acutangula ssp. wilsonii was inconsistent between nDNA- and cpDNA-derived phylogenies (as an early branching lineage in nDNA phylogeny vs. forming a lineage with Y. acutangula ssp. acutangula in cpDNA phylogeny)(Fig. 5A and B). When nDNA and cpDNA was combined (Fig. 5C), the topology of tree was mostly congruent with cpDNA results. The major difference between the above species is fruit morphology (oblong to oblong-linear in Y. acutangula ssp. acutangula vs. globose in Y. acutangula ssp. wilsonii). Y. exiensis Y.H.Zhang (Zhang, 1993), which was treated as a synonym of Y. zayuensisby Al-Shehbaz et al.(1998), formed an independent clade within Yinshania (Fig. 5). The two species are similar in all other characters except for differences in infructescence rachis (flexuous in Y. exiensis vs. straight in Y. zayuensis)and leaf-surface trichomes (flat and bifurcate trichomes on abaxially and simple trichomes on adaxially in Y. exiensis vs. forked and simple trichomes on both surfaces in Y. zayuensis)(Zhang, 2003). Based on our molecular analyses, Y. exiensis should be maintained as an independent species. Although Y. henryiand Y. zayuensisare nested together (Fig. 5), the two species show many differences in morphology. The lobes of Y. henryiare ovate to suborbicular, while those of Y. zayuensisare oblong to linear. Furthermore, Y. henryiis pubescent with straight simple trichomes, while Y. zayuensisis pubescent with forked trichomes. The lack of resolution within a given genus also occurs in other genera in Brassicaceae, such as Cardamine L.(Carlsen et al., 2009) and Draba L.(Jordon-Thaden et al., 2010). This is often interpreted as the outcome of an early rapid radiation in the family (Bailey et al., 2006; Carlsen et al., 2009; Franzke et al., 2009).
4.3 Systematic position, infrageneric relationships of the reinstated genus HilliellaThe species of Hilliella were originally placed in genus Cochlearia as Sect. Hilliella (Schulz, 1923), but the section was excluded from Cochlearia by Pobedimova (1970, 1971) and was raised to generic rank by Zhang (1986). All species of Hilliella are endemic to S to E China (Fig. 2), though H. paradoxa spreads further into North Vietnam (Zhou et al., 2001). Our molecular studies on Hilliella suggest that it forms a moderately to strongly supported lineage (Figs. 3 and 4) distinct from the other tribes and is embedded in the Expanded Lineage II described by Franzke et al.(2011). The sister group of Hilliella is not yet clear because of the unresolved backbone relationships of Brassicaceae resulting from early rapid diversification (Bailey et al., 2006; Franzke et al., 2009; Warwick et al., 2007) associated with polyploidization events (Lysak et al., 2005; Mandáková et al., 2010; Mandáková and Lysak, 2008). However, many recent phylogenetic studies utilizing transcriptome data (e.g., Huang et al., 2016) show substantial promise, though they have yet to include family-wide tribal representation.
Monophyly of the reinstated Hilliella is supported by our analyses (Figs. 3 and 4), but its sister group was not resolved when we used two nuclear and four chloroplast markers and S. limprichtiana, P. scapiflorum, and E. heterophylhum as outgroups. Within Hilliella, three clades (Fig. 6, A-C) were resolved. H. fumarioides forms an independent early branching lineage (Clade A) and is sister to the remaining species of the genus. This species is distributed in E China (Zhejiang and N Fujian) and is clearly distinguished within the genus by erect stems, small leaf blade (<2 cm), and plump suborbicular fruit with long inflated papillae on the valves. The species was the basis for the establishment of monotypic genus Cochleariella (Zhang, 1985; Zhang and Cai, 1989). The B Clade includes H. yixianensis, H. lichuanensis , and H. paradoxa , and the first species, which is only found in Yixian in C China, is sister to the widespread latter two. The C Clade includes H. hui , H. hunanensis , H. rupicola , H. rivulorum , and H. sinuata . The systematic position of H. hui showed a conflict between nDNA- and cpDNA-derived phylogenies (Fig. 6A and B). Morphologically, it resembles H. hunanensis in having thick rhizomes, stems branched from base, and compressed elliptic to suborbicular fruits, and it resembles H. sinuata in having decumbent stems and simple leaves. H. hui may have originated by hybridization between H. hunanensis and H. sinuata, and further studies are needed to fully elucidate this possibility. The holotype of H. hui at Berlin was most likely destroyed in World War II (Zhang, 2003), and the species was originally described as an annual herb (Schulz, 1923) and later followed by Zhang (1986), Kuan (1987), and Al-Shehbaz et al.(1998). However, during a recent field investigation, we found that H. hui is a perennial species with thick rhizomes up to 3 mm in diam (Fig. 1G).
4.4 Taxonomic treatmentBased on the above molecular phylogenetic analyses, in addition to morphological, and karyological evidence, we place Hilliella in the new tribe Hilliellieae.
Hillielleae H.L.Chen, T.Deng, J.P.Yue, Al-Shehbaz & H.Sun, trib. nov. Type genus: Hilliella (O.E.Schulz) Y.H.Zhang & H.W.Li.
Herbs annual, biennial, or perennial; trichomes simple or absent; stems erect or decumbent; basal leaves simple, trifoliolate, or pinnately compound; cauline leaves compound or rarely simple; racemes few to many flowered; petals obovate or spatulate; fruits oblong, elliptic, ovoid, or suborbicular; replum rounded; septum absent; stigma entire; seeds ovate, slightly flattened, tuberculate; cotyledons incumbent or rarely accumbent.
Distribution and habitat. - China (Anhui, Chongqing, Fujian, Guangdong, Guangxi, Hunan, Jiangxi, Taiwan, Zhejiang), North Vietnam. Streamsides, roadsides, wet shady slopes, rock cliffs; 100-1700 m.
5 ConclusionsThe previously recognized tribe Yinshanieae is not monophyletic and is divided herein into two remotely related unigeneric tribes: Hillielleae and Yinshanieae s.str. The sister group of Hillielleae is not clear. Within Hilliella, there are three clades (A-C), but species relationships within Yinshanieae s.str. remain unresolved. To clarify the infratribal relationships of the two tribes, additional molecular markers and extensive taxon sampling of critical species are needed.
Acknowledgements
We thank the National Natural Science Foundation of China for the grant NSFC-31170181 to J.P. Yue, and the Major Program of National Natural Science Foundation of China (31590823 to Hang Sun). We are also grateful to KUN and PE for providing specimens materials, to D.G. Zhang for collecting samples, and to Dr. Yang Niu for assistance on creating graphics.
Appendix A. Taxa and GenBank accession numbers for the ITS and (trnL-F sequences downloaded from GenBank and used in the phylogenetic analyses (ITS, (trnL-F).
CLEOMACEAE. Cleome lutea (AF137588, -); Cleome viridiflora (-, AY122441); BRASSICACEAE. Aethionema arabicum (AY254539, DQ180218); Aethionema saxatile (GQ284853, AY122451); Alyssopsis mollis (-, FJ188227); Alyssopsis trinervis (GQ497846, -); Alyssum sibiricum (GQ284890, -); Anastatica hierochuntica (GQ424524, -); Anchonium billardierei (DQ357512, -); Aphragmus oxycarpus (DQ165337, DQ518350); Aphragmus nepalensis (DQ165335, -); Arabis alpina (DQ060111, EF449513); Asta stricta (HQ541172, -); Asta schaffneri (HQ541168, -); Barbarea vulgaris (AJ232915, -); Biscutella auriculata (DQ452057, -); Biscutella laevigata (DQ452056, -); Bivonaea lutea (HQ327490, JF826129); Boechera holboellii (-, DQ013055); Boechera retrofracta (GQ166472, -); Brassica oleracea (AY722423, -); Brassica rapa (-, AY752717); Brayopsis calycina (KM376249, KM376287); Bunias erucago (GQ497885, -); Bunias orientalis (-, FN677645); Calepina irregularis (DQ249822, AY751760); Calymmatium draboides (FM958512, -); Camelina alyssum (KC172842, -); Camelina microcarpa (KC172843, DQ821412); Carinavalva glauca (GQ424527, -); Catenulina hedysaroides (GQ424607, -); Chorispora bungeana (-, FN677730); Chorispora tenella (DQ357526, -); Cithareloma lehmannii (DQ357528, -); Clausia aprica (DQ357529, -); Clypeola jonthlaspi (EF514644, -); Cochlearia officinalis (HQ268642, HQ268697); Cochlearia pyrenaica (-, HQ268698); Coluteocarpus vesicaria (GQ497857, -); Conringia perfoliata (AY722505, -); Conringia planisiliqua (-, AY751762); Crambe filiformis (AY722435, -); Cremolobus peruvianus (-, KF662808); Crucihimalaya lasiocarpa (AF137556, -); Crucihimalaya rupicola (-, FN677737); Crucihimalaya wallichii (-, DQ310520); Cusickiella douglasii (-, AF307557); Cyphocardamum aretioides (GQ497859, -); Didymophysa fedtschenkoana (EF514648, -); Diplotaxis tenuifolia (-, EU310491); Diptychocarpus strictus (-, FN677717); Dontostemon glandulosus (FN821612, -); Draba araboides (AF146505, -); Draba incana (-, DQ467003); Eremoblastus caspicus (-, FN677643); Eruca sativa (-, AY751765); Erysimum canescens (-, EU170623); Erysimum cheiranthoides (-, EU170622); Erysimum cyaneum (KJ417998, -); Erysimum czernjajevi (KJ417999, -); Eudema rupestris (KM376254, -); Euclidium syriacum (KJ623477, EF426780); Galitzkya potaninii (-, FN677635); Goldbachia laevigata (DQ357546, -); Halimolobos diffusa (AF307645, -); Halimolobos parryii (-, AF307539); Heliophila coronopifolia (DQ249846, -); Heliophila variabilis (HE806278 and HE806279, -); Hesperis matronalis (DQ357547, AY546166); Hesperis sibirica (-, EU170624); Hormathophylla purpurea (-, FN677738); Hornungia petraea (KF022705, -); Hornungia alpina (-, DQ310515); Iberis amara (AJ440311, -); Iberis oppositifolia (-, AY122456); Iberis spathulata (AJ440312, -); Ionopsidium abulense (HQ268661, HQ268716); Isatis minima (-, DQ821409); Isatis tinctoria (GQ131323, DQ479874 and DQ518370); Kernera saxatilis subsp, saxatilis (AJ440313, -); Lepidium apetalum (JF976768, DQ821406); Lepidium sisymbrioides (-, DQ997068); Litwinowia tenuissima (-, FN677714); Macropodium nivale (-, FN677638); Macropodium pterospermum (GU182055, -); Mancoa pubens (-, AF307546); Mathewsia foliosa (KC174388, -); Mathewsia peruviana (-, EU620362); Menonville flexuosa (KF662771, KF662776); Menonvillea pinnatifida (KF662738, KF662815); Microlepidium pilosulum (GQ497869, -); Microstigma deflexum (-, FN677641); Mostacillastrum stenophyllum (EU620305, EU620364); Murbeckiella huetii (GQ424546, -); Neslia paniculata (-, DQ310518); Noccaea bulbosa (-, AY154798); Noccaea fendleri (AY154824, -); Noccaea jankae (-, AY154796); Notothlaspi australe (AF100689, -); Notothlaspi rosulatum (AF100690, -); Olimarabidopsis pumila (-, DQ310519); Onuris hauthalii (-, KM376275); Oreophyton falcatum (GQ424549, -); Pachycladon exilis (-, EF015658); Pachycladon latisiliqua (-, EF015656); Parrya nudicaulis (DQ249842, -); Paysonia stonensis (AF137585, -); Pennellia longifolia (-, AF307549); Pennellia micrantha (AF307629, -); Physaria pruinosa (AF137584, -); Polypsecadium solidagineum (-, EU620373); Pugionium cornutum (JF978166, -); Pugionium dolabratum (JF978171, -); Rhammatophyllum kamelinii (-, FN677742); Rhizobotrya alpina (AJ440315, -); Rorippa palustris (-, EF426789); Sandbergiawhitedii (DQ399119, -); Schimpera arabica (GQ424556, -); Schizopetalon brachycarpum (KC174406, -); Schizopetalon walkeri (-, EU620378); Sisymbrium officinale (AB856333, -); Sisymbrium orientale (AB856332, -); Sisymbrium strictissimum (-, AY958566); Sisymbrium volgense (-, AY958568); Smelowskia porsildii (EU489556, -); Smelowskia sisymbrioides (-, JF298539); Sterigmostemum ramosissimum (DQ357596, -); Stevenia axillaris (-, FN677639); Stevenia canescens (KF022716, -); Strigosella africana (-, DQ4798770; Tchihatchewia isatidea (GQ497882, -); Thelypodium flexuosum (KF730217, -); Thlaspi arvense (KJ623518, -); Transberingia bursifolia (DQ399110, -); Turritis glabra (DQ249853, DQ649082); Turritis laxa (KF547126, -); Xerodraba patagonica (-, KM376264); Zuvanda crenulata (DQ357606, -).
| Al-Shehbaz I.A, 2012. A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon, 61, 931 -954. | ||
| Al-Shehbaz I.A, Yang G, Lu L.L, et al., 1998. Delimitation of the Chinese genera Yinshania, Hilliella, and Cochleariella (Brassicaceae). Harv. Pap. Bot., 3, 79 -94. | ||
| Al-Shehbaz I.A, Beilstein M.A, Kellogg E.A, 2006. Systematics and phylogeny of the Brassicaceae (cruciferae): an overview. Plant Syst. Evol., 259, 89 -120. DOI:10.1007/s00606-006-0415-z | ||
| Al-Shehbaz I.A, German D.A, Mummenhoff K, et al., 2014. Systematics, tribal placements, and synopses of the Malcolmia s. l. segregates (Brassicaceae). Harv. Pap. Bot., 19, 53 -71. DOI:10.3100/hpib.v19iss1.2014.n4 | ||
| An, Z.X., 1987. Sisymbrieae. In: Cheo, T.Y. (Ed.), Cruciferae. Fl. Reipubl. Popularis Sin. 33Science Press, Beijing, pp. 396-453. | ||
| Bailey C.D, Koch M.A, Mayer M, et al., 2006. Toward a global phylogeny of the Brassicaceae. Mol. Biol. Evol., 23, 2142 -2160. DOI:10.1093/molbev/msl087 | ||
| Baldwin B.G, Markos S, 1998. Phylogenetic utility of the external transcribed spacer (ETS) of 18S-26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae). Mol. Phylogen. Evol., 10, 449 -463. DOI:10.1006/mpev.1998.0545 | ||
| Beilstein M.A, Al-Shehbaz I.A, Kellogg E.A, 2006. Brassicaceae phylogeny and trichome evolution. Amer. J. Bot., 93, 607 -619. DOI:10.3732/ajb.93.4.607 | ||
| Beilstein M.A, Al-Shehbaz I.A, Mathews S, et al., 2008. Brassicaceae phylogeny infrerred from phytochrome A and ndhF sequence data: tribes and trichomes revisited. Amer. J. Bot., 95, 1307 -1327. DOI:10.3732/ajb.0800065 | ||
| Carlsen T, Bleeker W, Hurka H, et al., 2009. Biogeography and phylogeny of Cardamine (Brassicaceae). Ann. Mo. Bot. Gard, 215 -236. | ||
| Couvreur T.L, Franzke A, Al-Shehbaz I.A, et al., 2010. Molecular phylogenetics, temporal diversification and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol., 27(1), 55 -71. DOI:10.1093/molbev/msp202 | ||
| Darriba D, Taboada G.L, Doallo R, et al., 2012. jModelTest 2: more models. new heuristics and parallel computing. Nat. Methods, 9, 772 -772. | ||
| Farris J.S, Kallersjo M, Kluge A.G, et al., 1994. Testing significance of incongru-ence. Cladistics, 10, 315 -319. DOI:10.1111/cla.1994.10.issue-3 | ||
| Felsenstein J, 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39, 783 -791. DOI:10.2307/2408678 | ||
| Franzke A, German D, Al-Shehbaz I.A, et al., 2009. Arabidopsis family ties: mo-lecular phylogeny and age estimates in Brassicaceae. Taxon, 58, 425 -437. | ||
| Franzke A, Lysak M.A, Al-Shehbaz I.A, et al., 2011. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends. Plant. Sci., 16, 108 -116. | ||
| German D, Friesen N, 2014. Shehbazia (Shehbazieae, Cruciferae), a new monotypic genus and tribe of hybrid origin from Tibet. Turczaninowia, 17(4), 17 -23. DOI:10.14258/turczaninowia.17.4.3 | ||
| German D.A, Friesen N, Neuffer B, et al., 2009. Contribution to ITS phylogeny of the Brassicaceae, with special reference to some Asian taxa. Pl. Syst. Evol., 283, 33 -56. DOI:10.1007/s00606-009-0213-5 | ||
| Hall, T.A., 1998. BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic Acids Symposium Series. | ||
| Huang C.H, Sun R.R, Hu Y, et al., 2016. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol., 33(2), 394 -412. DOI:10.1093/molbev/msv226 | ||
| Jordon-Thaden I, Hase I, Al-Shehbaz I.A, et al., 2010. Molecular phylogeny and systematics of the genus Draba (Brassicaceae) and identification of its most closely related genera. Mol. Phylogen. Evol., 55, 524 -540. DOI:10.1016/j.ympev.2010.02.012 | ||
| Kelchner S.A, Thomas M.A, 2007. Model use in phylogenetics: nine key questions. Trends. Ecol. Evol., 22, 87 -94. DOI:10.1016/j.tree.2006.10.004 | ||
| Koch M, Al-Shehbaz I.A, 2000. Molecular systematics of the Chinese Yinshania (Brassicaceae): evidence from plastid and nuclear ITS DNA sequence data. Ann. Mo. Bot. Gard., 87, 246 -272. DOI:10.2307/2666163 | ||
| Koch M.A, Dobes C, Kiefer C, et al., 2007. Supernetwork identifies multiple events of plastid trnF (GAA) pseudogene evolution in the Brassicaceae. Mol. Biol. Evol., 24, 63 -73. | ||
| Kuan, K.C., 1987. Lepidieae. In: Cheo, T.Y. (Ed.), Cruciferae. Fl. Reipubl. Popularis Si, vol. 33. Science Press, Beijing, pp. 44-109. | ||
| Lysak M.A, Koch M.A, Pecinka A, et al., 2005. Chromosome triplication found across the tribe Brassiceae. Genome Res., 15, 516 -525. DOI:10.1101/gr.3531105 | ||
| Ma Y.C, Chao I.T, 1979. Yinshania, a new genus of Chinese cruciferae. Acta Phy- totax.Sin., 17, 113 -114. | ||
| Mandáková T, Joly S, Krzywinski M, et al., 2010. Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell., 22, 2277 -2290. DOI:10.1105/tpc.110.074526 | ||
| Mandáková T, Lysak M.A, 2008. Chromosomal phylogeny and karyotype evolution in x= 7 crucifer species (Brassicaceae). Plant Cell., 20, 2559 -2570. DOI:10.1105/tpc.108.062166 | ||
| Mayrose I, Friedman N, Pupko T, 2005. A Gamma mixture model better accounts for among site rate heterogeneity. Bioinformatics, 21, ii151 -ii158. | ||
| Miller, M., Pfeiffer, W., Schwartz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Gateway Computing Environments Workshop (GCE), 2010. IEEE, pp. 1-8. | ||
| Mummenhoff K, Franzke A, Koch M, 1997. Molecular phylogenetics of Thlaspi sl (Brassicaceae) based on chloroplast DNA restriction site variation and se-quences of the internal transcribed spacers of nuclear ribosomal DNA. Canad. J. Bot., 75, 469 -482. DOI:10.1139/b97-051 | ||
| O′Kane S.L, Al-Shehbaz I.A, 2003. Phylogenetic position and generic limits of Arabidopsis (Brassicaceae) based on sequences of nuclear ribosomal DNA. Ann. Mo. Bot. Gard., 90, 603 -612. DOI:10.2307/3298545 | ||
| Pobedimova E, 1970. Revisio generis Cochlearia L.. 1. Nov. Sist. Vyssh. Rast, 6, 67 -106. | ||
| Pobedimova E, 1971. Revisio generis Cochlearia L.. 2. Nov. Sist. Vyssh. Rast, 7, 167 -195. | ||
| Ren F, Tanaka H, Yang Z.H, 2005. An empirical examination of the utility of codon-substitution models in phylogeny reconstruction. Syst. Biol., 54, 808 -818. DOI:10.1080/10635150500354688 | ||
| Ronquist F, Teslenko M, van der Mark, P, et al., 2012. MrBayes 3. 2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol., 61, 539 -542. | ||
| Schulz O.E, 1923. Eine neue Sektion der Gattung Cochlearia L. Not. Bot. Gart. Berlin- Dahlem, 8, 544 -546. | ||
| Shaw J, Lickey E.B, Schilling E.E, et al., 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Amer. J. Bot., 94, 275 -288. DOI:10.3732/ajb.94.3.275 | ||
| Shaw J, Small R.L, 2005. The tortoise and the hare II: relative utility of 21 non-coding chloroplast DNA sequences for phylogenetic analysis. Amer. J. Bot., 92, 142 -166. DOI:10.3732/ajb.92.1.142 | ||
| Stamatakis A, 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22, 2688 -2690. DOI:10.1093/bioinformatics/btl446 | ||
| Stamatakis A, 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312 -1313. DOI:10.1093/bioinformatics/btu033 | ||
| Swofford D.L, 2003. PAUP*, Phylogenetic Analyses Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA, USA . | ||
| Taberlet P, Gielly L, Pautou G, et al., 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Pl. Mol. Biol., 17, 1105 -1109. DOI:10.1007/BF00037152 | ||
| Tao S, Crawford D.J, Stuessy T.F, et al., 1997. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Amer. J. Bot., 84, 1120 -1136. DOI:10.2307/2446155 | ||
| Tian Y, 1990. A Taxonomic Study on the Genus Yinshania and its Affinities (Cru- ciferae). Biology Department of Inner Mongolia University. Inner Mongolia University, pp.7 -8. | ||
| Warwick S.I, Al-Shehbaz I.A, Sauder C.A, 2006. Phylogenetic position of Arabis arenicola and generic limits of Aphragmus and Eutrema (Brassicaceae) based on sequences of nuclear ribosomal DNA. Botany, 84, 269 -281. | ||
| Warwick S.I, Mummenhoff K, Sauder C.A, et al., 2010. Closing the gaps: phylo-genetic relationships in the Brassicaceae based on DNA sequence data of nu-clear ribosomal ITS region. Pl. Syst. Evol., 285, 209 -232. DOI:10.1007/s00606-010-0271-8 | ||
| Warwick S.I, Sauder C.A, Al-Shehbaz I.A, et al., 2007. Phylogenetic relationships in the tribes Anchonieae, Chorisporeae, Euclidieae, and Hesperideae (Brassi- caceae) based on nuclear ribosomal ITS DNA sequences. Ann. Mo. Bot. Gard., 94, 56 -78. DOI:10.3417/0026-6493(2007)94[56:PRITTA]2.0.CO;2 | ||
| Weeks A, Daly D.C, Simpson B.B, 2005. The phylogenetic history and biogeog-raphy of the frankincense and myrrh family (Burseraceae) based on nuclear and chloroplast sequence data. Mol. Phylogen. Evol., 35, 85 -101. DOI:10.1016/j.ympev.2004.12.021 | ||
| White T.J, Bruns T, Lee S.J.W.T, et al., 1990. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. PCR Protocols: a Guide to Methods and Applications. Academic Press, NY, pp. 315 -322. | ||
| Yang Z.H, Rannala B, 1997. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol. Biol. Evol., 14, 717 -724. DOI:10.1093/oxfordjournals.molbev.a025811 | ||
| Yue J.P, Sun H, Li J.H, et al., 2008. A synopsis of an expanded Solms-laubachia (Brassicaceae), and the description of four new species from western China 1. Ann. Mo. Bot. Gard., 95, 520 -538. DOI:10.3417/2006214 | ||
| Zhang Y.H, 1985. Cochleariopsis - a new genus of Chinese Cruciferae. Acta Bot. Yunnan, 2, 143 -145. | ||
| Zhang Y.H, 1986. Hilliella, a new genus of Cruciferae. Acta Bot. Yunnan, 4, 397 -406. | ||
| Zhang Y.H, 1993. A new species of Yinshania with a discussion on the evolution and origin of the genus. Acta Bot. Yunnan, 4, 364 -368. | ||
| Zhang Y.H, 1995. A comparison of chromosome numbers and peroxidase zymo-grams of Yinshania and Hilliella. J. Pl. Resour. Environm, 4(2), 27 -31. | ||
| Zhang Y.H, 1996. A study on the genus Yinshania. Bull. Bot. Res., 16, 445 -454. | ||
| Zhang Y.H, 2003. Delimitation and revision ofHilliella and Yinshania (Brassicaceae). Acta Phytotax. Sin., 41, 305 -349. | ||
| Zhang Y.H, Cai J.J, 1989. Observation on the genera Yinshania, Hilliella, Cochleariella and Cochlearia (Cruciferae) by SEM. Acta Bot. Boreal.-Occid. Sin., 9, 224 -231. | ||
| Zhang Y.H, Ma G.C, 2001. The chromosome numbers of two species in Brassica- ceae. J. Wuhan. Bot. Res., 4, 345 -346. | ||
| Zhou G.L, Wei Y, 2001. Comparative observation on venation about four genera in Cruciferae. J. Wuhan. Bot. Res., 20, 258 -262. | ||
| Zhou, T.Y., Lu, L.L., Yang, G., et al., 2001. Brassicaceae. In: Wu, C.Y., Raven, P.H. (Eds.), Flora of China (8), p. 58. Science Press (Beijing) and Missouri Botanical Garden (St. Louis). |



