b. Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, China
Paris polyphylla Smith var. yunnanensis, a member of the Liliaceae family, is a commercially important species due to its exceptional medicinal properties. Unfortunately, slow growth and over-harvesting for the past several years have led to a significant decline in population and now the species faces the risk of extinction (Zhang et al., 2011). The rhizome of this plant is an important ingredient of certain Chinese patented medicines, such as Biyan Qingdu Keli, which is widely used in southern China to treat chronic rhinitis and nasopharyngeal cancer (Guo et al., 2006; Han et al., 2009). Recent pharmacological studies have also demonstrated that the plant has hemostatic, antitumor, uterine contractile, analgesic, and sedative effects (Huang et al., 2007; Guo et al., 2008). The main active ingredients of the plant are steroidal saponins (Wu et al., 2004; Zhang, 2007), at least 30 of which have been isolated through phytochemical methods (Liu et al., 2006; Xu et al., 2007; Zhao et al., 2007, 2009). These saponins have various effects, including cytotoxicity (Zhao et al., 2009). Furthermore, they are potential anti-cancer agents that destroy multidrug-resistant hepatocarcinoma cells (Lee et al., 2009). They have also shown protective effects on ethanol-or indomethacin-induced gastric mucosal lesions in rats (Matsuda et al., 2003), platelet agonist activity (Fu et al., 2008), and contractile agonist activity in the uterus (Guo et al., 2008).
Steroidal saponins are synthesized via the mevalonic acid (MVA) pathway in the cytoplasm (Haralampidis et al., 2002), or through the non-mevalonate pathway (MEP) in plastids (Rohdich et al., 2001; Rohmer, 2003). Cyclization of 2, 3-oxidosqualene leads to the formation of various saponins, catalyzed by oxidosqualene cyclase, combined with modifications on steroid skeletons such as hydroxylations. According to the proposed pathway (Kumar et al., 2012), some specific CYP450s and UDP-glycosyltransferases (UGTs) may catalyze the conversion of cycloartenol to various steroidal saponins. To date, several oxidosqualene cyclase genes have been cloned from various plant systems (Corey et al., 1993; Herrera et al., 1998). However, little is known about the molecular mechanisms of the biosynthetic pathway downstream of cyclization. Despite the pharmacological importance of P. polyphylla var. yunnanensis, the very limited information on its transcriptome and genome greatly hinders investigations of the mechanism of steroidal saponin biosynthesis.
Usually, older roots contain more saponins and therefore have more commercial value than younger roots. Thus, determining the biosynthetic pathway for saponins and the underlying mechanism of gene regulation of P. polyphylla var. yunnanensis could be of great significance. Expressed sequence tag (EST) analysis is a useful tool for revealing information about genomes, especially in non-model plants for which no reference genome sequences are available (Margulies et al., 2005). In addition, next-generation sequencing is a very useful technique for providing large amounts of expression data to expedite the understanding of metabolic pathways and identify genes (Morozova et al., 2009; Shendure and Ji, 2008). Although huge amounts of parallel-sequence short reads are yielded by Illumina high-throughput sequencing technology, many de novo assembly tools have been developed to analyze the short read sequences (Wang et al., 2010a, b), facilitating the analysis of these short read sequences in the absence of any reference sequences. RNA-seq transcriptome analysis has become an attractive alternative for in-depth analysis at high resolution. Compared with 454 pyrosequencing, Illumina sequencing has been shown to yield more accurate contigs despite the shorter read-lengths (Luo et al., 2012), owing to substantially more extensive sequence coverage.
In this paper, we characterized the transcriptomes of P. polyphylla var. yunnanensis roots of different ages, using the de novo Illumina sequencing platform. In order to determine the candidate genes that encode enzymes involved in the saponin biosynthetic pathway, we focused on the transcripts involved in saponin biosynthesis. To our knowledge, this study is the first transcriptome resource for the endangered species P. polyphylla var. yunnanensis' roots. Our data and findings will contribute to future studies on the functional genomics and biogeography of this species.
2 Materials and methods 2.1 Plant material8-year-old and 4-year-old roots of P. polyphylla var. yunnanensis plants cultivated on farms are routinely harvested for medical purposes. The plants used in this study were collected from the fields of Kunming City, Yunnan Province, China. They were cleaned, cut into small pieces, immediately frozen in liquid nitrogen, and stored at -80 ℃ until further processing.
2.2 RNA extraction, library preparation, and sequencingFor each year, the RNA was extracted from a mixture of several old plants. Total RNA was extracted from roots using the RNeasy Plant Mini Kit (Qiagen), according to the manufacturer's instructions. The RNA quality was tested using a 1% ethidium bromide-stained (EtBr-stained) agarose gel, and the concentration was assessed using a GeneQuant100 spectrophotometer (GE Healthcare, UK) before processing. The RNA samples were treated with DNase I (TURBO DNase; Ambion, USA) at a concentration of 1.5 units/l g of total RNA prior to cDNA synthesis. The transcriptome library for sequencing was generated using the Illumina TruSeqTM RNA Sample Preparation Kit (Illumina, San Diego, USA) following the manufacturer's recommendations. The clustering of the indexcoded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer's instructions. After cluster generation was complete, the library preparations were sequenced on an Illumina Hiseq 2000 platform and 100 bp paired-end reads were generated.
2.3 Data processing, assembly, and annotationThe sequencing-derived raw images were transformed by base calling into raw reads using CASAVA (http://www.illumina.com/support/documentation.ilmn). The raw reads were cleaned by removing reads containing adapter and poly-N, and low-quality reads. In addition, the Q20, Q30, GC-content, and sequence duplication level of the clean data were calculated for high-quality downstream analysis. The reads with quality Q-value over 20 and sequence length longer than 100 bp were assembled with Trinity software (Grabherr et al., 2011). De novo assembly followed Li et al. (2013). The longest transcript of each subcomponent was defined as the “unigene” for functional annotation. In order to identify the putative mRNA functions, all the assembled unigenes were searched against the nr database using the BLASTalgorithm (Bedell et al., 2003) with an E-value cut-off of 10-5. In addition, GO terms were extracted from the best hits obtained from the BLASTx against the nr database using the Blast2GO program (GÖtz et al., 2008). The € BLAST algorithm was also used to align unique sequences to the nr and SWISS-PROT databases, with E-value cut-off of 10-5, to predict possible functional classifications and molecular pathways.
2.4 Gene expression pattern analysisRSEM was used to estimate gene expression levels by mapping clean reads to the Trinity transcripts for each sample (Li and Dewey, 2011). Then the RPKM method (Mortazavi et al., 2008) was used to normalize and calculate the abundance of all genes using uniquely mapped reads. Differential expression analysis of the two samples was performed by modeling count data with negative binomial distributions described in the DEGseq method (Anders and Huber, 2010; Wang et al., 2010a). The P-value was adjusted using the qvalue (Storey and Tibshirani, 2003), and q-value b 0.005 & fold change|log2| > 1 was set as the threshold for significantly differential expression. The identified DEGs were used for GO and KO enrichment analyses. GO enrichment analyses were performed using GOseq (Young et al., 2010), based on the Wallenius noncentral hypergeometric distribution, to map all DEGs to terms in the GO database (P-value≤0.05), looking for significantly enriched GO terms in DEGs compared with the genome background. KEGG pathway enrichment analysis of the DEGs was done using KOBAS (Mao et al., 2005).
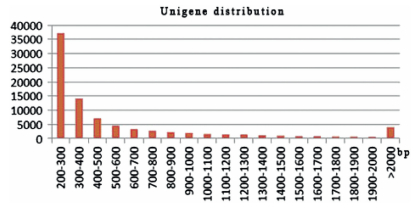
3 Results 3.1 Sequencing and reads assemblyTo obtain an overview of the root transcriptome, two sequencing libraries were prepared from the two tissues and sequenced with the Illumina paired-end technique (Accession No: SRS1413148). We generated 13, 040, 000 raw reads from a 4-year-old root and 13, 309, 941 from an 8-year-old root (Table 1). Of the raw reads, 98.37% bases from the 4-year-old root and 98.50% of those from the 8-year-old root had a Q value≥30. The percentages of GC pairs were 50.14% and 49.02% for the 4-year-old and 8-yearold root, respectively. These were used for de novo assembly. Trinity software generated 87, 577 all-unigenes (Table 2) with an average length of 614 bp and an N50 of 972 bp. Of these, 37, 170 (42.44%) contained 200-300 bp, 14, 008 (16.00%) contained 300-400 bp, 4027 (4.50%) were >2 000b, and the remaining 4462 (37.06%) contained 400-2000 bp (Fig. 1).
| Sample | Raw reads | Bases | Q20 | Q30 | Avg. Quality |
| R1 (4 years root) | 13040000 | 1.09(GB) | 1 | 0.9837 | 37.5 |
| R2 (4 years root) | 13040000 | 1.06(GB) | 1 | 0.9789 | 37.38 |
| R1 (8 years root) | 13309941 | 1.12(GB) | 1 | 0.985 | 37.62 |
| R2 (8 years root) | 13309941 | 1.09(GB) | 1 | 0.979 | 37.45 |
| Assembly | Statistics |
| Number of unigene | 87577 |
| Large unigene ( > =1000bp) | 14481 |
| Maximum unigene length (bp) | 15211 |
| Mean unigene length (bp) | 614 |
| N50 length (bp) | 972 |
| Total bases (MB) | 53.77 |
|
| Fig. 1 Overview of Paris polyphylla var. yunnanensis transcriptome assembly (Size distribution of the unigenes was produced from further assembly of contigs after clustering.). |
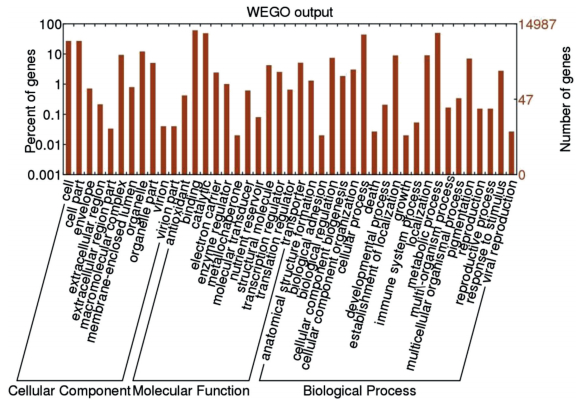
The 87, 577 unigene sequences were first searched against the National Center for Biotechnology Information's (NCBI's) nonredundant database of protein sequences (nr); SWISS-PROT, a manually annotated and reviewed protein sequence database; and the Clusters of Orthologous Groups (COG) database using the BLAST algorithm (E-value cut off of 10-5). As a result, 65, 535 unigenes (65.51%) were annotated. The number of unigenes with significant similarity to sequences in the nr, SWISS-PROT, and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were 30, 507 (34.83%), 16, 396 (33.72%) and 8345 (18.69%), respectively. We used Gene Ontology (GO) to classify the functions of the annotated genes. Using the GO classification, 47, 042 sequences (53.71%) were categorized in three different GO trees (cellular component, molecular function, and biological process). Within cellular components, the unique sequences were further classified in 11 subclassifications, of which the most represented cellular components were cell (22.11%) and cell part (21.91%). Within molecular function, the categories were further classified in 12 subclassifications. The largest subcategory of the molecular function was binding (28.37%), and the second largest was catalytic activity (28.14%). Within biological processes, the unique sequences were further classified in 20 subclassifications, of which the most represented biological processes were metabolic process (26.15%) and cellular process (30.47%) (Fig. 2).
|
| Fig. 2 Gene Ontology (GO) classification of the Paris polyphylla var. yunnanensis transcriptome. GO term assignments to unigenes were based on significant plant species hits against the non-redundant database. Results presented in three main GO categories (cellular component, molecular function, and biological process) and 39 subcategories. |
To further understand the transcriptome data, we carried out pathway analysis with the KEGG database, which contains a systematic analysis of inner-cell metabolic pathways and functions of gene products. Pathway-based analyses help to further determine the biological function of genes. A total of 3934 genes were assigned to five KEGG biochemical pathways: metabolism (3171 unigenes), organism system (750), cellular processes (484), genetic information processing (471), and environmental information processing (150). Metabolism was the largest group and was most associated with amino acid metabolism (905), carbohydrate metabolism (885), nucleotide metabolism (510), energy metabolism (332), lipid metabolism (245), and metabolism of cofactors and vitamins (160). Pathways related to genetic information processing composed the second-largest group and included genes involved in translation (129), folding (147), replication and repair (144), and transcription (75). Organismal systems comprised the third-largest group, with a majority of proteins involved in the nervous (92) and endocrine (132) systems. Pathways related to cellular processes and environmental information processing were also well represented by unigenes. These results provide a valuable resource for investigating metabolic pathways in P. polyphylla.
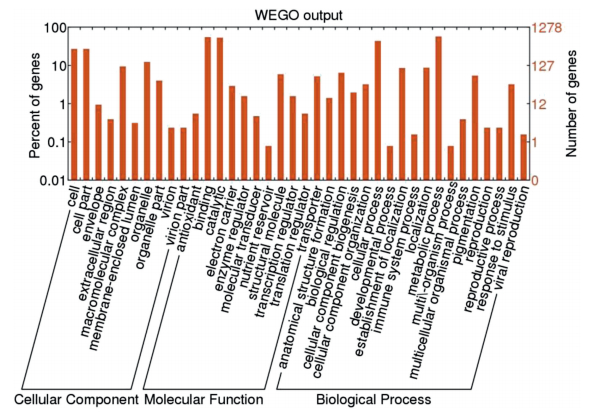
3.3 Differential gene expression between roots of different agesTaking the sequencing depth and gene length on read count into account, we calculated gene expression according to the method of RPKM (reads per kilobase of transcript per million reads mapped). A total of 3082 genes (3.51% of all genes) were significantly differentially expressed genes (DEGs) between the roots of different ages, on the basis of the applied criteria [q-value < 0.001 and log2 (fold change) > 1]. These DEGs included 1518 upregulated genes (accounting for 49.25% of all significant DEGs) and 1564 downregulated genes (accounting for 50.75% of all significant DEGs) in the 8-year-old root. After mapping them to terms in the GO database and comparing them to the whole transcriptome background, we were able to assign 1278 of the DEGs a GO ID and categorize them into 26 functional groups in three main categories: cellular component, molecular function, and biological process (Fig. 3). To further investigate the pathways with which these DEGs were involved, we mapped all DEGs to terms in the KEGG database and compared the results with the whole transcriptome background. We assigned a KO ID to 491 genes out of all the DEGs and categorized them into 243 pathways. Specifically, 11 pathways were significantly enriched (corrected P-value≤0.05), and tyrosine metabolism, chloroalkane and chloroalkene degradation, and metabolism of xenobiotics by cytochrome P450 were the most significantly enriched.
|
| Fig. 3 Gene Ontology (GO) classification of the transcriptome of Paris polyphylla var. yunnanensis roots of different ages. GO term assignments to P. polyphylla var. yunnanensis unigenes were based on significant plant species hits against the non-redundant database. Results are presented in three main GO categories (cellular component, molecular function, and biological process) and 38 sub-categories. |
One goal of this study was to establish deep transcriptome databases for Paris species producing a variety of bioactive compounds. With the development of RNA-seq, transcriptome analysis has become an attractive alternative for in-depth analysis at high resolution. Compared with 454 pyrosequencing, Illumina sequencing has been shown to yield more accurate contigs despite the shorter read-lengths (Luo et al., 2012), owing to substantially more extensive sequence coverage. However, until now, no genomic data were available for P. polyphylla' root. In this study, we carried out de novo transcriptome assembly using short-read (Illumina) sequencing. Despite the shorter reads, our assembly is comparable to other published transcriptomes using 454 pyrosequencing (Barakat et al., 2009; Wall et al., 2009).
As shown in Fig. 3, more than 4.6% of unigenes were greater than 2 kb, and 15.9% of unigenes were greater than 500 bp. These results demonstrated that the assembly effectively captured a large portion of the transcriptome. Another useful metric is the BLAST hit corresponding to each proportion of the unigenes. Due to the lack of genomic resources for Paris, the proportions of unigenes that were significantly similar to known genes in GenBank were considered the “gold standard” reference in our studies. Nearly 65.51% of our unigenes had matches in the nr database, as many as in other de novo assemblies (Sutapa et al., 2011). The large quantity of unique sequences should cover the vast majority of genes from P. polyphylla var. yunnanensis root and, for the first time, provides a powerful gene resource for this medicinal plant. Furthermore, these results indicate that for transcript profiling and gene discovery Illumina sequencing represents a good alternative to 454 pyrosequencing, which is time consuming, labor intensive, and expensive.
4.2 Gene expression differences between roots of different agesThe root tissue is the main site of steroid saponin biosynthesis in P. polyphylla var. yunnanensis; thus, the cDNA library was created from this tissue. We predicted protein functions using the annotation searched in BLASTX against the nr database. Of the 87, 577 sequences, 65% coded for proteins whose functions were known. According to the GO database, 59%, 29%, and 12% of genes could be classified as having molecular, biological, and cellular functions, respectively. Within molecular functions, the most represented category of the unique sequences, genes were associated with binding, catalytic enzymes, structural molecule activity, and transporting. The second most represented category was biological process; these unique sequences were associated with metabolic processes, cellular processes, localization, pigmentation, and response to stimulus. These results aligned with the view that external stimuli can alter plants' growth and development (Jeter and Roux, 2006). Under the cellular compound category, cell and cell part were significantly represented. More ESTs were present in the 8-year-old than in the 4-year-old roots.
The KEGG pathway database contains information on networks of intracellular molecular interactions and their organism-specific variations (Kanehisa et al., 2008). To identify the biological pathways in P. polyphylla var. yunnanensis, we mapped the annotated sequences to the reference by KEGG. The annotation information enabled gene functions and metabolic pathways to be determined. Only 7721 unique sequences (16.41% of the total) were assigned EC numbers and had 243 unique mappings to KEGG biochemical pathways representing six categories and 36 subcategories. Metabolism, the most represented category, contained the subcategories of carbohydrate metabolism, lipid metabolism, energy metabolism, secondary metabolism, amino acid metabolism, glycan metabolism, and vitamin cofactor metabolism. The transcripts for glycolysis/glyconeogenesis are the largest subcategory in carbohydrate metabolism. Glycolysis/glyconeogenesis is known to be very important for plant and non-plant species development, including nutrient limitation and osmotic and non-osmotic stress (Roef et al., 2003). The second subcategory is lipid metabolism. Fatty acids are important for the stabilization of plant cells and responsible for plants' cold stress tolerance and antifungal activity (Kargiotidou et al., 2008).
The second major category is environmental information processing. Transcripts encoding ATP-binding cassette transporters found in this category have long been recognized as a class of ubiquitously distributed proteins, which has often been cited as one of the largest found in nature (Henikoff et al., 1997). The main function of this protein superfamily is to mediate the energy-driven transport across membranes of a multitude of substances (Nagata et al., 2008). Generally, ATP-binding cassette transporters are characterized by the alternating occurrence of two transmembrane domains and two nucleotide binding domains in tandem (Higgins, 1992; Higgins et al., 1986). Mitogen-activated protein kinase pathways serve as highly conserved central regulators of growth, death, differentiation, proliferation, stress responses, metabolism photosynthesis, fatty acid oxidation, and provision of energy to bacteroids in root nodules (Nakagami et al., 2005).
Generally, both upregulation and downregulation of gene expression occur during different stages of development. Huijun et al. found that the number of upregulated and downregulated genes may not correlate perfectly with the stage of development (2014). In that study, between bud and senescence libraries, 3616 differentially expressed transcripts were found, including 1444 upregulated transcripts and 2172 downregulated transcripts. A total of 3711 DEGs were upregulated and 1751 DEGs were downregulated in the open flower stage, compared with the senescent flower stage. In our study, 3082 DEGs were identified in P. polyphylla var. yunnanensis. The relative genomic proportions are unknown due to the lack of genome resources. About 49% of genes increased in abundance and more than 51% were decreased in abundance. Among the DEGs in P. polyphylla var. yunnanensis, over 50% of them had no homologues in the NCBI nr database. Some of these genes might represent new transcripts that have not been reported in previous studies.
Gene expression was calculated using RPKM. On the basis of the applied criteria, 3082 genes (6.55% of all genes) were identified as significant DEGs between the roots of different ages. The most abundant P. polyphylla var. yunnanensis transcript expressed in the 8-year-old root was annotated as UDP-glucuronosyl/UDPglucosyltransferase. Glucuronosyltransferases are responsible for glucuronidation, a major process in phase II metabolism. In corn, the two UGTs like BX8 and BX9 specifically glucosylate benzoxazinoids (Rad et al., 2001). The most abundant P. polyphylla var. yunnanensis transcript expressed in the 4-year-old root was annotated as a HSP20-like chaperone. Heat shock proteins are a group of proteins that are induced by heat shock (De Maio, 1999). Production of high levels of heat shock proteins can also be triggered by exposure to different kinds of environmental stress conditions, such as infection, inflammation, exercise, exposure of the cell to toxins, starvation, hypoxia, nitrogen deficiency (in plants), or water deprivation. As a consequence, the heat shock proteins are also referred to as stress proteins and their upregulation is sometimes described more generally as part of the stress response (Santoro, 2000).
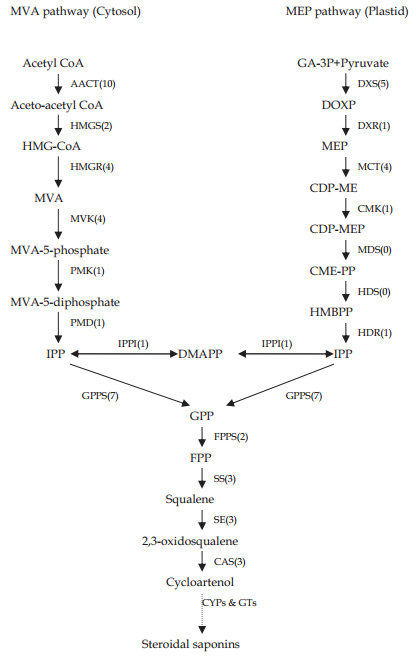
4.3 Candidate genes involved in the carotenoid biosynthesis pathwaySaponins are major therapeutic components in P. polyphylla var. yunnanensis. The two isoprenoid pathways in plants are the backbone of saponin synthesis. The first of these pathways is the cytosol MVA pathway, producing the end product isopentenyl pyrophosphate (IPP); the other is the plastidial deoxyxylulose-5-phosphate pathway, with the end products IPP and dimethylallyl pyrophosphate (DMAPP) (Liu et al., 2005). The MVA pathway starts with the condensation of acetyl-CoA (Qureshi and Porter, 1981), whereas the MEP pathway needs pyruvate and glyceraldehydes 3-phosphate (Eisenreich et al., 1998). By sequential head to tail addition of IPP and its allelic isomer DMAPP (Wise and Croteau, 1998), geranyl pyrophosphate is synthesized and then leads to farnesyl diphosphate. Next, the biosynthesis of squalene from farnesyl diphosphate constitutes the first commitment of carbon from the isoprenoid pathway toward triterpene biosynthesis. The next step is the oxidation of squalene, which leads to 2, 3-oxidosqualene synthesis. 2, 3-oxidosqualene can serve as the substrate for the synthesis of saponins by cyclization to cycloartenol and a dammarane-type triterpene skeleton. As shown in Fig. 4, oxidosqualene is a precursor that is common to the biosynthesis of both steroids and triterpenoids in higher plants (Haralampidis et al., 2002). The conversion of protopanaxadiol to protopanaxatriol is then catalyzed by a specific CYP450. Finally, one or multiple monosaccharides are added to triterpene aglycones by UGTs, leading to the production of various ginsenosides. In the P. polyphylla var. yunnanensis transcriptome dataset, most of the candidate genes involved in the MVA, MEP, and saponin biosynthesis pathways were present (Table 3). In almost all the cases, more than one unique sequence was annotated as the same enzyme. These unique sequences may represent different fragments of a single transcript, different members of a gene, or both. The results highlight the immense capacity of high-throughput sequencing to discover genes involved in metabolic pathways.
|
| Fig. 4 Paris polyphylla var. yunnanensis unigenes involved in saponin biosynthesis. The number in the bracket following each gene name indicates the number of corresponding unigenes. |
| Gene name | EC number | Transcript ID | Total ESTsa |
| Acetyl Co-acetyl transferase | 2.3.1.9 | comp83111_c0_seq1 comp87428_c1_seq2 comp87428_c0_seq1 comp36197_c0_seq1 comp91473_c0_seq1 comp68676_c0_seq1 comp476453_c0_seq1 comp74794_c0_seq1 comp22481_c0_seq1 comp88621_c0_seq1 |
10 |
| HMG-CoA synthase | 2.3.3.10 | comp327490_c0_seq1 comp94328_c0_seq8 |
2 |
| HMG-CoA reductase | 1.1.1.34 | comp94358_c2_seq4; comp91672_c0_seq1 comp94358_c2_seq3 comp94358_c2_seq2 |
4 |
| Mevalonate kinase | 2.7.1.36 | comp90803_c0_seq1 comp90803_c0_seq3 comp90803_c0_seq5 comp482484_c0_seq1 |
4 |
| Phosphomevalonate kinase | 2.7.4.2 | comp90359_c0_seq1 | 1 |
| Mevalonate diphosphate decarboxylase | 4.1.1.33 | comp95348_c0_seq4 | 1 |
| 1-deoxy-D-xylulose-5-phosphate synthase | 2.2.1.7 | comp75138_c0_seq1 comp197365_c0_seq1 comp82529_c1_seq1 comp82529_c0_seq1 comp92318_c0_seq2 |
5 |
| 1-deoxy-D-xylulose-5-phosphate reductoisomerase | 1.1.1.267 | comp89994_c0_seq1 | 1 |
| 2-C-methyl-D-erythritol 4-phosphate cytidylyl transferase | 2.7.7.60 | comp89603_c0_seq4 comp89603_c0_seq2 comp89603_c0_seq1 comp89603_c0_seq3 |
4 |
| 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase | 2.7.1.148 | comp485602_c0_seq1 comp95163_c0_seq2 |
2 |
| 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | 1.17.1.2 | comp94925_c0_seq3 | 1 |
| Isopentenyl diphosphate isomerase | 5.3.3.2 | comp89875_c0_seq1 | 1 |
| Geranylgeranyl diphosphate synthase | 2.5.1.29 | comp62094_c0_seq1 comp86532_c0_seq1 comp93332_c0_seq1 comp93332_c0_seq2 comp57192_c0_seq1 comp92260_c0_seq1 comp86814_c0_seq2 |
7 |
| Farnesyl diphosphate synthase | 2.5.1.10 | comp79513_c0_seq1 comp88440_c0_seq2 |
2 |
| Squalene synthase | 2.5.1.21 | comp36497_c0_seq1 comp241115_c0_seq1 comp90479_c2_seq1 |
3 |
| Squalene epoxidase | 1.14.13.132 | comp93612_c0_seq2 comp93612_c0_seq3 comp88004_c1_seq1 |
3 |
| Cycloartenol synthase | 5.4.99.8 | comp18960_c0_seq1 comp95850_c0_seq1 comp392455_c0_seq1 |
3 |
| a EST=expressed sequence tag. | |||
Most of the genes involved in the MVA and MEP pathways were found in our transcriptome, including acetyl CoA-acetyltransferase (EC 2.3.1.9, 10 unigenes), HMG-CoA reductase (EC 1.1.1.34, 4 unigenes), HMG-CoA synthase (EC 2.3.3.10, 2 unigenes), mevalonate kinase (EC 2.7.1.36, 4 unigenes), phosphomevalonate kinase (EC 2.7.4.2, 1 unigenes), mevalonate diphosphate decarboxylase (EC 4.1.1.33, 1 unigenes), 1-deoxy-D-xylulose-5-phosphate synthase (EC 2.2.1.7, 5 unigenes), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (EC 1.1.1.267, 1 unigenes), 2-C-methyl-D-erythritol 4-phosphate cytidylyl transferase (EC 2.7.7.60, 4 unigenes), 4-diphosphocytidyl-2-Cmethyl-D-erythritol kinase (EC 2.7.1.148, 2 unigene), and 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (EC 1.17.1.2, 1 unigenes). However, 2-C-methyl-D-erythritol, 2, 4-cyclodiphosphate synthase (EC 4.6.1.12), and 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (EC 1.17.7.1) were not found. Their absence indicates that plastids were not an important saponin biosynthesis site in P. polyphylla var. yunnanensis roots. These enzymes catalyze the synthesis of IPP, which is then converted to squalene by another series of enzymes that include isopentenyl diphosphate isomerase (EC 5.3.3.2, 1 unigenes), geranylgeranyl diphosphate synthase (EC 2.5.1.29, 7 unigenes), farnesyl diphosphate synthase (EC 2.5.1.10, 2 unigenes), and squalene synthase (EC 2.5.1.21, 3 unigenes). Subsequently, squalene is catalyzed to 2, 3-oxidosqualene squalene by monooxygenase (EC 1.14.13.132, 3 unigenes). Then, cycloartenol synthase converts 2, 3-oxidosqualene to cycloartenol, leading to the creation of sterols. Cycloartenol synthase breaks 11 bonds of 2, 3-epoxysqualene to form 11 new ones that transform the acarbocyclic epoxysqualene to the plant sterol precursor cycloartenol in Arabidopsis thaliana (Corey et al., 1993). This event is a branch point for sterol and triterpenoid biosynthesis in plants and is also the rate-limiting step (Park et al., 2005). There were also three cycloartenol synthase (E.C. 5.4.99.8, 3 unigenes) unigenes (Ohyama et al., 2009) in our P. polyphylla var. yunnanensis dataset. In the downstream reaction path, CYP450s are known to be involved in the synthesis of terpenoids and sterols, including other secondary metabolites, such as fatty acids, hormones, and defense-related phytoalexins (Morant et al., 2003). By comparison, more than 100 CYP450 unique sequences were found in our dataset. Although we cannot be certain exactly which CYP450 candidates are involved in saponin biosynthesis in P. polyphylla var. yunnanensis, this large number of CYP450 candidates provides a potential gene pool to identify the correct candidates. Saponin is often glycosylated before bioactivation. eGlycosylation occurs through the transfer of activated saccharides to an aglycone substrate and is often the last step in the biosynthesis of natural plant products. It plays an important role in stabilizing the product and altering its physiological activity (Hefner et al., 2002). In this study, more than 200 GT unique sequences were found in the P. polyphylla var. yunnanensis root transcriptome. This large number of GT candidates also provides a potential gene pool to identify the gene involved in saponin biosynthesis in P. polyphylla var. yunnanensis.
Our de novo analysis identified 3082 DEGs between the 8-yearold and 4-year-old root. Many of these genes were involved in saponin biosynthesis pathways in both transcriptomes. A comparison of saponin biosynthesis in the roots of different ages showed that some genes involved in saponin biosynthesis were upregulated in the 8-year-old root, an understandable finding considering that the amount of saponin is higher in older roots. For example, squalene epoxidase (EC 1.14.99.7) was expressed at a higher level in the 8-year-old root. This membrane-associated enzyme, active in the middle stage of the sterol biosynthetic pathway, catalyzes the conversion of squalene to 2, 3-oxidosqualene. The inducibility of saponin biosynthesis has in most cases been analyzed using in vitro cultures. For example, the exposure of plant cell cultures of Panax ginseng and Glycyrrhiza glabra to methyl jasmonate or other elicitors showed that squalene synthase is rapidly induced (Hayashi et al., 2004; Hu et al., 2003). In addition, CYP450, which is required for saponin epsilon-ring hydroxylation activity in Arabidopsis (Tian et al., 2004) was higher in the 8-year-old root.
Plant secondary metabolites are one of the most important sources of new drugs. However, due to the lack of genetic and genomic information for most plants, relatively little is known about the related biosynthetic genes and their mechanisms. Our study created a dataset for P. polyphylla var. yunnanensis and provides significant resources for gene discovery in this species that will pave the way to characterize the biosynthetic pathways of saponins.
AcknowledgmentsThis study was supported by the National Natural Science Foundation of China (81473310, 31260075, 31560085).
| Anders S., Huber W., 2010. Differential expression analysis for sequence count data[J]. Genome Biol. 11, 11, 106 . DOI:10.1186/gb-2010-11-2-106 | ||
| Barakat A., DiLoreto D.S., Zhang Y., Smith C., Baier K., Powell W.A., Wheeler N., Sederoff R., Carlson J.E., 2009. Comparison of the transcriptomes of American chestnut, Castanea dentata and Chinese chestnut, Castanea mollissima in response to the chestnut blight infection[J]. BMC Plant Biol. 9, 9, 51 . DOI:10.1186/1471-2229-9-51 | ||
| Bedell, J., Korf, I., Yandell, M., 2003. BLAST: an Essential Guide to the Basic Local Alignment Search Tool. O'Reilly and Associates, Sebastopol, CA. | ||
| Corey E.J., Matsuda S.P.T., Bartel B., 1993. Isolation of an Arabidopsis thaliana gene encoding cycloartenol synthase by functional expression in a yeast mutant lacking lanosterol synthase by the use of a chromatographic screen[J]. Proc. Natl. Acad. Sci. U. S. A, 90, 11628 -11632. DOI:10.1073/pnas.90.24.11628 | ||
| De Maio A., 1999. Heat shock proteins: facts, thoughts, and dreams[J]. Shock, 11, 1 -12. | ||
| Eisenreich W., Schwarz M., Cartayrade A., Arigoni D., Zenk M.H., Bacher A., 1998. The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms[J]. Chem. Biol, 5, 221 -233. DOI:10.1016/S1074-5521(98)90002-3 | ||
| Fu Y.L., Yu Z.Y., Tang X.M., Zhao Y., Yuan X.L., Wang S., Ma B.P., Cong Y.W., 2008. Pennogenin glycosides with a spirostanol structure are strong platelet agonists: structural requirement for activity and mode of platelet agonist synergism[J]. J. Thromb. Haemost, 6, 524 -533. DOI:10.1111/jth.2008.6.issue-3 | ||
| GÖtz S., Garcia-Gomez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., Robles M., Talon M., Dopazo J., Conesa A., 2008. High-throughput functional annotation and data mining with the Blast2GO suite[J]. Nucleic Acids Res, 36, 3420 -3435. DOI:10.1093/nar/gkn176 | ||
| Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome[J]. Nat. Biotechnol, 29, 644 -652. DOI:10.1038/nbt.1883 | ||
| Guo C.K., Zhang S., Kong W.J., 2006. Clinical study of Bi Yan Qing Du granule and Bi Yan Shu oral liquid in treatment patients with nasopharyngeal carcinoma after radiotherapy[J]. Chin. Cancer, 15, 113 -115. | ||
| Guo L., Su J., Deng B.W., Yu Z.Y., Kang L.P., Zhao Z.H., Shan Y.J., Chen J.P., Ma B.P., Cong Y.W., 2008. Active pharmaceutical ingredients and mechanisms underlying phasic myometrial contractions stimulated with the saponin extract from Paris polyphylla Sm. var. yunnanensis used for abnormal uterine bleeding[J]. Hum. Reprod, 23, 964 -971. DOI:10.1093/humrep/den001 | ||
| Han H., Shen X.L., Cui Y., Xu H.Q., Su X.M., 2009. Inhibitory effect of Biyan Qingdu Keli on nasopharyngeal carcinoma cells in vivo[J]. Guangdong Med. J, 30, 1244 -1245. | ||
| Haralampidis K., Trojanowska M., Osbourn A.E., 2002. Biosynthesis of triterpenoid saponins in plants[J]. Adv. Biochem. Eng. Biotechnol, 75, 31 -49. | ||
| Hayashi H., Huang P., Takada S., 2004. Differential expression of three oxidosqualene cyclase mRNAs in Glycyrrhiza glabra[J]. Biol. Pharm. Bull, 27, 1086 -1092. DOI:10.1248/bpb.27.1086 | ||
| Hefner T., Arend J., Warzecha H., Siems K., Stochigt J., 2002. Arbutin synthase, a novel member of the NRD1beta glycosyltransferase family, is a unique multifunctional enzyme converting various natural products and xenobiotics[J]. Bioorg. Med. Chem, 10, 1731 -1741. DOI:10.1016/S0968-0896(02)00029-9 | ||
| Henikoff S., Greene E.A., Pietrokovski S., Bork P., Attwood T.K., Hood L., 1997. Genome families: the taxonomy of protein paralogs and chimeras[J]. Science, 278, 609 -614. DOI:10.1126/science.278.5338.609 | ||
| Herrera J.B., Bartel B., Wilson W.K., Matsuda S.P., 1998. Cloning and characterization of the Arabidopsis thaliana lupeol synthase gene[J]. Phytochemistry, 49, 1905 -1911. DOI:10.1016/S0031-9422(98)00366-5 | ||
| Higgins C.F., 1992. ABC transporters: from microorganisms to man[J]. Annu. Rev. Cell Biol, 8, 67 -113. DOI:10.1146/annurev.cb.08.110192.000435 | ||
| Higgins C.F., Hiles I.D., Salmond G.P.C., Gill D.R., Downie J.A., Evans I.J., Holland I.B., Gray L., Buckel S.D., Bell A.W., Hermodson M.A., 1986. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria[J]. Nature, 323, 448 -450. DOI:10.1038/323448a0 | ||
| Hu X.Y., Neill S., Cai W.M., Tang Z.C., 2003. Hydrogen peroxide and jasmonic acid mediate oligogalacturonic acid-induced saponin accumulation in suspensioncultured cells of Panax ginseng[J]. Physiol. Plant, 118, 414 -421. DOI:10.1034/j.1399-3054.2003.00124.x | ||
| Huang Y., Cui L.J., Zhan W.H., Dou Y.H., Wang Y.L., Wang Q.Z., Zhao D., 2007. Separation and identification of steroidal compounds with cytotoxic activity against human gastric cancer cell lines in vitro from rhizomes of Paris polyphylla varr. chinensis[J]. Chem. Nat. Compd, 43, 672 -677. DOI:10.1007/s10600-007-0225-8 | ||
| Jeter C.R., Roux S.J., 2006. Plant responses to extracellular nucleotides: cellular processes and biological effects[J]. Purinergic Signal, 2, 443 -449. DOI:10.1007/s11302-005-3981-6 | ||
| Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., Katayama, Tokimatsu T., Yamanishi, Y., 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480-D484. PubMed: 18077471. | ||
| Kargiotidou A., Deli D., Galanopoulou D., Tsaftaris A., Farmaki T., 2008. Low temperature and light regulate delta 12 fatty acid desaturases, FAD2. at a transcriptional level in cotton, Gossypium hirsutum.[J]. J. Exp. Bot, 59, 2043 -2056. DOI:10.1093/jxb/ern065 | ||
| Kumar S., Kalra S., Kumar S., Kaur J., Singh K., 2012. Differentially expressed transcripts from leaf and root tissue of Chlorophytum borivilianum: a plant with high medicinal value[J]. Gene, 511, 79 -87. DOI:10.1016/j.gene.2012.09.046 | ||
| Lee R.K.Y., Ong R.C.Y., Cheung J.Y.N., Li Y.C., Chan J.Y.W., Lee M.M.S., 2009. Poly-phyllin Dea potential anti-cancer agent to kill hepatocarcinoma cells with multi-drug resistance[J]. Curr. Chem. Biol, 3, 89 -99. DOI:10.2174/2212796810903010089 | ||
| Li B., Dewey C.N., 2011. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome[J]. BMC Bioinforma. 12, 12, 323 . DOI:10.1186/1471-2105-12-323 | ||
| Li C.Q., Wang Y., Huang X.M., Li J., Wang H.C., Li J., 2013. De novo assembly and characterization of fruit transcriptome in Litchi chinensis Sonn and analysis of differentially regulated genes in fruit in response to shading[J]. BMC Genom. 14, 14, 552 . DOI:10.1186/1471-2164-14-552 | ||
| Liu Y., Wang H., Ye H.C., Li G.F., 2005. Advances in the plant isoprenoid biosynthesis pathway and its metabolic engineering[J]. J. Integr. Plant Biol, 47, 769 -782. DOI:10.1111/jipb.2005.47.issue-7 | ||
| Liu H., Zhang T., Chen X.Q., 2006. Steroidal saponins of Paris polyphylla Smith var. yunnanensis[J]. Chin. J. Nat. Med, 4, 265 -267. | ||
| Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y., Tang J., Wu G., Zhang Y., Shi Y., Liu Y., Yu C., Wang B., Lu Y., Han C., Cheung D.W., Y S.M., Peng S., Zhu X., Liu G., Liao X., Li Y., Yang H., Wang J., Wang J., 2012. SOAPdenovo2: an empirically improved memory-efficient shortread de novo assembler[J]. Gigascience, 1, 18 . DOI:10.1186/2047-217X-1-18 | ||
| Mao X., Cai T., Olyarchuk J.G., Wei L., 2005. Automated genome annotation and pathway identification using the KEGG Orthology, KO as a controlled vocabulary[J]. Bioinformatics, 21, 3787 -3793. DOI:10.1093/bioinformatics/bti430 | ||
| Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., 2005. Genome sequencing in microfabricated high-density picolitre reactors[J]. Nature, 37, 376 -380. | ||
| Matsuda H., Pongpiriyadacha Y., Morikawa T., Kishi A., Kataoka S., Yoshikawa M., 2003. . yunnanensis on ethanolor indomethacin-induced gastric mucosal lesions in rats: structural requirement for activity and mode of action[J]. Bioorg. Med. Chem. Lett, 13, 1101 -1106. DOI:10.1016/S0960-894X(03)00052-0 | ||
| Morant M., Bak S., Moller B.L., Werck-Reichhart D., 2003. Plant cytochromes P450: tools for pharmacology, plant protection and phytoremediation[J]. Curr. Opin. Biotech, 14, 151 -162. DOI:10.1016/S0958-1669(03)00024-7 | ||
| Morozova O., Hirst M., Marra M.A., 2009. Applications of new sequencing technologies for transcriptome analysis[J]. Annu. Rev. Genom. Hum. Genet, 10, 135 -151. DOI:10.1146/annurev-genom-082908-145957 | ||
| Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq[J]. Nat. Methods, 5, 621 -628. DOI:10.1038/nmeth.1226 | ||
| Nagata T., Iizumi S., Satoh K., Kikuchi, 2008. Comparative molecular biological analysis of membrane transport genes in organisms[J]. Plant Mol. Biol, 66, 565 -585. DOI:10.1007/s11103-007-9287-z | ||
| Nakagami H., Pitzschke A., Hirt H., 2005. Emerging MAP kinase pathways in plant stress signalling[J]. Trends Plant Sci, 10, 339 -346. DOI:10.1016/j.tplants.2005.05.009 | ||
| Ohyama K., Suzuki M., Kikuchi J., Saito K., Muranaka T., 2009. Dual biosynthetic pathways to phytosterol via cycloartenol and lanosterol in Arabidopsis[J]. Proc. Natl. Acad. Sci. U. S. A, 106, 725 -730. DOI:10.1073/pnas.0807675106 | ||
| Park J., Rhee D., Lee Y., 2005. Biological activities and chemistry of saponins from Panax ginseng C[J]. A. Meyer. Phytochem. Rev, 4, 159 -175. DOI:10.1007/s11101-005-2835-8 | ||
| Qureshi, N., Porter, W., 1981. Conversion of acetyl-coenzyme A to isopentenyl pyrophosphate. In: Porter, J.W., Spurgeon, S.L. (Eds.), Biosynthesis of Isoprenoid Compounds, vol. 1. John Wiley, New York. | ||
| Rad U., Huttl R., Lottspeich F., Gierl A., Frey M., 2001. Two glucosytransferases are involved in detoxification of benzoxazinoids in maize[J]. Plant J, 28, 633 -642. | ||
| Roef M.J., de Meer K., Kalhan S.C., Straver H., Berger R., Reijngoud D.J., 2003. Gluconeogenesis in humans with induced hyperlactatemia during lowintensity exercise[J]. Am. J. Physiol. Endocrinol. Metab., 284, E1162 -E1171. DOI:10.1152/ajpendo.00425.2002 | ||
| Rohdich F., Kis K., Bacher A., Eisenreich W., 2001. The nonmevalonate pathway of isoprenoids: genes, enzymes and intermediates[J]. Curr. Opin. Chem. Biol, 5, 535 -540. DOI:10.1016/S1367-5931(00)00240-4 | ||
| Rohmer M., 2003. Mevalonate-independent methylerythritol phosphate pathway for isoprenoid biosynthesis. Elucidation and distribution[J]. Pure Appl. Chem, 75, 375 -387. | ||
| Santoro M.G., 2000. Heat shock factors and the control of the stress response[J]. Biochem. Pharmacol, 59, 55 -63. DOI:10.1016/S0006-2952(99)00299-3 | ||
| Shendure J., Ji H., 2008. Next-generation DNA sequencing[J]. Nat. Biotechnol, 26, 1135 -1145. DOI:10.1038/nbt1486 | ||
| Storey J.D., Tibshirani R., 2003. Statistical significance for genomewide studies[J]. Proc. Natl. Acad. Sci. U. S. A, 100, 9440 -9445. DOI:10.1073/pnas.1530509100 | ||
| Sutapa D., Giriraj K., Bikram P.S., Deepak K.G., Sangeeta S., Vivek D., Kishor G., Tilak D., 2011. Development of genic-SSR markers by deep transcriptome sequencing in pigeon pea, Cajanus cajan, L[J]. Millspaugh. BMC Plant Biol. 11, 11, 17 . DOI:10.1186/1471-2229-11-17 | ||
| Tian L., Musetti V., Kim J., Magallanes-Lundback M., Dellapenna D., 2004. The arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid epsilon-ring hydroxylation activity[J]. Proc. Natl. Acad. Sci. U. S. A, 101, 402 -407. DOI:10.1073/pnas.2237237100 | ||
| Wall P.K., Leebens-Mack J., Chanderbali A.S., Barakat A., Liang H., 2009. Comparison of next generation sequencing technologies for transcriptome characterization[J]. BMC Genomics 10, 10, 347 . DOI:10.1186/1471-2164-10-347 | ||
| Wang X.W., Luan J.B., Li J.M., Bao Y.Y., Zhang C.X., Liu S.S., 2010a. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development[J]. BMC Genomics 11, 11, 400 . DOI:10.1186/1471-2164-11-400 | ||
| Wang L., Feng Z., Wang X., Zhang X., 2010b. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data[J]. Bioinformatics, 26, 136 -138. DOI:10.1093/bioinformatics/btp612 | ||
| Wise, M.L., Croteau, R., 1998. Monoterpene biosynthesis. In: Cane, D.E. (Ed.), Comprehensive Natural Products Chemistry, vol. 2. Pergamon Press, Oxford. | ||
| Wu S.S., Gao W.Y., Duan H.Q., Jia W., 2004. Advances in studies on chemical constituents and pharmacological activities of Rhizoma paridis[J]. Chin. Tradit. Herb. Drugs, 35, 344 -347. | ||
| Xu T.H., Ma X.X., Xu Y.J., 2007. New steroidal saponin from Paris polyphylla Sm. var. yunnanensis, France[J]. Hand-Mazz Chem. J. Chin. U, 28, 2303 -2306. | ||
| Young M.D., Wakefield M.J., Smyth G.K., Oshlack A., 2010. Gene ontology analysis for RNA-seq: accounting for selection bias[J]. Genome Biol., 11, R14 . DOI:10.1186/gb-2010-11-2-r14 | ||
| Zhang S.P., 2007. Research progress on chemical constituents and pharmacological effect of genus Paris[J]. Strait Pharm. J, 19, 4 -7. | ||
| Zhang M., Li Y.W., Li Z.Y., Huang X., Zhu D., Liu Q., 2011. Progress on studies of endangered ethno-medicine of Rhizoma Paris[J]. J. Cent. Univ. Natl. Nat. Sci. Ed, 20, 65 -69. | ||
| Zhao Y., Kang L.P., Liu Y.X., Zhao Y., Xiong C.Q., Ma B.P., Dong F.T., 2007. Three new steroidal saponins from the rhizome of Paris polyphylla[J]. Magn. Reson. Chem, 45, 739 -774. DOI:10.1002/(ISSN)1097-458X | ||
| Zhao Y., Kang L.P., Liu Y.X., Liang Y.G., Tan D.W., Yu Z.Y., Cong Y.W., Ma B.P., 2009. Steroidal saponins from the rhizome of Paris polyphylla and their cytotoxic activities[J]. Planta Med, 75, 356 -363. DOI:10.1055/s-0028-1088380 |



