b. University of the Chinese Academy of Sciences, Beijing 100039, China;
c. College of Life Science, Henan University, 85 Minglun Street, Kaifeng 475001, Henan, China
Plants synthesize numerous secondary metabolites that are beneficial for adaptation to biotic or abiotic stresses and thus contribute to the fitness of species (Pichersky and Lewinsohn, 2011). Flavonoids are an important class of plant secondary metabolites. In the model plant Arabidopsis thaliana, at least 54 flavonoid molecules (35 flavonols, 11 anthocyanins and 8 proanthocyanidins) have been identified (Saito et al., 2013). Genes that play a role in flavonoid modification, including glycosylation, methylation and acylation (Grotewold, 2006; Lepiniec et al., 2006), constitute large families and thus greatly increase the structure and genetic diversity of flavonoids in plants.
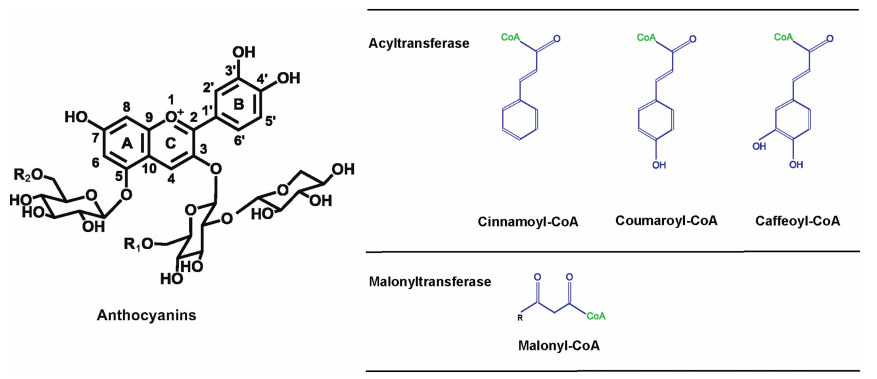
Anthocyanins are mostly red, purple, or blue water-soluble pigments derived from a branch of the flavonoid pathway. As antioxidants and pigments, anthocyanins have applications in agriculture, food products, and human health. Biosynthesis of anthocyanins requires acylation which is carried out by a group of flavonoid-specific, acyl CoA-dependent acyltransferases in the BAHD family, named according to the first letter of the first four characterized members in the family (BEAT, AHCT, HCBT, DAT) (St-Pierre and De Luca, 2000). Over 60 BAHD acyltransferases have been assigned functions in plants based on the genetic mutants and/or biochemical assays (Tuominen et al., 2011). Aromatic acylation and aliphatic acylation are two major types of acylation: the former usually involves the addition of cinnamoyl, coumaroyl, caffeoyl groups and the latter is normally malonylation (Winkel-Shirley, 2001) (Fig. 1). The acyltransferases that have anthocyanins as acceptor substrates constitute a superclade of phylogenetic tree but functional predictions on the catalytic versatility is difficult to infer from the primary sequences alone (Luo et al., 2007).
|
| Fig. 1 The substrate and product versatility of anthocyanin acyltransferase and anthocyanin malonyltransferase that catalyze the acylation and malonylation of 3-O-glucoside and 5-O-glucoside in Arabidopsis. R1, the 6" position of the 3-O-glucoside; R2, the 6"" position of the 5-O-glucoside. 3AT and 5AT catalyze the acylation of cinnamoyl, coumaroyl, or caffeoyl groups in the R1 and R2 positions, respectively. 3MAT and 5MAT add the malonyl group to the R1 and R2 positions, respectively. |
Horizontal gene transfer (HGT) refers to the transfer of genetic material to non-offspring genomes (Bock, 2010; Zhang et al., 2014b). In plants, most HGTs involve mitochondrial DNA transferred to nuclear or mitochondrial genomes (Davis and Xi, 2015; Rice et al., 2013; Xi et al., 2013). Such transfer events have been considered neutral or to confer no advantage to the recipients because the majority of transferred genes evolve into pseudogenes (Bock, 2010). Gene transfer between plant nuclear genomes is important because the newly introduced genes may endow the recipients with adaptive advantages. Nevertheless such transfers are rare in plants. Furthermore, nuclear transfers that have been reported mostly involve parasitic plants with a narrow host range (Xi et al., 2012). Very few nuclear genes were found to have been transferred between parasitic plants with a broad host range and their hosts. Two notable exceptions include albumin 1 and strictosidine synthase-like (SSL) genes from Fabaceae and Brassicaceae hosts to both Phelipanche aegyptiaca and Cuscuta australis, respectively (Zhang et al., 2013, 2014a).
Plants in the genus Cuscuta (Convolvulaceae) are world-wide obligate stem holoparasites. There are about 200 species of this genus and most have broad host ranges (Garcia et al., 2014), including Brassicaceae, Leguminosae, Solanaceae, Apiaceae, Asteraceae, and Cucurbitaceae. By establishing intimate connections with their hosts through haustoria, Cuscuta can take up water, nutrients, and macromolecules, including proteins and mRNAs (Jiang et al., 2013; Kim and Westwood, 2015). Relatively high frequency genetic material exchanges enhance the occurrence of horizontal gene transfer. Surprisingly, except for the foreign albumin 1 and SSL gene, no nuclear gene transfer events have been reported in Cuscuta to date. Utilizing the next-generation sequencing technology, we generated eight transcriptomes from different tissues and developmental stages of C. australis. Multiple Cuscuta pentagona transcriptomes are also publicly accessible, offering reliable data sources for screening for foreign genes at the transcriptional level. In this study, we show that one putative anthocyanin acyltransferase gene (AT-like) has been transferred from Fabaceae to Cuscuta, although the possibility of convergent evolution of these genes can not be absolutely ruled out.
2 Materials and methods 2.1 Transcriptome screening for foreign genes in C. australisTo identify HGT events between host and parasitic plant, we screened C. australis transcriptomes for foreign genes according to the procedure described previously (Zhang et al., 2014a). In brief, a transcriptome assembly was obtained by combining all the RNAseq datasets from eight tissues, seeds, germinated seeds, seedlings, pre-haustoria, stems, buds, flowers, and capsules using Trinity v 2.0.6 (Grabherr et al., 2011). The predicted coding regions with a contiguous amino acid length≥100 were screened using AlienG (Tian et al., 2011). The alien origin of the candidate genes was predicted if the score ratio of the first non-Convolvulaceae hit to the first Convolvulaceae species hit was more than 1.2. These candidates were then manually identified as horizontally transferred genes.
2.2 Gene identification and homolog search for putative foreign genes in C. australis genome assembly and the transcriptomes of C. pentagonaWe produced 73 Gb (about 170X coverage) of high quality Illumina paired-end reads from a library with~270-bp insertion fragments. The genome assembly was performed using the SOAPdenovo package (Li et al., 2010) with a K-mer size of 41. The transcriptome contigs encoding foreign genes were used as queries to search our local genome assembly by BLASTn to obtain the corresponding DNA and homology sequences. The gene structures in the homology sequences were determined using our transcriptome data and by BLAST search (BLASTx) against the nr database. The candidate foreign genes were used as queries to search online against the RNA-seq datasets of C. pentagona deposited in the NCBI SRA database using BLASTn and the read pairs with more than 94% identity were extracted as the evidence of transcription.
2.3 Phylogenetic analyses of the anthocyanin acyltransferase gene in C. australisTo investigate the origin of the AT-like (anthocyanin acyltransferases-like) gene in Cuscuta, protein homologs were extracted from representative species with genome sequences (Supplementary Table 1), and our homemade genome assembly of C. australis by BLAST search using the candidate anthocyanin acyltransferase gene from C. australis as queries. To obtain insight into the putative function of the anthocyanin acyltransferase gene in Cuscuta, the putative genes or characterized proteins belonging to the clade I (Luo et al., 2007) and clade Ia (Tuominen et al., 2011) from two previous studies, together with the candidate BAHD acyltransferase gene and homologs in C. australis, were also collected. The sequences were aligned with ClustalX v2.1 (Thompson et al., 1997), visually inspected, and manually refined. Gaps and ambiguous sites were removed from the alignment. ModelGenerator (v_851) (Keane et al., 2006) was used to find the best-fitting model of protein substitution. The protein phylogenetic tree was inferred under maximum likelihood optimization using RAxML v 8.0.0 with the parameters “-f a, -m PROTGAMMAJTTF, -# 100” (Stamatakis, 2006). Trees were viewed and edited using MEGA7 (Kumar et al., 2016).
2.4 Expression level estimation of the candidate BAHD acyltransferase gene in C. australis and C. pentagonaThe expression levels of the AT-like gene in different tissues at different developmental stages of C. australis were estimated using RSEM (Li and Dewey, 2011). We mapped all clean reads to the Trinity assembly, and to obtain normalized expression levels we used the fragments per kilo base of exon per million fragments mapped (FPKM) values of the sequences. The mean FPKM values of the AT-like gene from 10 accessible public RNA-seq datasets in C. pentagona (Ranjan et al., 2014), from the tissues of seeds, seedlings, stems, pre-haustroria, haustoria, and flowers, were calculated using the RPKM formula=(10^9 * C)/(N * L), where C is the number of read pairs mapped to a gene, N refers to the total mapped read pairs in the experiment and here is approximate to the total number of raw read pairs, L is the exon length in base-pairs for a gene. The SRA accession numbers of the 10 public RNA-seq datasets of C. pentagona were SRX345073, SRX345113, SRX345282-3, SRX345400-1, and SRX345411-4.
3 Results 3.1 The abnormal affiliation of one anthocyanin acyltransferase gene from Cuscuta with the homologs from FabaceaeUsing a pipeline we developed to screen transcriptomes for foreign genes, we found that a 1853-bp sequence in C. australis transcriptomes exhibited as high as 56% and 66% identities to the Fabaceae coumaroyl-CoA:anthocyanidin 3-O-glucoside-6"-O-coumaroyltransferases at the amino acid and nucleotide levels, respectively. However, this sequence in C. australis transcriptomes was highly divergent from the most similar sequences in the Convolvulaceae species (the highest identity was 24% at the amino acid level with Ipomoea and there was no significant similarity at the nucleotide level). We then searched the transcriptome datasets of C. pentagona deposited in NCBI SRA database and found 22 read pairs mapped to the C. australis transcript with high identity (Supplementary File 1). Therefore, the corresponding genes are likely present in C. pentagona as well. Using the 73 Gb high quality Illumina paired-end reads, we used SOAPdenovo to generate a genome assembly of 343.9 Mb, which covers 81.7% of the genome given that the genome size of C. australis was estimated to be~421 Mb by flow cytometer in our lab. We searched the genome assembly using the above transcriptome contig and found a 4165-bp scaffold that contained a complete open reading frame (ORF) of 1503 bp (500 amino acids) without introns. Notably, the homologous genes in Fabaceae are intron-less. Furthermore, the transcription of this gene can be detected in each of the transcriptomes from different tissues of C. australis and C. pentagona. The presence of this gene in the genome assembly of C. australis and multiple transcriptomes of C. australis and C. pentagona makes the possibility of contamination from other organisms very low.
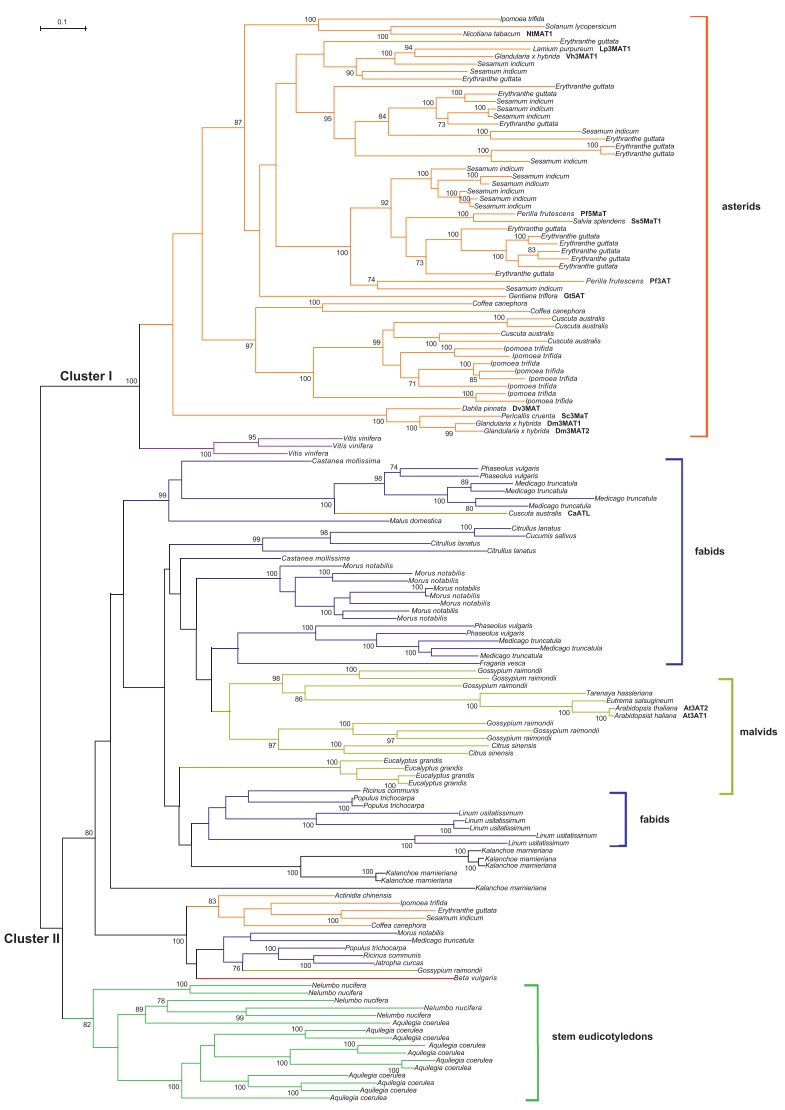
To confirm the possible transfer of this gene from Fabaceae to Cuscuta, we constructed a phylogeny of the BAHD acyltransferase genes by collecting homologs from 37 species whose genomes have been sequenced and those with genetic and biochemical evidence (Supplementary Table 1). Multiple copies were found in most species and the homologs from monocots clustered at the base of the phylogenetic tree, which had been illustrated in other studies (Luo et al., 2007; Tuominen et al., 2011). To construct a more reliable alignment and phylogenetic trees, the homologs from monocots were excluded from further analysis and only those from dicots were used. This gene tree formed two clusters (Fig. 2). Cluster Ⅰ contained homologs from asterids and Vitis, including the published anthocyanin acyltransferase and anthocyanin malonyltransferase activity proteins, which corresponded to the subclade 2 of the anthocyanin superclade (Luo et al., 2007). Cluster Ⅱ contained proteins from 28 of the 31 representative dicot species, which greatly expanded subclade 1 of the anthocyanin superclade (Luo et al., 2007). Both cluster Ⅰ and Ⅱ were included in the clade Ia by Tuominen et al. (2011).
The A. thaliana enzymes in cluster Ⅱ have been shown to possess anthocyanin acyltransferase activity (Luo et al., 2007). Therefore, proteins in this cluster are likely to be anthocyanin acyltransferases. Five genes were found in C. australis, four in cluster Ⅰ, which probably have anthocyanin acyltransferases or anthocyanin malonyltransferase activity. In addition, one gene in cluster Ⅱ likely possesses AT-like activity (named CaATL in this study for convenience, accession number KX019005). As expected, CaATL formed a sister branch with the homologs from Fabaceae and the abnormal affiliation was supported by 100% bootstrap value, which suggested that horizontal gene transfer probably occurred between Fabaceae and the ancestor of Cuscuta. No other branches with obvious abnormal species affiliation were found in the gene tree of the BAHD family proteins, although some main branches contained species from multiple linages, which may be caused by gene loses or convergent evolution as speculated by other researchers (Luo et al., 2007).
3.2 Domain structure and motif comparison of CaATL with other closely related functional BAHD membersAnalysis of domain structures in the anthocyanin acyltransferases (AT), anthocyanin malonyltransferase (MAT), and CaATL by InterPro (Quevillon et al., 2005) indicted that all these proteins possess two tandemly arranged chloramphenicol acetyltransferase-like domains (IPR023213), each~205 aa long. Both ATs and MATs can catalyze the acylation of the 6" position of the 3-O-glucoside (3ATs and 3MATs, respectively) and the 6"" position of the 5-O-glucoside (5ATs and 5MATs, respectively) (Luo et al., 2007). Therefore, these previously characterized motifs were tested by aligning the known representatives of 3ATs, 3MATs, 5ATs and 5MATs, CaATL with their closely related putative 3ATs in Medicago truncatula (Fig. 3). The three conserved motifs of the BAHD family, consensuses of PXLKXSLSX(T/A)L around position 69-79, QX(T/A)XFP(N/G)XG around position 173-181, and (D/Z)FGXG(K/R)(P/A)XK around position 452-460, were detected by WEBLOGO (http://weblogo.berkeley.edu/logo.cgi) (Fig. 3, Supplementary File 2). Two specific motifs, P(L/V)(T/S)F(F/ L)D around position 34-39 and PXXYFGNC adjacent to position 369-376, were also detected (Fig. 3, Supplementary File 2). Interestingly, one amino acid deletion occurred in the conserved HXXXD motif, which was changed into HXXD in the homologs of some fabaceous plants and CaATL (Fig. 3). These lines of evidence support the hypothesis that CaATL is a BAHD family member that likely uses anthocyanins as substrates, and is closely related to the homologs from Fabaceae.

|
| Fig. 3 Multiple sequence alignment of representative ATs, MATs, CaATL and its homologs in Fabaceae. The conserved and specific motifs are indicated in green and yellow boxes. The HXXD motif in CaATL and some Fabaceae is shown in red boxes. The sequence names are displayed in the order of abbreviation names, accession numbers, genera, and species names. Dashes indicate that the sequences are incomplete or gaps introduced in the alignment. Background colors indicate the degree of conservation of the sites. |
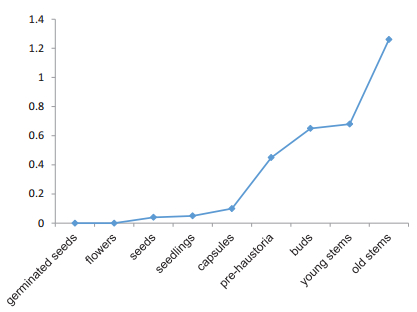
The presence of the ATL genes in the RNA-seq data of C. australis indicates that they are actively transcribed. To determine how the expression level changes in different tissues at different developmental stages, we mapped all clean paired-end reads to our own Trinity assembly of the C. australis transcriptome. The FPKM values of ATL varied within tissues at different developmental stages. No transcription was detected in germinated seeds or flowers (Fig. 4). Seeds, seedlings, and capsules had similar transcriptional levels with FPKM values less than 0.1. Pre-haustoria, young stems, and buds exhibited higher expression levels with FPKM values from 0.45 to 0.68. Notably, the FPKM value reached a maximum of 1.26 in old stems, which is 1.8-fold that of young stems. The high transcriptional activity in old tissues is consistent with the function of the gene in secondary metabolism, namely, anthocyanin modification.
|
| Fig. 4 The expression levels of the CaATL gene in C. australis in different tissues at different developmental stages. The normalized transcriptional levels were estimated by FPKM in Illumina RNA-seq datasets. |
In C. pentagona, the transcriptional levels of ATL were found to be relatively low. Only 22 read pairs were found in the 10 published paired-end RNA-seq datasets, which contain 63.45 million read pairs from seeds, seedlings, stems, pre-haustoria, haustoria, and flowers. Given that the current cDNA length is 1503 bp, the mean FPKM value was estimated to be 0.23 using the FPKM equation described in the Materials and Methods. The transcriptional activity indicates that ATL exists in C. pentagona and may still have its normal function.
4 DiscussionAnthocyanins are a class of flavonoids and are widely present in flowers, fruits, and leaves. Rich in phenolic hydroxyl groups, anthocyanins have high antioxidant properties and protect plants from reactive oxygen species produced in metabolism thus improving resistance to abiotic stresses, such as UV, drought, and low temperature, as well as biotic stresses like pathogens and herbivorous insects (Xie et al., 2013). In plants, anthocyanin acylation is catalyzed by BAHD family proteins, which show great versatility in substrate specificity (Rinaldo et al., 2015) and are able to evolve new substrate specificities rapidly (Luo et al., 2007). These traits are consistent with the phylogeny showing that cluster Ⅰ is composed of 3AT, 5AT, 3MAT and 5MAT and the 3ATs are both present in cluster Ⅰ and Ⅱ (Fig. 2).
By searching the genome assembly, five genes belonging to the BAHD family, having anthocyanin as substrates, were identified in C. australis. Four of them formed a sister branch with the homologs from the relative Ipomoea and should be native copies. CaATL and the Fabaceae homologs constituted a clade with 100% bootstrap support, and this gene very likely was transferred from a fabaceous plant to the common ancestor of Cuscuta since C. pentagona also possesses this gene. The reverse transfer direction is impossible since no other asterid genes were present in cluster Ⅱ. The identity of the Fabaceae donor was unclear from the phylogeny given that CaATL forms a basal branch of the homologs of different Fabaceae species. The occurrence of HGT is probably mediated by direct contact via haustoria of Cuscuta as Fabaceae is its preferred hosts.
The acylation of anthocyanins affects its stability and light absorption in solution. In Arabidopsis, At3AT1 and At3AT2 influence the stability of anthocyanins at neutral pH and the anthocyanin absorption maxima. The higher transcription level in old compared to young stem tissues suggested this gene might still perform its functions in secondary metabolism by stabilizing anthocyanins at neutral pH and thus improving the biotic and abiotic resistance of Cuscuta to stresses such as UV radiation and insect herbivory. Considering the separation of cluster I with cluster Ⅱ in the phylogeny, we speculate that the horizontally acquired CaATL probably endowed Cuscuta with certain adaptive characters, such as changes in expression pattern under stresses or varied substrate specificity. Further investigations and functional analysis are needed to understand the benefits of HGT on the evolution of this special holoparasitic plant.
AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (No. 31301037 and 31470012 to G.S.), the Natural Science Foundation of Yunnan Province of China (No. 2013FB068 to G.S.), the Young Academic and Technical Leader Raising Foundation of Yunnan Province (No. 2014HB046, to G.S.), the Western Light Talent Culture Project of the Chinese Academy of Sciences (to G.S.), and the Yunnan Recruitment Program of Experts in Sciences (2012HA016 to J.W.).
Appendix A. Supplementary dataSupplementary data related to this article can be found at http://dx.doi.org/10.1016/j.pld.2016.04.002.
| Bock R., 2010. The give-and-take of DNA: horizontal gene transfer in plants[J]. Trends Plant Sci, 15, 11 -22. | ||
| Davis C.C., Xi Z., 2015. Horizontal gene transfer in parasitic plants[J]. Curr. Opin. Plant Biol, 26, 14 -19. DOI:10.1016/j.pbi.2015.05.008 | ||
| Garcia M.A., Costea M., Kuzmina M., Stefanovic S., 2014. Phylogeny, character evolution, and biogeography of Cuscuta (dodders; Convolvulaceae) inferred from coding plastid and nuclear sequences[J]. Am. J. Bot, 101, 670 -690. DOI:10.3732/ajb.1300449 | ||
| Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome[J]. Nat. Biotechnol, 29, 644 -652. DOI:10.1038/nbt.1883 | ||
| Grotewold E., 2006. The genetics and biochemistry of floral pigments[J]. Annu. Rev. Plant Biol, 57, 761 -780. DOI:10.1146/annurev.arplant.57.032905.105248 | ||
| Jiang L., Qu F., Li Z., Doohan D., 2013. Inter-species protein trafficking endows dodder (Cuscuta pentagona) with a host-specific herbicide-tolerant trait[J]. New Phytol, 198, 1017 -1022. DOI:10.1111/nph.12269 | ||
| Keane T.M., Creevey C.J., Pentony M.M., Naughton T.J., McLnerney J.O., 2006. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified[J]. BMC Evol. Biol. 6, 6, 29 . DOI:10.1186/1471-2148-6-29 | ||
| Kim G., Westwood J.H., 2015. Macromolecule exchange in Cuscuta-host plant interactions[J]. Curr. Opin. Plant Biol, 26, 20 -25. DOI:10.1016/j.pbi.2015.05.012 | ||
| Kumar, S., Stecher, G., Tamura, K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. http://dx.doi.org/10.1093/molbev/msw054. | ||
| Lepiniec L., Debeaujon I., Routaboul J.M., Baudry A., Pourcel L., Nesi N., Caboche M., 2006. Genetics and biochemistry of seed flavonoids[J]. Annu. Rev. Plant Biol, 57, 405 -430. DOI:10.1146/annurev.arplant.57.032905.105252 | ||
| Li, B., Dewey, C.N., 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma. 12, 323. http://dx.doi.org/10.1186/1471-2105-12-323. | ||
| Li R., Zhu H., Ruan J., Qian W., Fang X., Shi Z., Li Y., Li S., Shan G., Kristiansen K., Li S., Yang H., Wang J., Wang J., 2010. De novo assembly of human genomes with massively parallel short read sequencing[J]. Genome Res, 20, 265 -272. DOI:10.1101/gr.097261.109 | ||
| Luo J., Nishiyama Y., Fuell C., Taguchi G., Elliott K., Hill L., Tanaka Y., Kitayama M., Yamazaki M., Bailey P., Parr A., Michael A.J., Saito K., Martin C., 2007. Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana[J]. Plant J, 50, 678 -695. DOI:10.1111/j.1365-313X.2007.03079.x | ||
| Pichersky E., Lewinsohn E., 2011. Convergent evolution in plant specialized metabolism[J]. Annu. Rev. Plant Biol, 62, 549 -566. DOI:10.1146/annurev-arplant-042110-103814 | ||
| Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., Lopez R., 2005. InterProScan: protein domains identifier[J]. Nucleic Acids Res., 33, W116 -W120. DOI:10.1093/nar/gki442 | ||
| Ranjan, A., Ichihashi, Y., Farhi, M., Zumstein, K., Townsley, B., David-Schwartz, R., Sinha, N.R., 2014. De novo assembly and characterization of the transcriptome of the parasitic weed Cuscuta pentagona identifies genes associated with plant parasitism. Plant Physiol. http://dx.doi.org/10.1104/pp.113.234864. | ||
| Rice D.W., Alverson A.J., Richardson A.O., Young G.J., Sanchez-Puerta M.V., Munzinger J., Barry K., Boore J.L., Zhang Y., dePamphilis C.W., Knox E.B., Palmer J.D., 2013. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella[J]. Science, 342, 1468 -1473. DOI:10.1126/science.1246275 | ||
| Rinaldo A.R., Cavallini E., Jia Y., Moss S.M.A., McDavid D.A.J., Hooper L.C., Robinson S.P., Tornielli G.B., Zenoni S., Ford C.M., Boss P.K., Walker A.R., 2015. A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins[J]. Plant Physiol, 169, 1897 -1916. | ||
| Saito K., Yonekura-Sakakibara K., Nakabayashi R., Higashi Y., Yamazaki M., Tohge T., Fernie A.R., 2013. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity[J]. Plant Physiol. Biochem, 72, 21 -34. DOI:10.1016/j.plaphy.2013.02.001 | ||
| St-Pierre B., De Luca V., 2000. Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism[J]. Evol. Metab. Pathw, 34, 285 -315. DOI:10.1016/S0079-9920(00)80010-6 | ||
| Stamatakis A., 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models[J]. Bioinformatics, 22, 2688 -2690. DOI:10.1093/bioinformatics/btl446 | ||
| Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucleic Acids Res, 25, 4876 -4882. DOI:10.1093/nar/25.24.4876 | ||
| Tian, J., Sun, G., Ding, Q., Huang, J., Oruganti, S., Xie, B., 2011. AlienG: an effective computational tool for phylogenetic identification of horizontally transferred genes. In: The Third International Conference on Bioinformatics and Computational Biology (BICoB): 23-25 March 2011; New Orleans, Louisiana. | ||
| Tuominen L.K., Johnson V.E., Tsai C.J., 2011. Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues[J]. BMC Genom. 12, 236 . | ||
| Winkel-Shirley B., 2001. Flavonoid biosynthesis[J]. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol, 126, 485 -493. | ||
| Xi, Z., Bradley, R.K., Wurdack, K.J., Wong, K., Sugumaran, M., Bomblies, K., Rest, J.S., Davis, C.C., 2012. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genom. 13, 227. http://dx.doi.org/10.1186/1471-2164-13-227. | ||
| Xi, Z., Wang, Y., Bradley, R.K., Sugumaran, M., Marx, C.J., Rest, J.S., Davis, C.C., 2013. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 9, e1003265. http://dx.doi.org/10.1371/journal.pgen.1003265. | ||
| Xie Y., Sun Y., Huang J., 2013. Anthocyanin modification in Arabidopsis[J]. Plant Physiol. J, 49, 101 -110. | ||
| Zhang, D.L., Qi, J.F., Yue, J.P., Huang, J.L., Sun, T., Li, S.P., Wen, J.F., Hettenhausen, C., Wu, J.S., Wang, L., Zhuang, H.F., Wu, J.Q., Sun, G.L., 2014a. Root parasitic plant Orobanche aegyptiaca and shoot parasitic plant Cuscuta australis obtained Brassicaceae-specific strictosidine synthase-like genes by horizontal gene transfer. BMC Plant Biol. 14 http://dx.doi.org/10.1186/1471-2229-14-19. | ||
| Zhang N., Han Z., Sun G., Hoffman A., Wilson I.W., Yang Y., Gao Q., Wu J., Xie D., Dai J., Qiu D., 2014b. Molecular cloning and characterization of a cytochrome P450 taxoid 9a-hydroxylase in Ginkgo biloba cells[J]. Biochem. Biophys. Res. Commun, 443, 938 -943. DOI:10.1016/j.bbrc.2013.12.104 | ||
| Zhang, Y.T., Fernandez-Aparicio, M., Wafula, E.K., Das, M., Jiao, Y.N., Wickett, N.J., Honaas, L.A., Ralph, P.E., Wojciechowski, M.F., Timko, M.P., Yoder, J.I., Westwood, J.H., dePamphilis, C.W., 2013. Evolution of a horizontally acquired legume gene, albumin 1, in the parasitic plant Phelipanche aegyptiaca and related species. BMC Evol. Biol. 13, 48. http://dx.doi.org/10.1186/1471-214813-48. |