2. School of Environmental and Biological Engineering, Nanjing University of Science and Technology, Nanjing 210094, China
3. University of Chinese Academy of Sciences, Beijing 100049, China
4. Laboratory of Tobacco Agriculture, Zhengzhou Tobacco Research Institute of CNTC, Zhengzhou 450001, China
Jasmonate(JA)is an essential hormone in higher plants where it acts as an endogenous growth regulator. Previous research indicates that JA acts as an important regulatory signal to influence multiple physiological processes, such as stomatal closure, plant resistance to cold, inhibition of Rubisco biosynthesis and so on. JA signaling is also involved in plant defense responses, including defense against mechanical damage and herbivorous insects. JA and its methly jasmonate(MeJA)are a class of fatty acid derivatives. The biosynthesis of JA and MeJA was elucidated by Vick and Zimmerman(1983) and Hamberg and Hughes(1988). With α-linolenic acid as substrate, molecular oxygen is inserted by a lipoxygenase(13-LOX)at carbon atom 13 leading to the formation of a fatty acid hydroperoxide 13 S-hydroperoxy -(9Z, 11E, 15)-octadecatrienoic acid. This compound is dehydrated by the allene oxide synthase(AOS)to an unstable allene oxide which can be either hydrolysed non-enzymatically to α- and γ -ketols or cyclized to racemic 12-oxo-phytodienoic acid(OPDA). In the presence of an allene oxide cyclase(AOC), preferential formation of the(9S, 13S)enantiomer of OPDA occurs. The AOC-catalysed step is regarded as the crucial step in octadecanoid and jasmonate biosynthesis because only this enantiomeric form is the substrate for the naturally occurring(+)-7-iso-JA, which is formed after reduction of OPDA by a specific OPDA reductase 3(12-oxophytodienoate reductase 3) and three cycles of β-oxidation. Treatment of plants with products of the AOS branch, the octadecanoids and jasmonates, has been shown to lead to accumulation of mRNAs coding for LOX, AOS, AOC and OPR3 suggesting a feedforward regulation in JA biosynthesis. Also, biotic or abiotic stresses, which result in endogenous increases of octadecanoids and jasmonates, are usually accompanied by a transcriptional up-regulation of AOS, AOC and OPR3.
Tobacco(Nicotiana tabacum)generates an array of alkaloids that play essential roles in the plant defense response against herbivore and insect attack(Kessler and Baldwin, 2002; Steppuhn et al., 2004). Nicotine is the main alkaloid produced by cultivated tobacco(N. tabacum L.), constituting approximately 0.6%-3% of the tobacco leaf dry weight. Nicotine is synthesized in the root from ornithine and arginine by way of putrescine. Putrescine is either metabolized to higher polyamines, such as spermidine and spermine, or conjugated with cinnamic acid derivatives or fatty acids in all higher plants; However, it is also converted into N-methylputrescine in plants that produces nicotine or tropane alkaloids. Thus, putrescine N-methyltransferase(PMT; EC 2.1.1.53)participates in the first committed step of alkaloid biosynthesis(Chattopadhyay and Ghosh, 1998; Chou and Kutchan, 1998). N-Methylputrescine is then oxidized by a diamine oxidase(EC 1.4.3.6) and cyclized spontaneously to the l-methyl-A′-pyrrolinium cation, which is condensed with nicotinic acid or its derivative. Quinolinic acid phosphoribosyltransferase(EC 2.4.2.19)serves as the entry-point enzyme in the pyridine nucleotide cycle, which supplies nicotinic acid. After biosynthesis in the tobacco root, nicotine is translocated to the leaf via the xylem and stored in the leaf vacuole with the help of a tonoplast-localized transporter. Nicotine can be demethylated in both leaves and roots, but is primarily demethylated in senescing leaves(Wagner et al., 1986; Chou and Kutchan, 1998). The accumulation of nicotine in tobacco is affected by environmental factors, cultural practices, and plant hormone levels. For example, the application of nitrogen fertilizer or jasmonate markedly increases nicotine biosynthesis(De Luca and St Pierre, 2000; Shoji et al., 2000; Goossens et al., 2003; Paschold et al., 2007). Zhang et al.(2012) reported that the tobacco transcription factors NtMYC2a and NtMYC2b formed a nuclear complex with NtJAZ1 to regulate jasmonate-induced nicotine biosynthesis, suggesting that NtMYC2a, NtMYC2b, and NtJAZ1 are involved in the regulatory pathway that controls nicotine biosynthesis.
To underst and how environmental factors affect nicotine biosynthesis in tobacco, we investigated the effects of multiple environmental stresses on nicotine content in tobacco. Our results show that salt stress increases the level of transcription factor NtMyc2a and the biosynthesis of nicotine, while blocking jasmonate biosynthesis and signaling after salt stress reduces plant nicotine biosynthesis. However, application of exogenous JA significantly induces nicotine accumulation. These results suggest that JA acts as an important signal that mediates salt-induced nicotine biosynthesis in tobacco.
1. Materials and Methods 1.1. Plant Growth and TreatmentSterilized tobacco(Nicotiana tabacum cv. Wisconsin 38)seeds were germinated and grown to seedlings under continuous illumination on half-strength Gamborg B5 medium solidified with 2% gellan gum and supplemented with 0.3% sucrose at 24℃. One-week-old plants were transferred to soil containing 3/5 peat soil, 1/5 vermiculite, 1/5 perlite, and grown for one week in the greenhouse at 24℃(16h light/8h dark, 1200 μmol m-2s-1 light intensity)before salt treatment. For salt treatment, 2-week-old seedlings were sprayed with 1/4 Hoagl and solution or a 100mmol·L-1 NaCl solution of Hoagl and for salt stress. The plants were irrigated every three days. For MeJA treatment, 2-week-old plants were sprayed with water, 10nmol·L-1, 50nmol·L-1, 100nmol·L-1 or 500nmol·L-1 MeJA solution. After various treatment times, tobacco leaves were collected for further analysis.
1.2. Nicotine content measurementAfter treatment, 0.5g of tobacco roots was collected and frozen in liquid nitrogen and lyophilized, before homogenization and resuspension in 4mL of 0.1mol·L-1 H2SO4. The homogenate was sonicated for 60 min and centrifuged at 2000 g for 15min. The supernatant pH was neutralized by adding 0.4mL 25% NH4OH, and the mixture was loaded onto an Extrelut-1 column(Merck) and eluted with 6mL of chloroform. The eluent was dried at 37℃, and the dry residues were dissolved in ethanol and analyzed by gas chromatography(Agilent 6890) and quadrupole mass spectrometry with electron impact ionization(Agilent 5973, Network, US). The column temperature was held at 100℃ for 10min, and then increased to 260℃, over a 35-min period, at a gradient of 8℃/min. Synthetic nicotine(Zhengzhou Tobacco Research Institute)was used as a st and ard. Nicotine content analysis was performed at Zhengzhou Tobacco Research Institute.
1.3. Jasmonate content analysisTo measure jasmonate contents, 2-week-old plants were treated with salt solution for different times. After treatment, root tissue was harvested and jasmonate was analyzed by ELISA using monoclonal antibodies of jasmonate according to Hu et al.(2013). Jasmonate antibody was purchased from Shanghai YUANYAN biological technology company. Weight about 1g tobacco roots tissue materials which was salt stress with different times, then using liquid nitrogen frozen ground into a powder. After the liquid nitrogen was volatilization, add 10mL 80% methanol(contain 100ug antioxidants butylatrd hydroxytouene)is extracted for the night, after NH4OH was add to supernatant fluid to pH 8.0, freeze-drying, then dissolved the extraction into 0.2mol·L-1 acetic acid for JA content measurement.
1.4. RNA extraction and quantitative real-time RT-PCR(qRT-PCR)Total RNA was extracted from 3-week-old tobacco seedling after different treatment using TRIzol reagent(Invitrogen), and qRT-PCR was performed as previously described(Zhang et al., 2012). Briefly, first-str and cDNA was synthesized from 1.5μg DNase-treated RNA in a 20μL reaction volume using M-MuLV reverse transcriptase(Fermentas)with oligo(dT)18 primer. qRT-PCR was performed using SYBR Green I Master Mix with a Roche LightCycler 480 real-time PCR machine, according to the manufacturer′s instructions. At least three biological replicates for each sample were used for qRT-PCR analysis, and at least two technical replicates were analyzed for each biological replicate. The ACTIN2 gene was used as an internal control of gene expression. Gene-specific primers used to detect the transcripts are listed as below:
NtMYC2a-F: 5′-cctcattgatggtgctcgtg-3′,
NtMYC2a-R: 5′-tttcggtgttcttgctcagc-3′;
NtLOX-F: 5′-ggacttgaaggatgttggtgc-3′,
NtLOX-R: 5′-tcacattaaacgtagcatctcct-3′;
NtAOS-F: 5′-gccaaacgcgaccttatgat-3′,
NtAOS-R: 5′-ccacaaaatcctttccggca-3′;
NtAOC-F: 5′-acagcttctacttcggcgat-3′,
NtAOC-R: 5′-tagcaactcagacggcagat-3′;
NtOPR-F: 5′-cctatgggcaaactgaagca-3′,
NtOPR-R: 5′-cgagatcagcatcaccttgt-3′.
1.5. Protein isolation and immunoblot analysisProteins were extracted from whole seedling with extraction buffer containing 50mmol·L-1 Tris-Cl, pH 7.5, 150mmol·L-1 NaCl, 1mmol·L-1 PMSF, 1×Complete Protease Inhibitor Cocktail(Roche), 5% glycerol, 1mmol·L-1 EDTA, and 1mmol·L-1 DTT. The protein concentration was determined using a commercial Bradford assay kit following the manufacturer′s instructions(Bio-Rad). The samples were mixed with equal volume of 10% sodium dodecyl sulfate(SDS)sample buffer and boiled for 3min before separation on a 10% SDS-PAGE gel. The fractionated proteins were then transferred to the PVDF membrane and immunoblot assays were performed as previously described(Zhang et al., 2012), using anti-PMT1 and anti-Actin(Abmart, Shanghai, China)antibodies at dilutions of 1∶3000 and 1∶2000, respectively. The anti-PMT1 antibody was prepared by immunizing rabbit with the synthesized peptide from the N-termination of tobacco PMT1 protein(MEVISTNTNGSTI).
2 . Results 2.1. Salt stress induced the accumulation of nicotineFor salt stress, we watered two-week-old plantlets with 1/4 Hoagl and with 50mmol·L-1 NaCl, and then measured nicotine content at different times. We found that salt stress induced nicotine accumulation in the tobacco root system(Fig. 1A). Compared with the control plants, nicotine levels increased 1 day after treatment, and rapidly accumulated 3 days after treatment in salt-stressed plants. Salt stress-induced nicotine content reached a peak after 7 days of treatment, and sustained a high level for 9 days. NtPMT1 is the key enzyme responsible for nicotine biosynthesis in tobacco(Chattopadhyay and Ghosh, 1998; Chou and Kutchan, 1998). Using NtPMT1 antibodies to analyze NtPMT1 levels in tobacco after salt stress, we found that salt stress increased NtPMT1 protein levels 24h after treatment, and detected sustained high protein levels 5-7 days after treatment(Fig. 1B). These results indicate that salt stress significantly induces nicotine biosynthesis and NtPMT1 protein accumulation in tobacco.
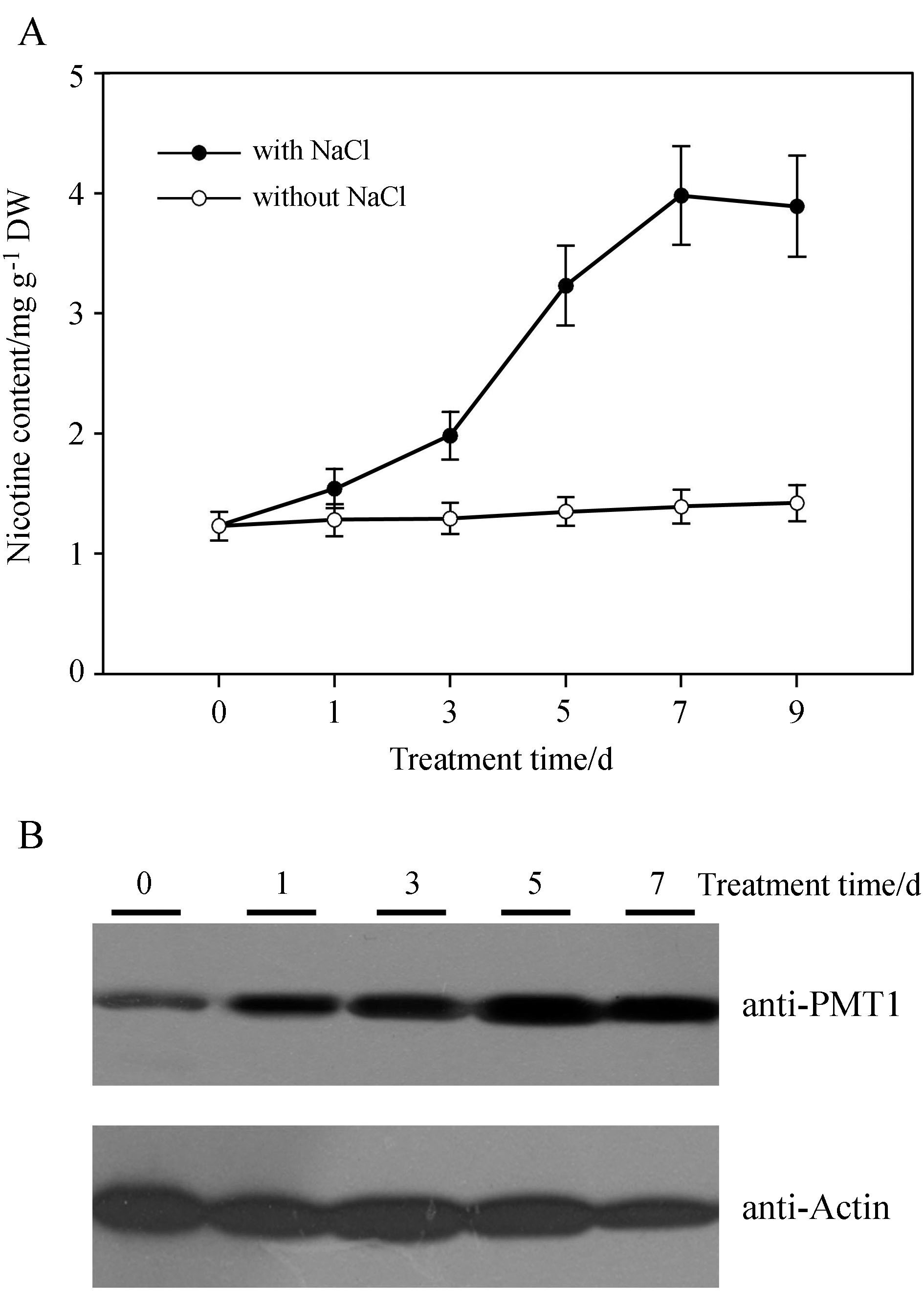
|
| Fig.1 Salt stress induced the accumulation of nicotine and NtPMT1. Two-week-old seedlings were treated with 50mmol·L-1 salt solution and the nicotine content was measured at indicated times(A); NtPMT1 protein levels were also examined using anti-PMT1 antibody and western blotting analysis. anti-Actin was used as a loading control. Data represent the mean of three replicate experiments(± SE). |
It has been reported that MYC2a transcriptional factors are involved in jasmonate-mediated biosynthesis of nicotine(Zhang et al., 2012). To underst and the role of MYC2a in salt-induced nicotine biosynthesis, we applied quantitative RT-PCR to detect transcriptional changes of MYC2a. As shown in Fig. 2, NtMYC2a transcript levels increased after 6 h of salt stress, reaching a peak after 36 h, with prolonged stress sustaining high levels of NtMYC2a transcripts. In contrast, NtMYC2a transcription levels did not change in control plants. These data suggest that salt treatment can significantly induce the transcriptional increase of NtMYC2a.
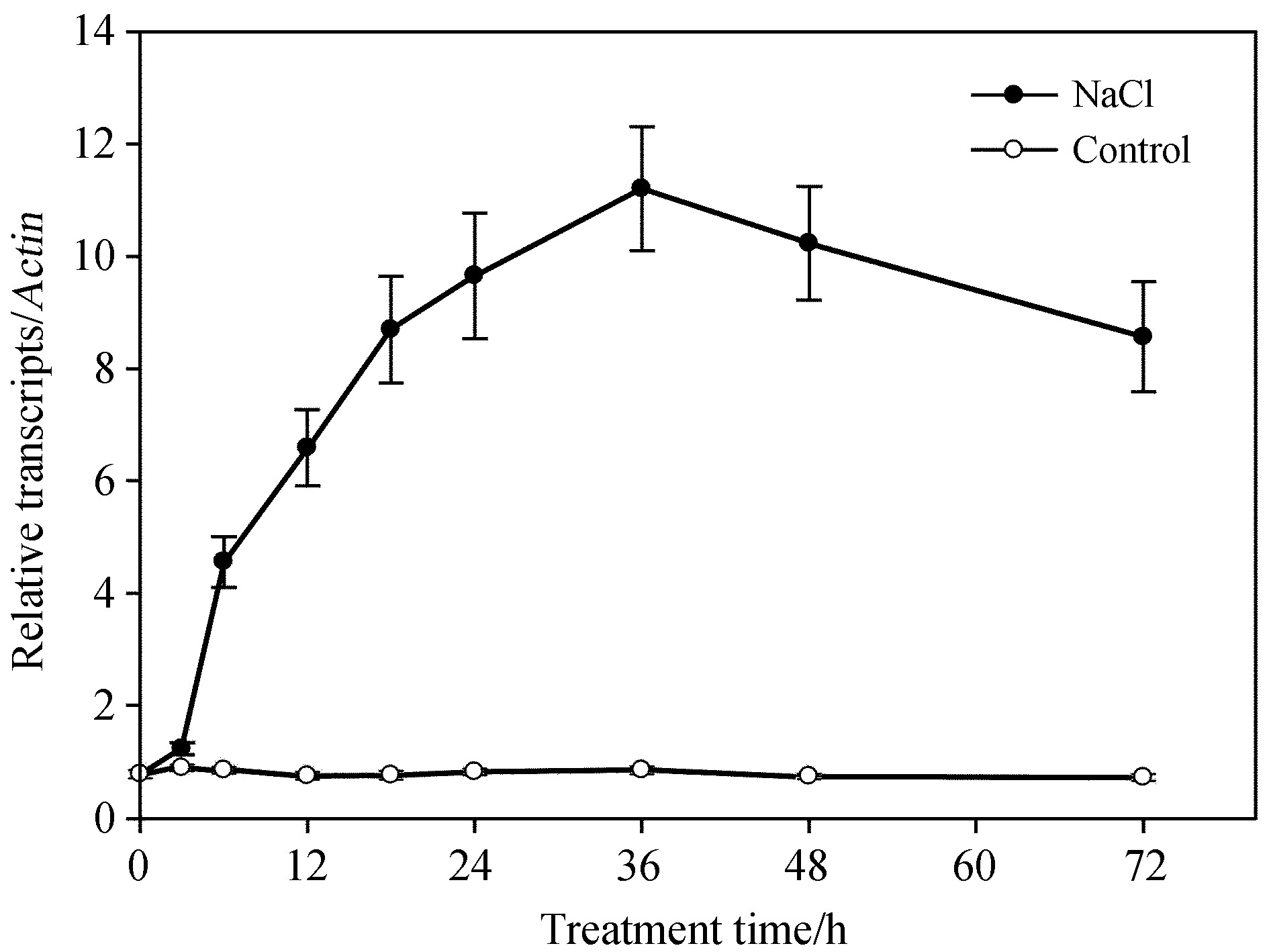
|
| Fig.2 Salt stress induced the transcriptional increase of NtMYC2a. Two-week-old seedlings were treated with 50mmol·L-1 salt solution for indicated time periods. Transcript levels were measured by qRT-PCR. Data represent the means of three replicate experiments(± SE). |
Previous results indicated that jasmonate acts as an important signal mediating the biosynthesis of nicotine in tobacco. Following jasmonate stimulation the transcription factor NtMYC2a specifically recognizes G-box(5′-CA(C/T)(G/A)T(T/A)-3′)sequences in the promoter of genes associated with nicotine biosynthesis. Here we first applied a bioinformatics approach to analyze promoter sequences of genes involved in jasmonate synthesis, including NtLOX, NtAOS, NtAOC, NtOPR. As shown in Fig. 3A, we identified several G-box elements in the promoter regions of these genes, suggesting that NtMYC2a may control their gene expression during salt stress. To test this possibility, we examined the expression of these genes after salt stress. As shown in Fig. 3B, we found that salt stress indeed induced substantial changes in the expression of these genes. At the same time, we also analyzed JA content in tobacco seedlings after salt stress. Salt treatment stimulated the accumulation of jasmonate, and JA content reached a peak after 7 days of salt stress(Fig. 4). These results indicate that salt treatment can increase jasmonate biosynthesis by inducing jasmonate synthesis-related gene transcription.
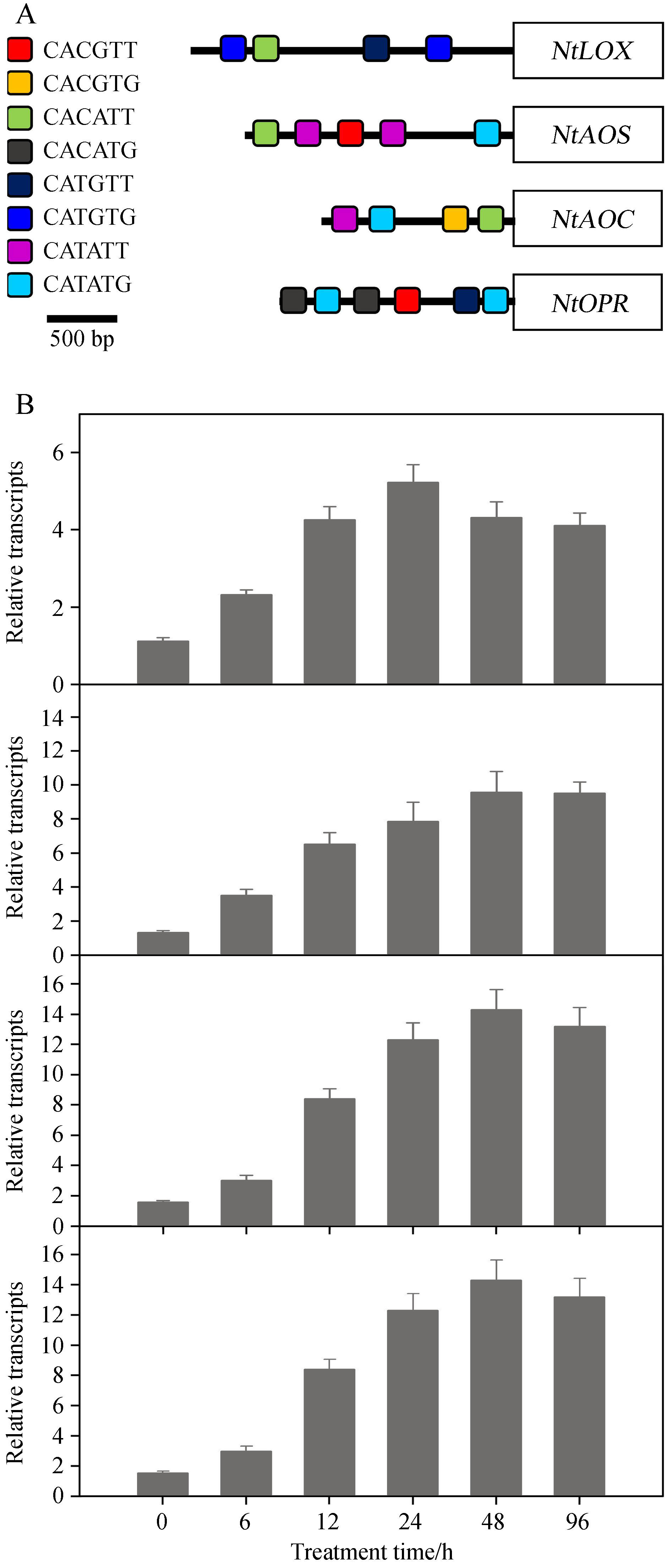
|
| Fig.3 Salt stress induced increased experession of NtLOX, NtAOS, NtAOC and NtOPR.(A)Bioinformatics were used to identify the G-box region in the promoter of NtLOX, NtAOS, NtAOC and NtOPR. Different colored boxes in the promoter region of these genes represent different motif sequences shown in the left panel.(B)Salt stress induced an increase in transcript levels of NtLOX, NtAOS, NtAOC and NtOPR. Two-week-old tobacco seedlings were treated with 50mmol·L-1 salt solution; the transcriptional levels of NtLOX, NtAOS, NtAOC and NtOPR were measured at indicated times. Data represent the means of three replicate experiments(± SE). |

|
| Fig.4 Salt stress induced jasmonate biosynthesis. Two-week-old seedlings were treated with a 50mmol·L-1 NaCl solution. Endogenous jasmoate levels were measured at various time. Data represent the means of three replicate experiments(± SE). |
Previous results indicate that exogenous application of JA can induce the accumulation of nicotine in tobacco suspension cells(Shoji et al., 2008). Our above results showed that salt stress induced JA and nicotine biosynthesis. To test whether salt-induced nicotine biosynthesis depends on JA signaling, we examined whether nicotine content of wild-type plants was affected by exogenous application of jasmonate, As shown in Fig. 5, nicotine biosynthesis was induced in plants sprayed with methyl jasmonate(MeJA; 10, 50, 100, 500nmol·L-1). Nicotine content was most effectively induced 3-6 days after treatment with 100nmol·L-1 MeJA. This result suggests that jasmonate acts as a signal factor that induces nicotine biosynthesis in tobacco plants.
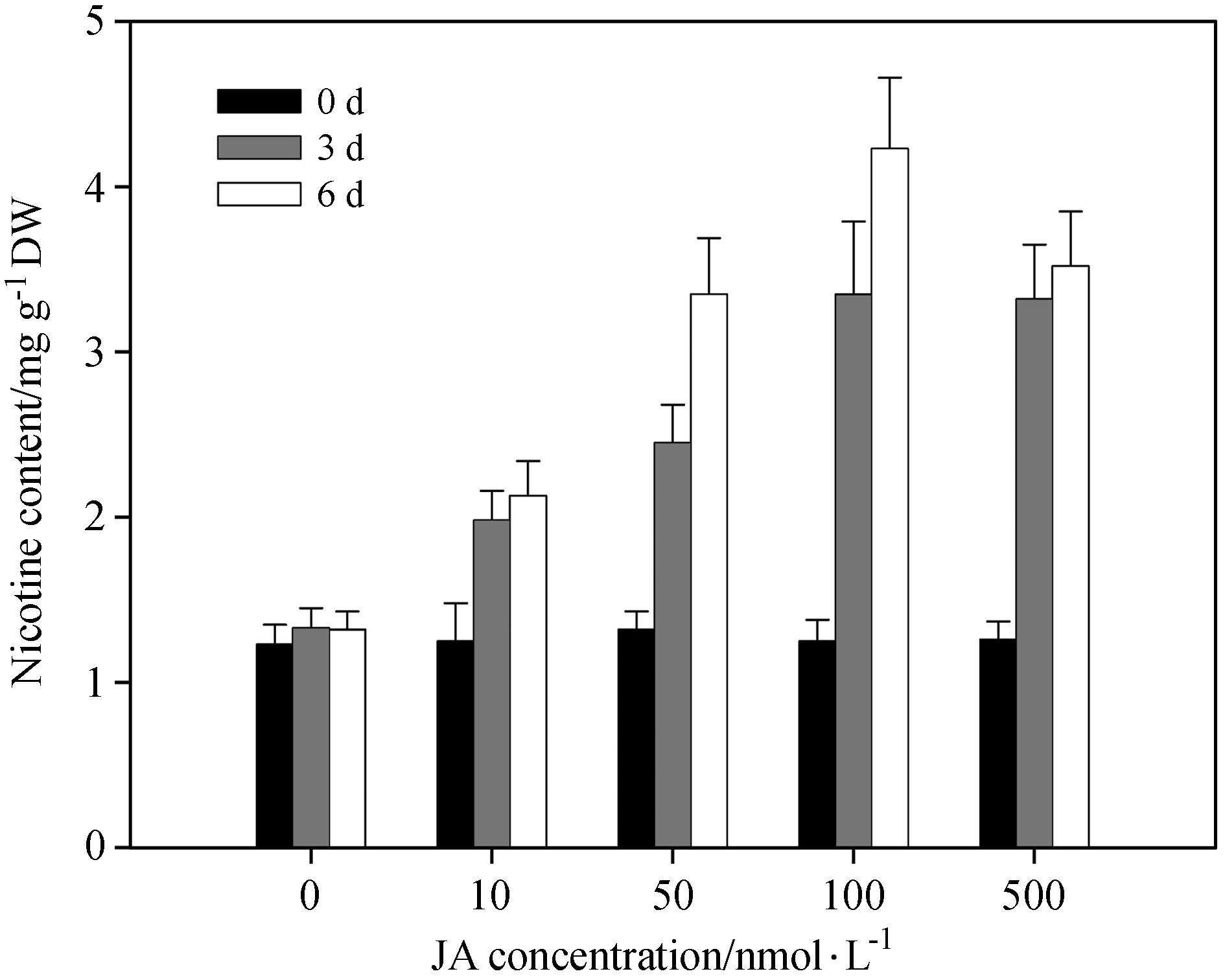
|
| Fig.5 Exogenous application of JA induced an increase in nicotine biosynthesis in tobacco root tissue. Two-week-old seedlings were sprayed with MeJA at indicated times, and nicotine content was measured in root tissue after 3 and 6 days of MeJA treatment. Data represent the means of three replicate experiments(± SE). The means denoted by different letters indicate significant differences according to Tukey′s test(P < 0.05). |
Having found that both salt and MeJA treatment directly induced nicotine biosynthesis, we then asked whether suppressing the accumulation of JA reduces nicotine biosynthesisunder salt stress. Sodium diethyldithiocarbamate(DIECA)is an inhibitor of LOX, a key enzyme in jasmonate synthesis. We treated tobacco seedlings with DIECA for 6h following salt stress. With or without jasmonate inhibition, salt-stressed plants had greater nicotine content than control plants; nicotine content in plants after DIECA treatment was lower than in untreated salt-stressed plants(Fig. 6). This data indicates that suppressing JA biosynthesis positively controls salt-induced nicotine biosynthesis in tobacco.
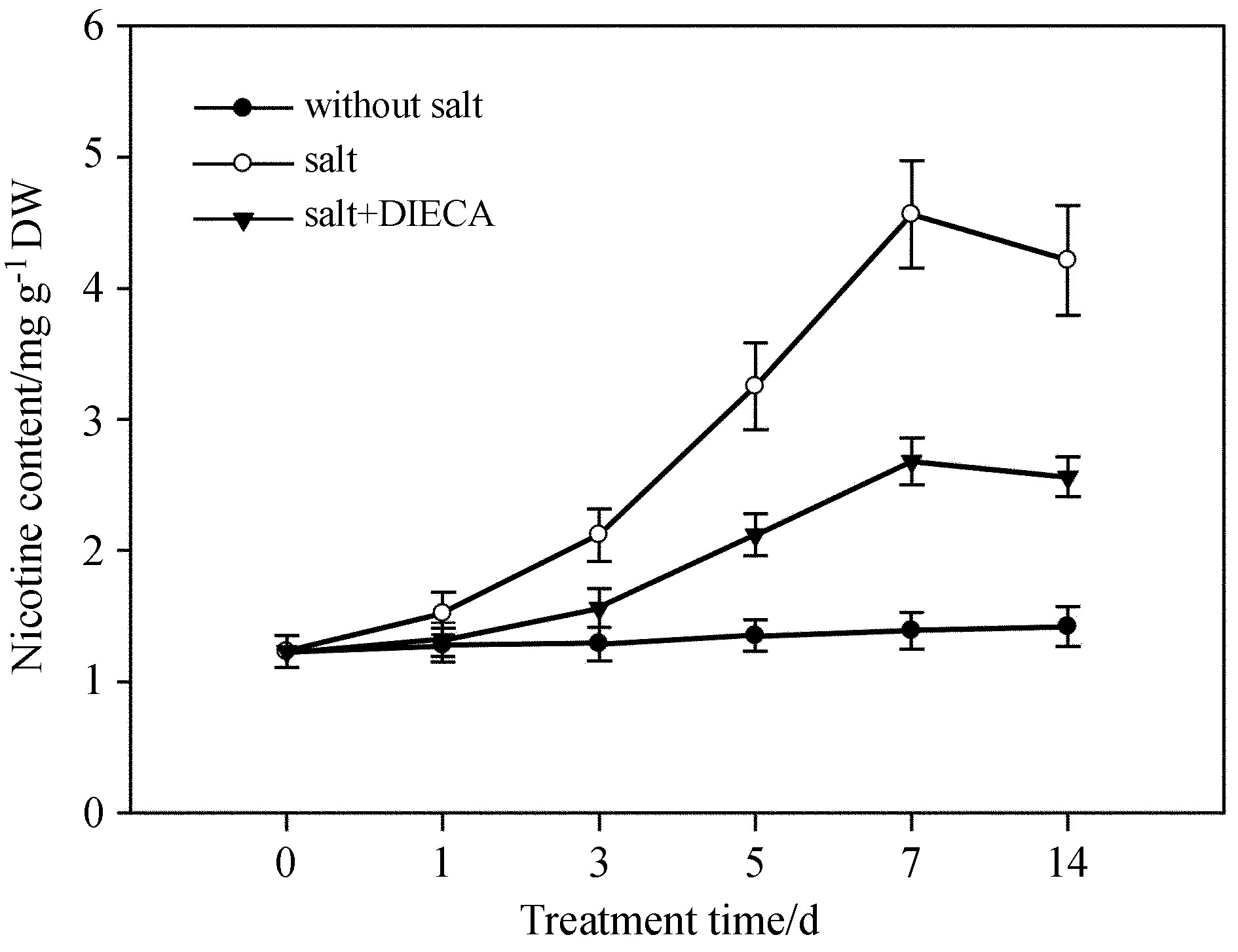
|
| Fig.6 The LOX enzyme inhibitor DIECA suppressed salt-induced nicotine biosynthesis. Two-week-old tobacco seedlings were treated with 100μmol·L-1 DIECA for 6h, following 50mmol·L-1 salt stress for indicated time, and nicotine content was measured. Data represent the means of three replicate experiments(± SE). |
NtCOI1 is a receptor of the JA signal and perceives JA stimulation in plants. Silencing NtCOI1 will lead to less nicotine after JA or less. To further underst and the role of NtCOI1 in salt-induced nicotine biosynthesis, we generated the transgenic line expressing antisense of NtCOI1 gene, these transgenic lines were termed anti-NtCOI1 lines. In three individual transgenic anti-NtCOI1 lines, we found that salt stress could not effectively induce nicotine accumulation, compared with the non-transgenic control lines(Fig. 7). This result indicates that NtCOI1 perception of the JA signal plays an essential role in salt-induced nicotine biosynthesis in tobacco.
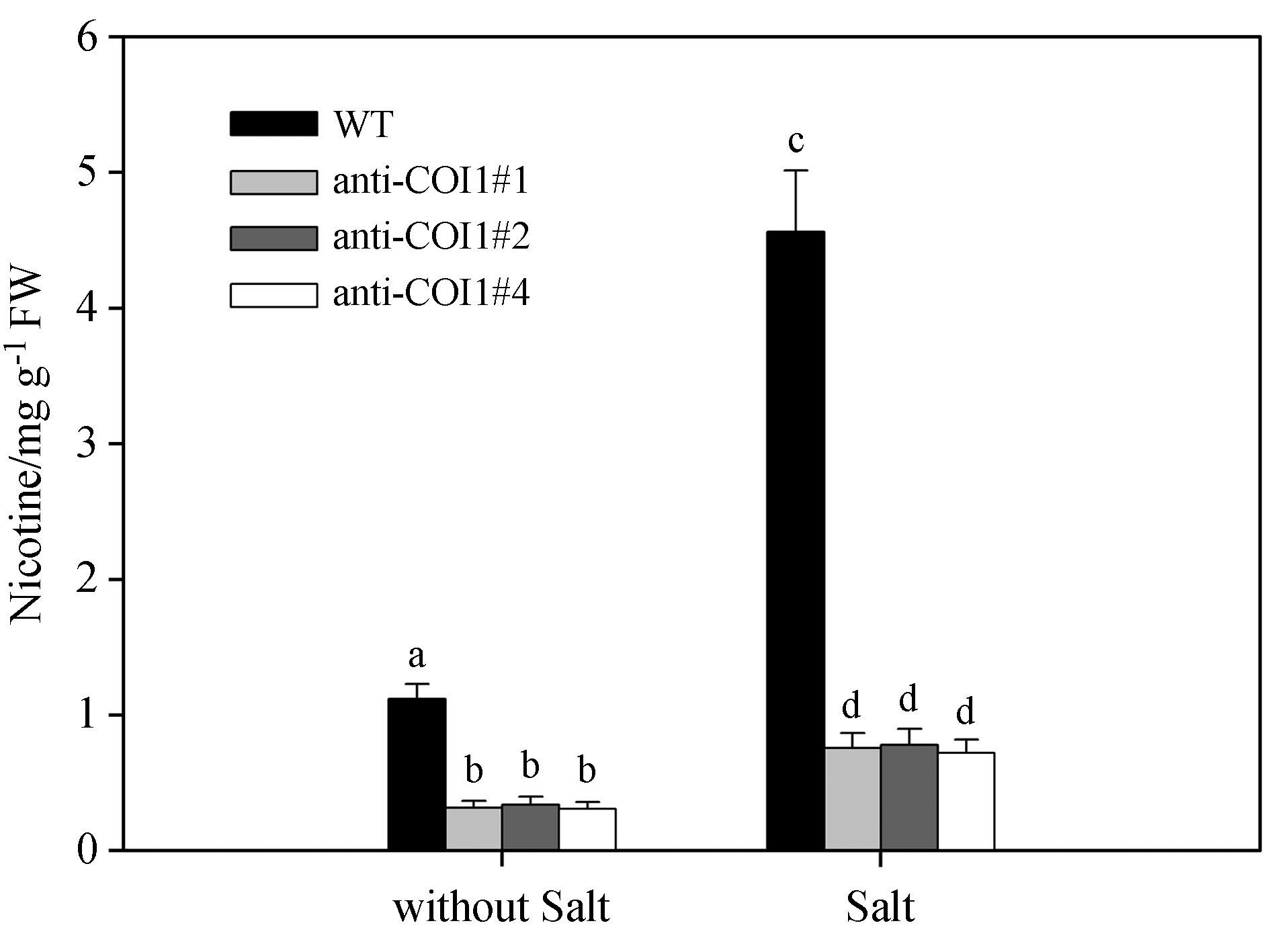
|
| Fig.7 Salt stress fails to induce nicotine biosynthesis in transgenic anti-COI1 tobacco seedlings(anti-COI1#1, anti-COI#2, anti-COI1#4). Two-week-old plants from three different coI1 mutant lines(colI1#1, coI1#2, coI1#4) and wild-type plants were treated with 50mmol·L-1 salt solution for 6 days. Nicotine content was measured. Data represent the means of three replicate experiments(± SE). The means denoted by different letters indicate significant differences according to Tukey′s test(P < 0.05). |
The plant hormone JA plays important roles in multiple processes of plant growth and development(Vick and Zimmerman, 1983; Hamberg and Hughes, 1988; Zhang et al., 2012). Previous results indicated that mechanical damage can induce the biosynthesisof JA. In tobacco plants, JA also participates in nicotine biosynthesis(Zhang et al., 2012). Our results show that salt stress leads to an increase in nicotine synthesis, accompanied by the accumulation of NtPMT1 gene transcripts(Fig. 1). At the same time, exogenous application of jasmonate can significantly induce nicotine biosynthesis in tobacco root tissue(Fig. 5), while the LOX enzyme inhibitor DIECA suppresses salt-induced nicotine biosynthesis(Fig. 6). These results suggest that jasmonate is an important signal molecule involved in regulation of nicotine biosynthesis after salt stress. In the jasmonate signaling pathway, MYC2 is an essential component that regulates the expression of a series of jasmonate-responsive genes(Hamberg and Hughes, 1988; Zhang et al., 2012). In the absence of jasmonate signaling, JAZ repressors bind to MYC2 transcription factors to suppress the activity of MYC2. Following jasmonate signal stimulation, JAZ repressors are degraded by ubiquitin proteins and MYC2 protein activity is restored. MYC2 specifically recognizes G-box cis-acting sequences found in target promoters to activate gene expression(Zhang et al., 2012). In this paper, we found that salt stress increased the transcriptional levels of NtMYC2a; therefore, we examined the expression of NtLOX, NtAOS, NtAOC, and NtOPR, genes which contain G-box sequences in their promoters and are known to be involved in JA biosynthesis in tobacco(Fig. 3A). We found salt stress treatment increased the transcriptional levels of NtLOX, NtAOS, NtAOC and NtOPR, indicating that salt stress induced the expression of NtMYC2 which subsequently activated the transcription of these genes and nicotine biosynthesis(Fig. 3B). We also noticed that short-term salt treatment(12h to 1 day)quickly led to a set of physiological responses, including increased JA and nicotine biosynthesis(Fig. 1, 4), while increasing the transcriptional levels of NtMYC2a and JA biosynthesis-related genes(Fig. 2). However, after long-term salt treatment(7 days), accumulation of JA and nicotine, and the transcriptional levels of NtMYC2a and JA biosynthesis-related genes dropped slightly compared with those following 5 days of treatment(Fig. 1, 2, 4), suggesting that JA and nicotine biosynthesis contribute to the short-term adaptation of tobacco response to salt stress. Once tobacco acclimatizes to salt stress, such as at 7 days of treatment, JA and nicotine biosynthesis may be regulated by a negative feedback loop, though the details of the regulatory mechanism require more investigations in the future.
A previous study indicated the COI1 protein, as the JA signal receptor in Arabidopsis, perceives JA stimulation. Arabidopsis coi1 mutants show insensitivity to JA stimulation, and transgenic antisense NtCOI1 tobacco plants also could not efficiently accumulate nicotine after JA treatment(Shoji et al., 2008). Our studies show that salt stress can induce an increase in jasmonate content to induce nicotine biosynthesis(Fig. 1, 4), and inhibition of jasmonate biosynthesis in hibits the salt stress-induced biosynthesis of nicotine(Fig. 6). However, in transgenic antisense NtCOI1 plants, salt stress could not efficiently induce nicotine biosynthesis(Fig. 7). This result indicates that tobacco NtCOI1 protein, as the jasmonate receptor, is involved in the nicotine biosynthesis induced by salt stress. Our above results showed that suppressing JA biosynthesis by DIECA compromised salt-induced nicotine biosynthesis. It seems that perceiving the JA signal by JA receptor NtCOI also plays an essential role in nicotine biosynthesis under salt stress.
In summary, our results show that JA acts as an essential signal mediating nicotine biosynthesis in tobacco under salt stress. During this process, salt treatment induced the expression of MYC2a transcription factors. MYC2a transcription factors then activated the expression of JA biosynthesis-related genes to induce JA generation. At the same time, the JA signal was sensed by COI1 proteins, inducing JAZ repressor protein degradation to release MYC2a activity, which then activated NtPMT1 expression, finally inducing nicotine synthesis.
Acknowledgments
This work is supported by the Major Science and Technology Program NOs. 110201101003-TS-03, 2011YN02 and 2011YN03. We thank Lin Huaming, Yang Qiuyun, BaiXuegui, Liu Tianmeng, Yin Xianlun, Zhang Wei(Kunming Institute of Botany, Chinese Academy of Sciences)for help with the tobacco leaf sample collection in the field for another tobacco project.
| Arnon D.I, Hoagland, D.R., 1940. Crop production in artificial culture solutions and in soils with special reference to factors influencing yields and absorption of inorganic nutrients. Soil Science, 50, 463-485. |
| Chattopadhyay and Ghosh, 1998. Molecular analysis of polyamine biosynthesis in higher plantsCurr. Sci, 74 (1998), 517-522. |
| Chini, A., Fonseca, S., Fernandez, G., et al., 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature, 448, 666-U664. |
| Chou, W.M., Kutchan, T.M., 1998. Enzymatic oxidations in the biosynthesis of complex alkaloids. Plant J, 15, 289-300. |
| De Luca, V., St Pierre, B., 2000. The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci, 5, 168-173. |
| De Vos, M., Van Zaanen, W., Koornneef, A., et al., 2006. Herbivore-induced resistance against microbial pathogens in Arabidopsis. Plant Physiol, 142, 352-363. |
| Dewey, R.E., Xie, J., 2013. Molecular genetics of alkaloid biosynthesis in Nicotiana tabacum. Phytochemistry, 94, 10-27. |
| Facchini, P.J., 2001. ALKALOID BIOSYNTHESIS IN PLANTS: Biochemistry, Cell Biology, Molecular Regulation, and Metabolic Engineering Applications. Annu. Rev. Plant Physiol. Plant Mol. Biol, 52, 29-66. |
| Farmer, E.E., Almeras, E., Krishnamurthy, V., 2003. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol, 6, 372-378. |
| Farmer, E.E., Caldelari, D., Pearce, G., et al., 1994. Diethyldithiocarbamic Acid Inhibits the Octadecanoid Signaling Pathway for the Wound Induction of Proteinase-Inhibitors in Tomato Leaves. Plant Physiol, 106, 337-342. |
| Gfeller, A., Liechti, R., Farmer, E.E., 2010. Arabidopsis jasmonate signaling pathway. Sci. Signal, 3, cm4. |
| Goossens, A., Hakkinen, S.T., Laakso, I., et al., 2003. A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad Sci. U.S.A, 100, 8595-8600. |
| Halim, V.A., Vess, A., Scheel, D., et al., 2006. The role of salicylic acid and jasmonic acid in pathogen defence. Plant Biol (Stuttg). 8, 307-313. |
| Hamberg, M., Hughes, M.A., 1988. Fatty acid allene oxides. III. Albumin-induced cyclization of 12, 13(S)-epoxy-9(Z), 11-octadecadienoic acid. Lipids, 23, 469-475. |
| Hu, Y., Jiang, L., Wang, F., et al., 2013. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. The Plant Cell, 25, 2907-2924. |
| Kazan, K., Manners, J.M., 2013. MYC2: the master in action. Mol. Plant, 6, 686-703. |
| Kessler, A., Baldwin, I.T., 2002. Plant responses to insect herbivory: the emerging molecular analysis. Annu. Rev. Plant Biol, 53, 299-328. |
| Khokon, M.A., Salam, M.A., Jammes, F., et al., 2015. Two guard cell mitogen-activated protein kinases, MPK9 and MPK12, function in methyl jasmonate-induced stomatal closure in Arabidopsis thaliana. Plant Biol (Stuttg). 17, 946-952. |
| Liechti, R., Farmer, E.E., 2006. Jasmonate biochemical pathway. Sci. STKE, 2006: cm3. |
| Memelink, J., 2009. Regulation of gene expression by jasmonate hormones. Phytochemistry, 70, 1560-1570. |
| Paschold, A., Halitschke, R., Baldwin, I.T., 2007. Co(i)-ordinating defenses: NaCOI1 mediates herbivore- induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J, 51, 79-91. |
| Pauwels, L., Goossens, A., 2011. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell, 23, 3089-3100. |
| Shan, X., Wang, J., Chua, L., et al., 2011. The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol, 155, 751-764. |
| Shoji, T., Hashimoto, T., 2011. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell. Physiol, 52, 1117-1130. |
| Shoji, T., Ogawa, T., Hashimoto, T., 2008. Jasmonate-induced nicotine formation in tobacco is mediated by tobacco COI1 and JAZ genes. Plant Cell. Physiol, 49, 1003-1012. |
| Spoel, S.H., Dong, X., 2008. Making sense of hormone crosstalk during plant immune responses. Cell. Host Microbe, 3, 348-351. |
| Steppuhn, A., Gase, K., Krock, B., et al., 2004. Nicotine′s defensive function in nature. PLoS Biology, 2, E217. |
| Vick, B.A., Zimmerman, D.C., 1983. The biosynthesis of jasmonic acid: a physiological role for plant lipoxygenase. Biochem Biophys Res Commun, 111, 470-477. |
| Wagner, R., Feth, F., Wagner, K.G., 1986. Regulation in tobacco callus of enzyme activities of the nicotine pathway : II. The pyridine-nucleotide cycle. Planta, 168, 408-413. |
| Xu, B.F., Timko, M.P., 2004. Methyl jasmonate induced expression of the tobacco putrescine N-methyltransferase genes requires both G-box and GCC-motif elements. Plant Mol. Biol, 55, 743-761. |
| Yamada, Y., Koyama, T., Sato, F., 2011. Basic helix-loop-helix transcription factors and regulation of alkaloid biosynthesis. Plant Signal Behav, 6, 1627-1630. |
| Yan, J., Zhang, C., Gu, M., et al., 2009. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell, 21, 2220-2236. |
| Zhang, H.B., Bokowiec, M.T., Rushton, P.J., et al., 2012. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol. Plant, 5, 73-84. |
| (Editor: Weiqi Li)Plant Diversity 1(2016)153-157 |



