2. School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, China
3. University of Chinese Academy of Sciences, Beijing 100049, China
Sulfur is involved in numerous biological functions and is necessary for a large variety of secondary metabolites, including sulfolipids, sulfated glucosides, glutathione, and coenzymes (Takahashi, 2010). Therefore, sulfur nutrition is crucial for plant growth and development. Plants obtain sulfur in the form of inorganic sulfate from the soil or as sulfur dioxide and hydrogen sulfide gases from the atmosphere. Given that inorganic sulfate has low redox potential and is nonreactive, it must be activated prior to its reduction and incorporation into organic compounds (Kopriva, 2006). The first step of the sulfate assimilation process is controlled by ATP sulfurylase, which catalyzes inorganic sulfate and ATP to form adenosine 5′-phosphosulfate (APS) and inorganic pyrophosphate (PPi). Subsequently, APS is utilized by two different sulfate assimilation pathways. In one, APS is further phosphorylated by APS kinase using ATP, resulting in the formation of 3′-phosphoadenosine 5′-phosphosulfate (PAPS), which is a high-energy SO42- donor for sulfation of macromolecules in plants. In the other, APS is reduced by adenosine 5′-phosphosulfate reductase (APR) to sulfite, which is further reduced and then incorporated into the amino acid skeleton of O-acetylserine (OAS). ATP sulfurylase activity was detected in plastids and the cytosol of spinach (Lunn et al., 1990; Renosto et al., 1993). The genes encoding cytosolic and chloroplastic APS sulfurylase have been identified in potato (Klonus et al., 1994). Subcellular fractionation results clearly showed that a cytosolic ATP sulfurylase exists in Arabidopsis (Rotte and Leustek, 2000). Arabidopsis contains four ATP sulfurylase-encoding genes, among which APS1 and APS4 were experimentally demonstrated to be localized to plastids (Leustek et al., 1994; Hatzfeld et al., 2000). Comparative protein sequence analysis revealed that the APS2 gene potentially encodes a cytosolic ATP sulfurylase (Hatzfeld et al., 2000).
Brassica species often have a high dem and for sulfur (Hawkesford and De Kok, 2006), which is associated with a high sulfate concentration in green tissue (Blake-Kalff et al., 1998). It is necessary to underst and the genetic and environmental control of sulfate accumulation to improve sulfur use efficiency in Brassica. By analyzing natural variation in shoot sulfate content in Arabidopsis, APR2 was identified to control sulfate concentration (Loudet et al., 2007). In the sulfate assimilation pathway, APR2 functions downstream of APS sulfurylase. Recently, APS1 was mapped and shown to control sulfate concentration (Koprivova et al., 2013). However, it is still unclear how and which APS gene (s) control sulfate concentration. In Arabidopsis roots, ATP sulfurylase activity is enhanced by sulfur deprivation and reduced following resupply of sulfate (Lappartient et al., 1999). In contrast, the transcript levels of APS genes are differentially affected by sulfur deprivation (Liang et al., 2010).
MicroRNAs (miRNAs), a class of 20-24nt length small RNAs, function to repress the expression of their target genes (Voinnet, 2009). Several miRNAs have been shown to regulate nutrient metabolism (Chiou et al., 2006; Abdel-Ghany et al., 2008; Liang et al., 2010, 2012), including miR395, which is involved in sulfur metabolism. It has been confirmed that three APS genes are regulated by miR395 at the post-transcriptional level during sulfate limitation (Jones-Rhoades and Bartel, 2004; Kawashima et al., 2009, 2011; Liang et al., 2010). Our previous results suggested that APS1, APS3, and APS4 are differentially regulated by sulfate starvation (Liang et al., 2010). miR395 is induced by sulfur starvation dependent on SLIM1, which plays a key role in mediating sulfur deficiency responses (Maruyama-Nakashita et al., 2006). Similarly, miR395 levels are also affected by treatments with metabolites regulating sulfate assimilation (Matthewman et al., 2012). Recent evidence suggests that over-expression of miR395 causes the constitutive repression of APS1, APS3, and APS4 under both normal and sulfate starvation conditions (Liang et al., 2010; Kawashima et al., 2011). Consequently, the sulfate concentration increases in miR395 over-expression plants (Liang et al., 2010). In this study, we further revealed that the sulfate concentration was positively correlated with abundance of miR395. Namely, elevated miR395 levels caused increased sulfate concentration, whereas down-regulation of miR395 led to reduced sulfate concentration. Correspondingly, over-expression of miR395-targeted APS genes also resulted in a decline of sulfate concentration. In contrast, cytosol-localized APS2 did not affect the sulfate concentration. Our results show that plastid-localized ATP sulfurylases are key regulators of sulfate concentration in Arabidopsis.
2. Methods 2.1. Plant growth conditionsArabidopsis thaliana seeds were surface sterilized with 20% bleach and washed three times with sterile water. Sterilized seeds were suspended in 0.1% agarose and plated on MS medium. Plates were vernalized in darkness for 2 d at 4 ℃ and then transferred to a tissue culture room at 22 ℃ under a 16-h-light/8-h-dark photoperiod. For the determination of sulfate content, seven-day-old seedlings were planted in the soil. Potted plants were grown in greenhouse at 22 ℃ and 75% humidity under a 16-h-light/8-h-dark photoperiod.
For sulfate starvation experiments, plants were grown in long-day conditions on modified MS/agar media, containing 0.7% agarose, in which the sulfate containing salts of the MS media were replaced with their chloride counterparts and the media supplemented with (NH4+) 2SO4 at the indicated concentration in figure legends. Root and shoot samples for RNA isolation were collected from 10-day-old seedlings grown vertically on MS media.
2.2. Plant materialsArabidopsis thaliana Columbia (Col-0) plants were used as wild type in this study. Putative aps2 T-DNA insertion mutant was obtained from the ABRC. The T-DNA line was in the Col background. Homozygous mutant was identified using gene-specific primers flanking the insertion site. 35S∶MIR395d, aps1-1, aps3, and APS4-RNAi5 plants were described previously by Liang (2010) .
2.3. Agrobacterium tumefaciens infiltration in Nicotiana benth-amianaThe constructs, 35S∶MIR395d/empty vector and 35S∶APS/35S∶mAPS, were co-expressed with 35S∶GUS in tobacco leaves by Agrobacterium tumefaciens strain EHA105. For co-infiltration experiments, the final OD600 value for Agrobacterium were 1.65 (35S∶MIR395d/empty vector), 0.25 (35S∶APS/35S∶mAPS), and 0.1 (35S∶GUS). The expression levels of APS or mAPS were normalized to GUS.
2.4. Northern blot and real-time RT-PCRTotal RNA was isolated from roots or shoots. For high-molecular-weight RNA gel blot analysis, 20 μg of total RNA was separated on a 1.5% agarose gel and transferred to Hybond-N+ membranes. Probes were labeled with [α-32P] dATP using Klenow fragment (Takara). Low-molecular-weight RNAs were separated by electrophoresis on denaturing 15% polyacrylamide gels, and miRNA gel blot hybridizations were performed as described (Lu et al., 2005). For small RNA gel blots, 20 μg of total RNA was separated on a 15% polyacrylamide gel with 7 M urea. DNA oligonucleotides complementary to miR395 were end-labeled using T4 polynucleotide kinase and used for hybridizations.
After total RNA was digested with the use of DNase I (Fermentas), reverse transcription was carried out according to the manufacturer′s protocol (Fermentas). One microgram of total RNA was used for cDNA synthesis. mRNAs were then converted into cDNAs using M-MLV reverse transcriptase (Fermentas) with an oligo (dT) 18 primer. For quantification, ten microliters of 2 X PreMix ExTaq (Takara) plus SYBR Green was used for a 20-mL qPCR reaction. The PCR program consisted of one cycle (95 ℃, 3 min), 45 cycles (95 ℃, 10 s; 60 ℃, 10 s; 72 ℃, 10 s), and one cycle (72 ℃, 5 min), followed by a melting curve program (55 to 90 ℃ in increasing steps of 1 ℃). Normalization was achieved using ACT2 amplification as constitutive controls. Three technical repeats were performed for each reaction. Two or three biological repeats were conducted and one representative biological repeat is shown.
2.5. Determination of sulfateRoots or shoots were harvested separately from 3-week-old plants grown in the soil and used for determination of sulfate content. Measurement of sulfate content was performed as described previously (Liang et al., 2010). Briefly, about 20 mg of fresh plant samples were immersed in 1 mL of 0.1 mol·L-1 HCl for two hours at room temperature. The supernatant was recovered and used for the determination of sulfate content according to the turbidimetric method (Tababai and Bremner, 1970). Appropriate calibration curves were constructed, using potassium sulfate as the st and ard.
2.6. Plasmid constructionThe STTM395 sequence was synthesized and subcloned between 35S promoter and polyA in the binary vector pOCA30 (Liang et al., 2010). For construction of APS gene over-expression vector, their corresponding genomic sequences were amplified and subcloned between 35S promoter and polyA in the binary vector pOCA30. The nucleotide substitution of the miR395 recognition site in the APS genes was conducted by over-lapping PCR. The cDNA of APS2 was amplified by RT-PCR and fused in frame to the 5′ terminal of GFP gene. For over-expression, the recombinant APS2-GFP sequence was subcloned between 35S promoter and polyA in the binary vector pOCA30. For transient expression, the APS2-GFP or GFP sequence was inserted into the downstream of 35S promoter in pGreeenⅡ 62-SK vector.
2.7. Accession numbersSequence data from this article can be found in The Arabidopsis Information Resource under accession numbers APS1 (AT3G22890), APS2 (AT1G19920), APS3 (AT4G14680), and APS4 (AT5G43780). The T-DNA insertion mutants used in this article: aps2 (SAIL_775_D12).
3. Results 3.1. Disruption of miR395 expression affects sulfate concentrationOur previous study showed that over-expression of miR395 caused over-accumulation of sulfate in Arabidopsis (Liang et al., 2010). Therefore, we hypothesized that a reduction of miR395 transcript levels would lead to reduced sulfate accumulation. To confirm this hypothesis, transgenic plants with reduced miR395 abundance were generated. The short t and em target mimic (STTM) strategy (Yan et al., 2012) was emplo-yed to repress the expression of miR395. As shown in Fig. 1A, two target mimic sequences, which were predicted to be bound by miR395a/d/e and miR395b/c/f, were separated by an em-pirically determined 48-nt DNA sequence linker. The STTM395 sequence was driven by the 35S promoter and introduced into Arabidopsis plants by Agrobacterium tumefaciens. Northern blot analysis was used to confirm that miR395 transcript levels were reduced in STTM395 plants. (Fig. 1B). Both wild type and transgenic seedlings were grown under sulfur starvation conditions for 10 days, and the shoots were harvested for RNA extraction and northern blot analysis. In all STTM395 plants examined, the miR395 abundance was significantly lower than that in the wild type, indicating that STTM395 effectively reduced the expression of miR395. Next, we determined the sulfate concentration in the roots and shoots of 3-week-old plants grown in the soil (Fig. 1C). In contrast to the high sulfate concentration in the shoots of miR395d over-expression plants (MIR395d-OX), STTM395 transgenic plants had a significantly lower sulfate concentration in comparison with wild type plants. Although the root sulfate concentration was low in MIR395d-OX plants compared with wild type plants, we did not observe a significant root sulfate concentration difference between wild type and STTM395 plants.

|
| Fig.1 Phenotypes of STTM395 plants. (A) Diagram of STTM395 structure showing the design strategy. A synthetic DNA sequence containing two miR395 mimic target sites and a 48-nt length spacer was inserted between 35S promoter and polyA; (B) Northern blot analysis of STTM395 transgenic plants. 10-day-old seedlings grown on sulfate-free medium were used for RNA extraction. rRNA and tRNA served as an internal control; (C) Measurement of sulfate concentration. Roots and shoots were harvested separately from 3-week-old plants grown in the soil and used for determination of sulfate content. The values indicated by an asterisk were significantly different from the wild-type value (P < 0.05, n=10, Student′s t-test). |
Here, we further investigated the expression of APS genes in wild type, MIR395d-OX and STTM395 plants (Fig. 2). In wild type plants, APS1 expression was not significantly affected by sulfur starvation. In contrast, APS1 was significantly induced by sulfur limitation in STTM395 plants. APS3 was up-regulated by sulfur limitation in the roots and shoots of wild type plants. APS3 expression in the roots of STTM395 plants was comparable with that in the roots of wild type plants. However, in sulfate-starved shoots, APS3 was repressed in STTM395 plants compared with wild type plants. As for APS4, transcript levels were suppressed by sulfate starvation in wild type plants. Although APS4 expression was increased in the roots and shoots of STTM395 plants compared with wild type plants independent of sulfate conditions, it was down-regulated in the roots of STTM395 plants by sulfate starvation and up-regulated in shoots. These data suggest that, in addition to regulation by miR395, APS1, APS3 and APS4 are also regulated by other factors.
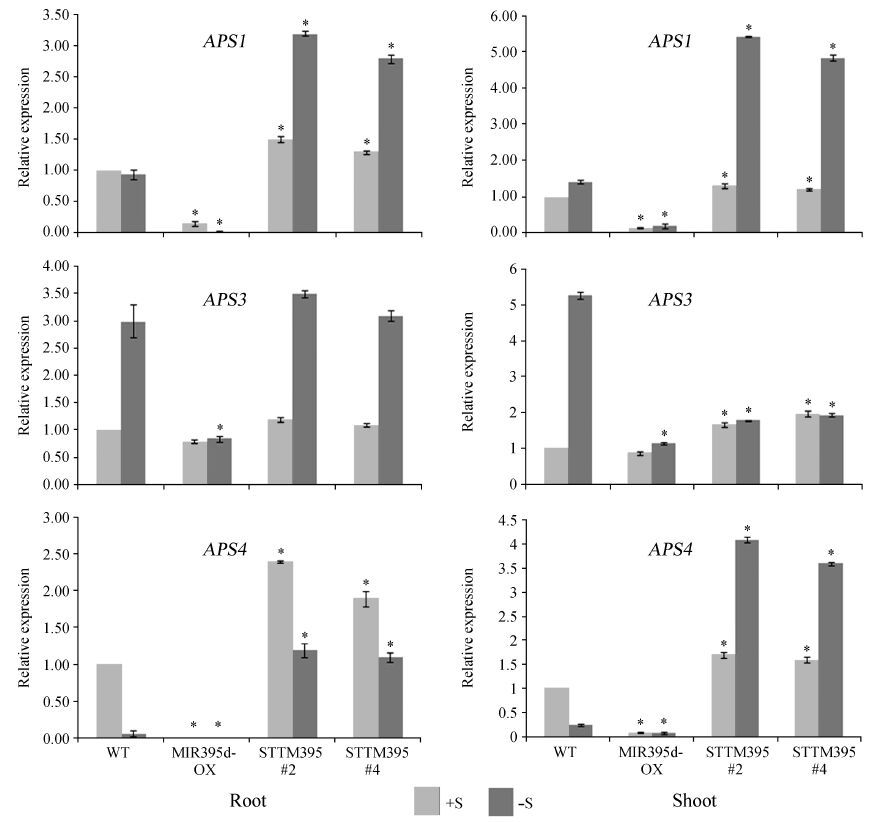
|
| Fig.2 Relative expression of APS1, APS3, and APS4 genes in wild type, MIR395d-OX, and STTM395 plants. Roots and shoots from 10-day-old seedlings grown on normal (+S) or sulfate-free (-S) MS medium were used for RNA extraction. The data represent means (± SE) of three biological repeats. Significant differences from the corresponding wild type are indicated by an asterisk (P < 0.05, n=3, Student′s t-test). ACT2 genes served as an internal control for qRT-PCR. |
Given that miR395 directly represses the APS1, APS3, and APS4 genes, which encode ATP sulfurylases responsible for sulfate activation, we attributed the decline of sulfate concentration in STTM395 plants to the up-regulation of these APS genes. To investigate this hypothesis, constructs in which the APS1, APS3, and APS4 genes were individually driven by the 35S promoter were created and 35S∶APS1, 35S∶APS3 and 35S∶APS4 transgenic plants were obtained (Fig. 3A). Sulfate concentration measurements indicated that sulfate levels in these transgenic plants were moderately lower than those in wild type plants (Fig. 3B).
Given that these APS genes are cleaved by miR395, we wanted to know whether miR395-resistant versions of the APS genes would cause a stronger decline in sulfate levels than wild type APS genes. Therefore, we introduced synonymous substitutions into the miR395 recognition regions of the APS genes and simultaneously produced a restriction enzyme recognition site for each target (Fig. 3C). To confirm whether these mutant APSs were insensitive to miR395 cleavage, we performed an Agrobacterium tumefaciens-mediated transient assay by co-expressing the miR395 gene (35S∶MIR395d) with the wild type APS genes (35S∶APS1, 35S∶APS3, or 35S∶APS4) or with their mutated versions (35S∶mAPS1, 35S∶mAPS3, or 35S∶mAPS4) (Fig. 3D-F). Two days after inoculation, the wild-type APS mRNAs were strongly reduced, whereas the mutant mRNAs were not affected by miR395. These results confirmed that the mutant APS mRNAs were completely resistant to miR395. To investigate the effect of the mutant APSs on sulfate concentration, 35S∶mAPS1, 35S∶mAPS3, and 35S∶mAPS4 transgenic plants were generated (Fig. 3A). To confirm that the existence of mutant APS transcripts, we amplified the full-length APS cDNAs. After digestion with the corresponding restriction enzymes, the mutated APS cDNAs were cleaved into two b and s while the wild type cDNAs were unaffected by the restriction enzymes (Fig. 3A). These results suggest that the mutant APS genes were present in the transgenic plants. Subsequently, the sulfate levels in these transgenic plants were determined (Fig. 3B). Except for 35S∶mAPS3, the mutant APS genes caused significantly lower sulfate levels than their wild type counterparts, indicating that elevated expression of miR395-targeted APS is sufficient to reduce the sulfate concentration.
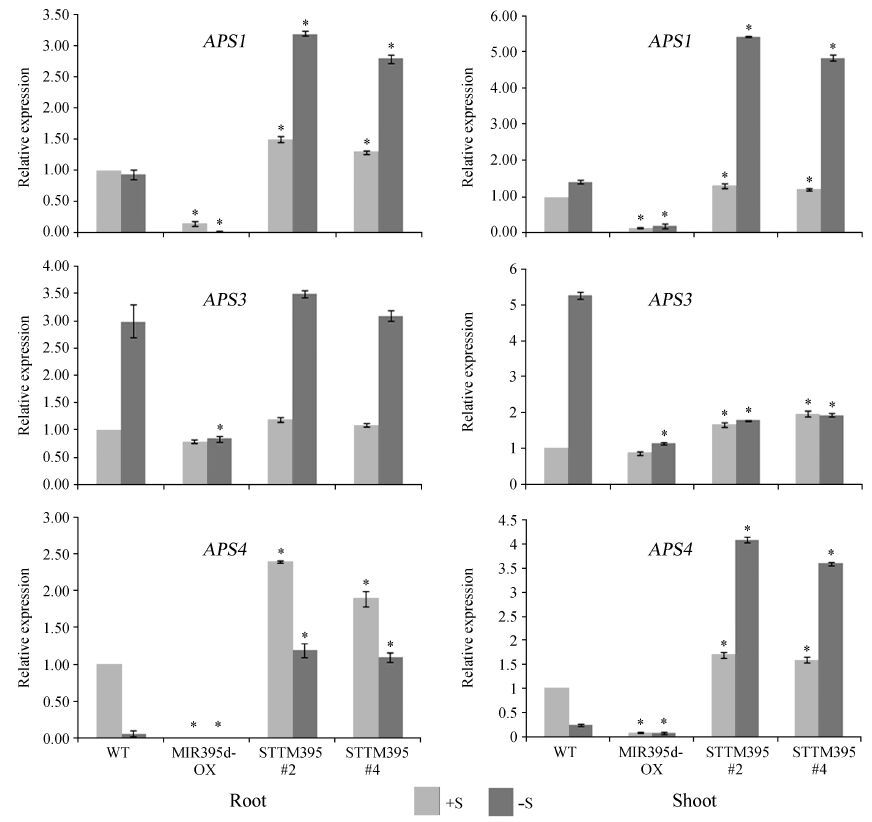
|
| Fig.3 Direct suppression of APS1, APS3, and APS4 by miR395. (A) Determination of different APS transgenic plants. Northern blot was used for determining the expression of different APS genes. Full-length APS cDNA from RT-PCR was digested with the corresponding restriction enzymes shown in Fig. 3A and then used for gel analysis; (B) Measurement of sulfate concentration. Shoots were harvested from 3-week-old plants grown in the soil and used for determination of sulfate content. An asterisk indicates significant difference from the wild-type (P < 0.05; n=10, Student′s t-test) ; (C) Design of miR395-insensitive version of APS genes. The characters underlined indicate mutated nucleotides.‘EcoRI’ and ‘PstI’ indicate the restriction enzyme recognition sites; (D), (E) and (F) Co-expression of various combinations of miR395 and APS expression constructs in N.benthamiana. 35S∶GUS construct was coexpressed as an internal control. Real-time RT-PCR quantifications were normalized to the expression of GUS gene. Error bars represent SE for three biological repeats. ‘+’ and ‘-’indicate the presence and absence of the corresponding constructs, respectively. |
We found that miR395 transcript abundance was positively correlated with sulfate concentration. Moreover, over-expression of its APS target genes was sufficient to down-regulate sulfate levels. Therefore, we hypothesized that the regulation of APS genes by miR395 accounts for the miR395-mediated sulfate concentration control. To confirm this hypothesis, 35S∶MIR395d-OX plants were crossed with 35S∶APS1, 35S∶mAPS1, 35S∶APS4, and 35S∶mAPS4 plants. Heterozygous progenies were identified by PCR. The qRT-PCR analysis suggested that wild type APS transcripts were significantly down-regulated by miR395 whereas mutant transcripts were not affected (Fig. 4A). The sulfate concentration in the heterozygous progenies was then measured. As shown in Fig. 4B, the sulfate concentration in 35S∶MIR395d-OX/35S∶APS1 and 35S∶MIR395d-OX/35S∶APS4 plants was comparable to that in 35S∶MIR395d-OX. In contrast, the sulfate concentration in 35S∶MIR395d-OX/35S∶mAPS1 plants was comparable to that in 35S∶mAPS1 plants. A similar case was also observed between 35S∶MIR395d-OX/35S∶mAPS4 and 35S∶mAPS4 plants. These results suggest that miR395-mediated sulfate concentration control is dependent on the repression of APS genes by miR395.
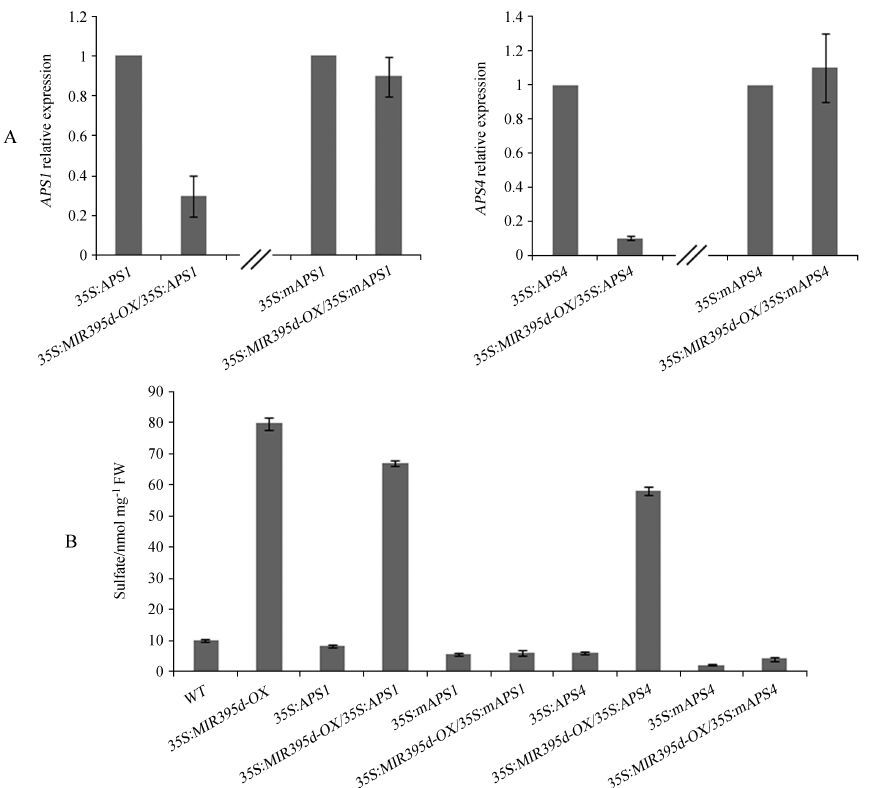
|
| Fig.4 mAPS1 and mAPS4 reverse the phenotypes of miR395 over-expression plants. (A) Relative expression of APS1 and APS4. Shoots of 3-week-old seedlings grown in the soil were used for RNA extraction. ACT2 served as an internal control for qRT-PCR; (B) Measurement of sulfate concentration. Shoots were harvested from 3-week-old plants grown in the soil and used for determination of sulfate content. |
Similar to the expression pattern of APS4 in response to sulfate limitation (Fig. 2), APS2 was repressed in both roots and shoots by sulfate deficiency (Fig. 5A). Its expression pattern negatively correlated with that of miR395. To confirm whether APS2 is regulated by miR395, its transcript levels were determined in miR395d-OX and STTM395 plants (Fig. 5B). In both the roots and shoots of examined plants, APS2 expression was independent of miR395 under both normal and sulfate starvation conditions. These results perfectly agree with the fact that APS2 is not targeted by miR395.
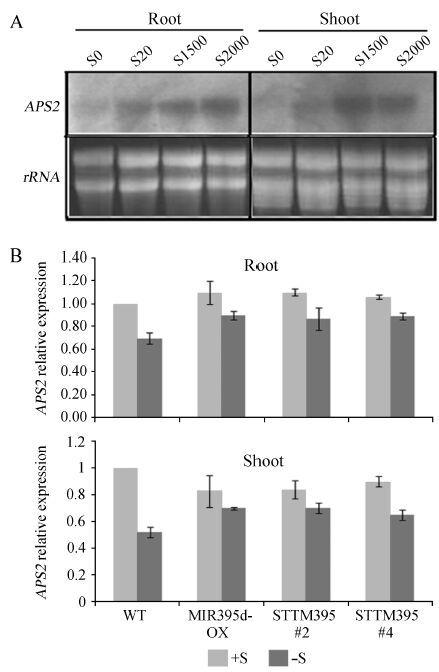
|
| Fig.5 APS2 gene expression. (A) Expression of APS2 gene in response to various sulfate concentrations. S0, S20, S1500, and S2000 indicate 0 μmol·L-1, 20 μmol·L-1, 1500 μmol·L-1, and 2000 μmol·L-1 sulfate, respectively; (B) Relative expression of the APS2 gene. Roots and shoots from 10-day-old seedlings grown on normal (+S) or sulfate-free (-S) MS media were used for RNA extraction. ACT2 served as an internal control for qRT-PCR. |
To investigate whether the APS2 protein contributes to the sulfate concentration in cells, we used available reverse genetics resources. One T-DNA insertion allele for the APS2 gene in the Columbia-0 genetic background was obtained from the Arabidopsis Biological Resource Center (ABRC) (Fig. 6A). A homozygous line was isolated by PCR. The RT-PCR results showed that no complete APS2 transcript was present in the aps2 mutant (Fig. 6A), which indicated that this mutant was an APS2 knock-out mutant. We subsequently determined the sulfate concentration in wild type plants, aps1-1, aps2 and aps3 mutants, and APS4-RNAi-5 transgenic plants. In agreement with previous results, the aps1-1 mutant and APS4-RNAi-5 transgenic plants accumulated significantly higher concentrations of sulfate compared with the wild type. In contrast, the sulfate concentration in the aps2 and aps3 mutants was similar to that in the wild type plants.

|
| Fig.6 Functional analysis of the APS2 gene. (A) Identification of the APS2 T-DNA mutant. A T-DNA insertion was established in the first intron of APS2 gene. RT-PCR confirmed that no APS2 full length cDNA was amplified from aps2 mutant; (B) Identification of APS2-GFP transgenic plants. qRTPCR confirmed the over-expression of the APS2 gene. RT-PCR confirmed the expression of the GFP gene; (C) Measurement of sulfate concentration. Shoots were harvested from 3-week-old plants grown in the soil and used for determination of sulfate content. |
Considering that the loss-of-function of APS2 might be compensated by other APS proteins, we generated 35S∶APS2-GFP transgenic plants (Fig. 6B). qRT-PCR analysis of APS2 transcript levels suggested that APS2 was highly expressed in transgenic plants. Correspondingly, GFP transcript levels were also determined by RT-PCR, suggesting that GFP transcript was only present in transgenic plants. Two independent transgenic lines were then used for sulfate concentration analysis. Unexpectedly, the sulfate concentration in both transgenic lines was comparable with wild type plants. These results implied that APS2 had no effect on sulfate concentration.
4. DiscussionSulfur nutrition is important for crop improvement because of a higher incidence of sulfur limitation by low sulfur deposition from the air. Sulfur deficiency is directly associated with crop yields and quality (McGrath et al., 1996; Jez, 2008). Thus, a better underst and ing of the control of sulfur metabolism is essential for improving crop sulfur nutrition, especially under low sulfur availability conditions. The investigation of sulfur assimilation is thus a prerequisite to optimize sulfur fluxes in these plants and improve sulfur use efficiency.
The first step of sulfate assimilation is the activation of sulfate by ATP sulfurylase. In Arabidopsis thaliana, four ATP sulfurylase encoding genes (APS1, 2, 3, 4) have been previously identified. In our study, these APS genes were differentially expressed in response to sulfate starvation (Fig. 2 and 5A). Previously, miR395 was shown to regulate the expression of APS1, APS3 and APS4 (Kawashima et al., 2009, 2011; Liang et al., 2010) and to repress APS1 expression irrespective of sulfate conditions. We found that miR395 was greatly induced by sulfate starvation whereas APS1 was not significantly responsive to sulfate starvation (Fig. 2), and therefore, when the induction of miR395 is blocked, APS1 should be induced by sulfate starvation. As expected, its expression was greatly up-regulated by sulfate starvation in STTM395 plants in which miR395 induction was disrupted (Fig. 1B). These results imply that APS1 is up-regulated by sulfate starvation at the transcriptional level, but down-regulated by miR395 at the post-transcriptional level. In both roots and shoots of wild type plants, APS3 was induced by sulfate starvation. It is worth noting that its induction was decreased in the shoots, but not in the roots, of STTM395 plants (Fig. 2). In contrast to APS3, APS4 was repressed by sulfate starvation in both the roots and shoots of wild type plants (Fig. 2). However, in STTM395 plants, APS4 was repressed by sulfate starvation in roots but induced in shoots (Fig. 2). Thus, shoot APS3 and root APS4 might be repressed by sulfate starvation via other unknown factors rather than miR395. Previous studies revealed that SLIM1 is a central transcription regulator of sulfur responses and metabolism (Maruyama-Nakashita et al., 2006), and that the induction of miR395 by sulfate starvation is dependent on SLIM1 (Kawashima et al., 2009). Similar to our results (Fig. 2), APS4 was also repressed in the roots of slim1 mutant plants by sulfate starvation (Maruyama-Nakashita et al., 2006), which implies that SLIM1 is not responsible for the repression of APS4 by sulfate starvation.
In contrast to miR395-targeted APS genes, which encode proteins localized to plastids, APS2 was predicted to encode a cytosol-localized APS protein (Hatzfeld et al., 2000). Bohrer et al. (2015) confirmed the cytosolic localization of APS2 but the function of APS2 remains unknown. The plastid form of ATP sulfurylase is most abundant in the youngest leaves and declines progressively during development whereas the cytosolic form increases with leaf age (Rotte and Leustek, 2000), which implies that the plastid and cytosolic forms of ATP sulfurylase may have specialized functions. Our data support this hypothesis because over-expression of plastid ATP sulfurylases (APS1, APS3 and APS4) facilitated the reduction of sulfate concentration whereas over-expression of cytosolic ATP sulfurylase (APS2) had no significant effect on sulfate levels. Inorganic sulfate entering the plant cell is mainly stored in vacuoles (Kaiser et al., 1989; Martinoia et al., 2000). This sulfate can be remobilized by sulfate transporters localized in the tonoplast, and enters the assimilation pathway in the plastid (Kataoka et al., 2004). The plastid is the major site for sulfate consumption (Leustek et al., 2000). This may be the reason why the elevated expression of plastid ATP sulfurylases is sufficient to reduce the sulfate concentration. Mutant analysis indicated that the lack of APS1 or APS4, but not APS2 or APS3, causes significant over-accumulation of sulfate, implying that APS1 and APS4 are the major regulators of sulfate concentration. Interestingly, although each of the plastid ATP sulfurylases could directly affect sulfate levels, only over-expression of APS4 led to a large reduction of sulfate concentration. It is likely that these plastid ATP sulfurylases have different sulfate activation efficiencies. Under sulfate starvation conditions, both the ATP sulfurylase activity and sulfate influx in the root are greatly increased (Lappartient et al., 1999). However, among the four APS genes, only APS3 is clearly up-regulated in the root by sulfate starvation. This implies that APS3 may play a key role under sulfate starvation conditions. Both over-expression and mutant analysis revealed that cytosolic APS2 has no significant effect on sulfate levels. Consistent with our results, the over-expression of APS2 in bright yellow 2 tobacco cells did not affect cellular sulfate levels (Hatzfeld et al., 1998). It is very likely that the proportion of cytosolic sulfate to the total sulfate of the whole cell is very low.
The identification of limiting factors for sulfate assimilation is crucial for improving the sulfur use efficiency of plants. Previous studies suggested that sulfite formation mediated by APS reductase is the limiting step in the assimilative sulfate reduction pathway of A.thaliana (Tsakraklides et al., 2002; Martin et al., 2005). In agreement with this, the loss-of-function mutant of Arabidopsis APR2 shows sulfate accumulation (Loudet et al., 2007). Over-expression of Arabidopsis APS1 increased assimilative sulfate reduction in Brassica juncea (Pilon-Smits et al., 1999), whereas over-expression of Arabidopsis APS2 had no effect on assimilative sulfate reduction in tobacco (Hatzfeld et al., 1998), implying that plastid ATP sulfurylases are limiting enzymes for sulfate assimilation. Consistent with this, the observed loss-of-function of plastid ATP sulfurylases also leads to sulfate accumulation, as previously shown by Liang et al. (2010) and Koprivova et al. (2013) . All this suggests that activation of sulfate by plastid ATP sulfurylases is a limiting step for the assimilative sulfate reduction pathway in A.thaliana.
AcknowledgementsWe thank the Arabidopsis Biological Resource Center for the support of T-DNA insertion mutants. We are grateful for the support of the National Natural Science Foundation of China [Grant No. 31100186]. We thank the anonymous reviewers for their constructive comments, which helped us to improve the manuscript.
| [1] | Abdel-Ghany, S.E., Pilon, M., 2008. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem, 283, 15932-15945. |
| [2] | Blake-Kalff, M.M., Harrison, K.R., Hawkesford, M.J. et al., 1998. Distribution of sulfur within oil seed rape leaves in response to sulfur deficiency during vegetative growth. Plant Physiol, 118, 1337-1344. |
| [3] | Bohrer, A.S., Yoshimoto, N., Sekiguchi, A., et al., 2015. Alternative translational initiation of ATP sulfurylase underlying dual localization of sulfate assimilation pathways in plastids and cytosol in Arabidopsis thaliana. Front. Plant Sci, 5, 750. doi: 10.3389/fpls.2014.00750 . |
| [4] | Chiou, T.J., Aung, K., Lin, S.I., et al., 2006. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell, 18, 412-421. |
| [5] | Hawkesford, M.J., DeKok, L.J., 2006. Managing sulfur metabolism in plants. Plant Cell Environ, 29, 382-395. |
| [6] | Hatzfeld, Y., Cathala, N., Grignon, C., et al., 1998. Effect of ATP sulfurylase overexpression in Bright Yellow 2 tobacco cells regulation of ATP sulfurylase and SO42- transport activities. Plant Physiol, 116, 1307-1313. |
| [7] | Hatzfeld, Y., Lee, S., Lee, M., et al., 2000. Functional characterization of a gene encoding a fourth ATP sulfurylase isoform from Arabidopsis thaliana. Gene, 248, 51-58. |
| [8] | Jez, J., 2008. Sulfur: A Missing Link between Soils, Crops, and Nutrition. Agronomy Monography. American Society of Agronomy. |
| [9] | Jones-Rhoades, M.W., Bartel, D.P., 2004. Computational identification of plant microRNAs and their targets, including a stress induced miRNA. Mol. Cell, 14, 787-799. |
| [10] | Kaiser, G., Martinoia, E., Schroppelmeier, G., et al., 1989. Active-transport of sulfate into the vacuole of plant cells provides halotolerance and can detoxify SO2. J. Plant Physiol, 133, 756-763. |
| [11] | Kataoka, T., Watanabe-Takahashi, A., Hayashi, N., et al., 2004. Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell, 16, 2693-2704. |
| [12] | Kawashima, C.G., Yoshimoto, N., Maruyama-Nakashita, A., et al., 2009. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J, 57, 313-321. |
| [13] | Kawashima, C.G., Matthewman, C.A., Huang, S., et al., 2011. Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J, 66, 863-876. |
| [14] | Klonus, D., Hfgen, R., Willmitzer, L., et al., 1994. Isolation and characterization of two cDNA clones encoding ATP sulfurylases from potato by complementation of a yeast mutant. Plant J, 6, 105-112. |
| [15] | Kopriva, S., 2006. Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot, 97, 479-495. |
| [16] | Koprivova, A., Giovannetti, M., Baraniecka, P., et al., 2013. Natural variation in the ATPS1 isoform of ATP sulfurylase contributes to the control of sulfate levels in Arabidopsis. Plant Physiol, 163, 1133-1141. |
| [17] | Lappartient, A.G., Vidmar, J.J., Leustek, T., et al., 1999. Inter-organ signaling in plants: regulation of ATP sulfurylase and sulfate transporter genes expression in roots mediated by phloem-translocated compound. Plant J, 18, 89-95. |
| [18] | Leustek, T., Murillo, M., Cervantes, M., 1994. Cloning of a cDNA encoding ATP sulfurylase from Arabidopsis thaliana by functional expression in Saccaromyces cerevisiae. Plant Physiol, 105, 897-902. |
| [19] | Leustek, T., Martin, M.N., Bick, J.A., et al., 2000. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Ann. Rev. Plant Physiol. Plant Mol. Biol, 51, 141-165. |
| [20] | Liang, G., Yang, F.X., Yu, D.Q., 2010. MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J, 62, 1046-1057. |
| [21] | Liang, G., He, H., Yu, D., 2012. Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS ONE, 7(11), e48951. |
| [22] | Loudet, O., Saliba-Colombani, V., Camilleri, C., et al., 2007. Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat. Genet, 39, 896-900. |
| [23] | Lu, S., Sun, Y.H., Shi, R., et al., 2005. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell, 17, 1378-1386. |
| [24] | Lunn, J., Droux, M., Martin, J., et al., 1990. Localization of ATP sulfurylase and O-acetylserine (thiol) lyase in spinach leaves. Plant Physiol, 94, 1345-1352. |
| [25] | Martin, M.N., Tarczynski, M.C., Shen, B., et al., 2005. The role of 5′-adenylylsulfate reductase in controlling sulfate reduction in plants. Photosynth. Res, 86, 309-323. |
| [26] | Martinoia, E., Massonneau, A., Frangne, N., 2000. Transport processes of solutes across the vacuolar membrane of higher plants. Plant Cell Physiol, 41, 1175-1186. |
| [27] | Maruyama-Nakashita, A., Nakamura, Y., Tohge, T., et al., 2006. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell, 18, 3235-3251. |
| [28] | Matthewman, C.A., Kawashima, C.G., Huska, D., et al., 2012. miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Lett, 586, 3242-3248. |
| [29] | McGrath, S.P., Zhao, F.J., Withers, P.J.A., 1996. Development of sulphur deficiency in crops and its treatment. Proceedings of the International Fertiliser Society. International Fertiliser Society. |
| [30] | Pilon-Smits, E.A.H., Hwang, S.B., Lytle, C.M., et al., 1999. Overexpression of ATP sulfurylase in Indian mustard leads to increased selenate uptake, reduction, and tolerance. Plant Physiol, 119, 123-132. |
| [31] | Renosto, F., Patel, H., Martin, H., et al., 1993. ATP sulfurylase from higher plants: kinetic and structural characterization of the chloroplast and cytosol enzymes from spinach leaf. Arch. Biochem. Biophys, 307, 272-285. |
| [32] | Rotte, C., Leustek, T., 2000. Differential subcellular localization and expression of ATP sulfurylase and 5′-adenylylsulfate reductase during ontogenesis of Arabidopsis leaves indicates that cytosolic and plastid forms of ATP sulfurylase may have specialized functions. Plant Physiol, 124, 715-724. |
| [33] | Tababai, M.A., Bremner, J.M., 1970. A simple turbidimetric method of determining total sulfur in plant material. Agron. J, 62, 805-806. |
| [34] | Takahashi, H., 2010. Regulation of sulfate transport and assimilation in plants. Int. Rev. Cell Mol. Biol, 281, 129-159 . |
| [35] | Tsakraklides, G., Martin, M., Chalam, R., et al., 2002. Sulfate reduction is increased in transgenic Arabidopsis thaliana expressing 5′-adenylylsulfate reductase from Pseudomonas aeruginosa. Plant J, 32, 879-389. |
| [36] | Voinnet, O., 2009. Origin, biogenesis, and activity of plant microRNAs. Cell, 136, 669-687. |
| [37] | Yan, J., Gu, Y., Jia, X., et al., 2012. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell, 24, 415-427. |



