2. Department of Agricultural Sciences, University of Bologna, Via Zamboni, 33, 40126 Bologna, Italy
1. Introduction
Restricted pollen and seed dispersal lead to non-r and om spatial distribution of genotypes in populations. Analysis of spatial genetic structure (SGS) at fine scale (i.e. within populations) allows detecting major gene dispersal distance, the knowledge of which is necessary for efficient species conservation management. According to the theory of isolation by distance, SGS arises from the interplay of limited gene flow and local genetic drift. The rate of decrease of genetic similarity with distance is a measure of strength of SGS (Loiselle et al., 1995; Rousset, 2000; Hardy, 2003) . How strength of SGS relates to the biology of the species and the degree of inter and intra-specific variation in this property is an important question of population genetics (Vekemans and Hardy, 2004; Jump et al., 2012). The extent of SGS within plant populations depends not only on seed and pollen dispersal distance but also on breeding type, level of self fertilization and effective plant density (Vekemans and Hardy, 2004) . In species with more restricted pollen dispersal, lower gene flow is expected to result in higher genetic differentiation, and therefore self-fertilizing species are expected to have both smaller effective populations sizes (Ingvarsson, 2002) and lower pollen movement leading to higher genetic structure than out-crossing species (Hamrick and Godt, 1996) . Higher SGS is also expected in more patchy and peripheral populations because of lower plant density, smaller population sizes, and lower intensity of gene flow (Doligez et al., 1998; Vekemans and Hardy, 2004) . Although still limited, there are an increasing number of reports on SGS in fragmented versus continuous (Williams et al., 2007; Born et al., 2008; De-Lucas et al., 2009) and core versus peripheral populations (Gapare and Aitken, 2005; Pandey and Rajora, 2012; Meeus et al., 2013; Volis et al., 2014) .
Here we investigate gene dispersal patterns in an annual grass Avena sterilis. This species is well suited for studying fine-scale gene flow and SGS in a predominantly self-fertilizing plant. It has a wide range with often almost continuous local distribution which allows sampling at specified distances. At the same time, while core populations are large and dense, peripheral populations are usually smaller, more isolated and patchily distributed. Our study employed (1) direct measurement of seed dispersal in a controlled environment; and (2) analyses of SGS in 3 natural populations of A.sterilis representing different ecological conditions and species range positions, sampled in linear transects at fixed increasing inter-plant distances.
Our hypotheses were: (i) since A.sterilis is predominantly (>95%) self pollinated with gravity dispersed seeds, this species is expected to have finescale within-population SGS; (ii) the peripheral populations of the species are expected to have higher levels of SGS than the core populations due to their lower population size and density; and potentially more restricted gene dispersal.
2. Materials and MethodsAvena sterilis L. is a winter annual, and a predominantly self-pollinating grass (Phillips et al., 1993) . This species is one of the major components of annual vegetation throughout Israel. It forms massive st and s in the mesic Mediterranean including open park-forests, maquis and hemicryptophytic/dwarf shrub formations, and also penetrates into favorable desert habitats (wadi beds and loessy depressions) (Harlan and Zohary, 1966; Zohary, 1983) .
In this species the inflorescence is a panicle. Upon maturation the whole spikelet (comprising two to four florets) disarticulates and acts as a drill-type dissemination device. The fallen spikelets either are impaled in the dry remnants of the dead mother plant or penetrate into soil cracks by the combined effects of wind and gravity (Volis personal observations) , where they remain until germination the following season. Spikelets are intensively harvested by ants and granivorous rodents (Volis personal observations) . Seeds that do not germinate in the autumn following dispersal either die or enter the soil seed bank where they can remain dormant for several years (Volis, 2012, 2014) .
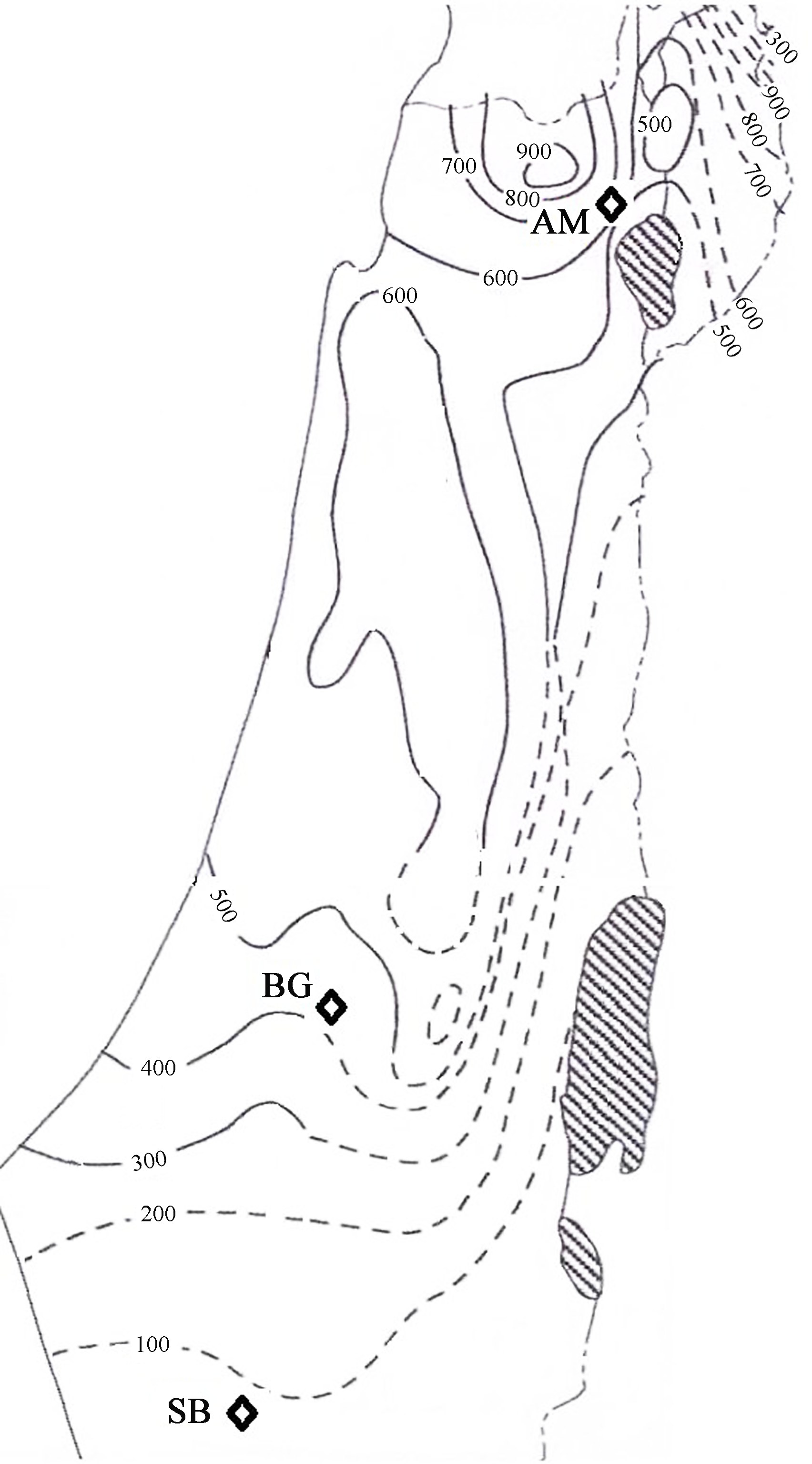
The sampling design followed Volis et al. (2010) . Seeds were sampled in nine linear transects at fixed distances (0, 1, 2, 5, 10, 20, 50, 100 and 400m) at three population locations in Israel, representing three distinct environments and vegetation communities: Mediterranean grassl and (AM) , shrub and semi shrub association called batha (BG) , and desert (SB) . The Mediterranean grassl and location (AM population) was in the Upper Galilee, 1km west of Kibbutz Ammiad (elevation 300m, annual precipitation around 600mm) . The batha location (BG population) was in Beit Guvrin National Park located in the Shefela Hills (elevation 300m, annual precipitation 400mm) . The desert location (SB population) was in a wadi in the Negev Desert (elevation 400m, annual precipitation 90mm) (Fig. 1) .
|
| Fig.1 Map of Israel showing isohyets of multiyear averages of annual rainfall amount (mm) and study populations. |
In addition, population locations differed in their position within a species distributional range, representing either species core (AM and BG) or periphery (SB) . For genetic analysis, seeds were germinated and grown to the two-leaf stage.
Average plant density was estimated in two of three populations (SB and BG) during 1996-1999. In 1996, six 10 m transects, distributed along a slope 20m apart, were marked in the BG location. Five 1 m2 plots 1 m apart were permanently marked along each transect. At this site, the distribution of A.sterilis was more or less continuous. At the SB location, where vegetation distribution was sparse and patchy, neither transects nor equidistant spacing was possible. Therefore, 1 m2 plots were marked in 1996 in each vegetation patch containing oat plants. Altogether, 30 and 50 plots were marked at the SB and BG locations, respectively. The higher number of plots at the SB location was due to the higher spatial heterogeneity at this location compared with the BG location. During the next four years, adult plants and number of seeds per plant were counted in each plot. In 1996, plant fecundity was estimated at both locations, but plants per plot were counted at SB location only. In 1999, because of very low amount of rainfall, no plant reached adulthood in the SB population, and there was no estimate of plant density.
The effects of wind and gravity on seed dispersal distance in A.sterilis were tested as described in Volis et al. (2010) . Two genotypes of SB and AM origin were planted in 2006 in a common environment to remove the environmental maternal effect. Next season, two seeds per genotype were germinated and planted in 15 l buckets; one plant in a greenhouse, and another one in a nethouse at the Campus Bergman of Ben Gurion University, Beer Sheva. The space around each plant was divided into circles of 20cm width. Upon maturation of spikelets and start of shattering, plants were visited daily for collection of shattered spikelets within respective distance classes. The AM plant placed in a greenhouse was infected by leaf rust and died. Therefore, we present results for SB greenhouse, SB nethouse and AM nethouse plants (421, 490 and 269 seeds, respectively) .
All the samples, 202 plants in total, were genotyped. DNA extraction followed modified CTAB protocol (Rogers and Benedich, 1985) . Six polymorphic nuclear microsatellite loci (Table 1) were amplified using primer-specific polymerase chain reaction (PCR) program as described in Table 2. Four dyes were used-FAM, NED, PET and VIC. The PCR products were detected and sized by the ABI PRISM 3700 DNA Analyzer at the Hebrew University, Jerusalem, Israel. The data were analyzed using Peak ScannerTM Software v1.0 (Applied Biosystems) .
| Locus | Repeat type | Populations | |||||||||||
| SB (n=75) | BG (n=63) | AM (n=64) | |||||||||||
| A | Ho | He | F1 | A | Ho | He | F1 | A | Ho | He | F1 | ||
| AM-1a | (AG)n(CAGAG)k | 7 | 0.013 | 0.380 | 0.965 | 4 | 0.019 | 0.586 | 0.968 | 7 | 0.016 | 0.809 | 0.980 |
| AM-3a | (AG)n | 13 | 0.013 | 0.790 | 0.983 | 10 | 0.000 | 0.793 | 1.000 | 10 | 0.016 | 0.842 | 0.981 |
| AM-22a | (AG)n | 5 | 0.107 | 0.709 | 0.850 | 5 | 0.132 | 0.774 | 0.829 | 5 | 0.141 | 0.771 | 0.818 |
| KSUM176a | (CTG)n | 4 | 0.013 | 0.728 | 0.982 | 4 | 0.075 | 0.542 | 0.861 | 3 | 0.063 | 0.461 | 0.864 |
| MAMA-4b | (TCTA)n | 17 | 0.027 | 0.896 | 0.970 | 13 | 0.000 | 0.808 | 1.000 | 12 | 0.063 | 0.735 | 0.915 |
| MAMA-6b | (TC)n(TCTA)k(TC)f | 4 | 0.093 | 0.570 | 0.836 | 5 | 0.132 | 0.677 | 0.805 | 5 | 0.047 | 0.675 | 0.931 |
| Locus mean | 8.33 | 0.044 | 0.679 | 0.931 | 6.833 | 0.050 | 0.697 | 0.910 | 7.000 | 0.057 | 0.715 | 0.915 | |
| (SE) | (2.216) | (0.018) | (0.074) | (0.028) | (1.536) | (0.025) | (0.046) | (0.036) | (1.390) | (0.019) | (0.056) | (0.026) | |
| a Source reference Fu et al. 2007; b Source reference Wight et al. 2003). | |||||||||||||
| A, number of alleles; Ho observed heterozygosity ;He, expected heterozygosity; F1, inbreeding coefficient. | |||||||||||||
| Steps | Primers | |||
| AM-1 | AM-3 | AM-22, KSUM176, MAMA-6 | MAMA-4 | |
| 1 | 94 ℃ 2 min | 94 ℃ 5 min | 94 ℃ 2 min | 94 ℃ 3 min |
| 2 | 94 ℃ 1 min | 94 ℃ 30 s | 94 ℃ 45 s | 94 ℃ 30 s |
| 3 | 64 ℃ 30 s decrease in 0.5 ℃ every cycle | 58 ℃ 1 min | 40.5 ℃ 45 s | 62 ℃ 1 min |
| 4 | 72 ℃ 1 min | 72 ℃ 45 s | 2 ℃ 1 min | 72 ℃ 2 min |
| 5 | 22 cycles | 40 cycles | 35 cycles | 35 cycles |
| 6 | 94 ℃ 1 min | 94 ℃ 30 s | 72 ℃ 10 min | 72 ℃ 7 min |
| 7 | 53 ℃ 1 min | 53 ℃ 45 s | 4 ℃ hold | 4 ℃ hold |
| 8 | 72 ℃ 1 min | 72 ℃ 45 s | ||
| 9 | 20 cycles | 10 cycles | ||
| 10 | 72 ℃ 10 min | 72 ℃ 10 min | ||
| 11 | 4℃ hold | |||
The pairwise genetic distances/relatedness coefficients were obtained for each transect separately. Analysis of spatial autocorrelation utilized the multivariate procedure in order to combine data from different transects (Smouse and Peakall, 1999) . The statistical significance of autocorrelation was tested for each sampling distance (from 1 to 400m) by 10, 000 r and om permutations and obtaining 95% confidence intervals after bootstrapping (10, 000 repeats) . The analysis was conducted for each population separately by GENALEX 6.5 software (Peakall and Smouse, 2006) .
We calculated the kinship coefficients for all pairs of individuals in transects using the statistic of Ritl and (1996) as implemented in GENALEX (Peakall and Smouse, 2006) . To estimate the regression slopes (b) , the multilocus kinship coefficients for all pairs of individuals in each population were plotted against the logarithm of geographic distance separating them. The extent of gene dispersal is estimated from this slope as Nb=- (1-F0) /b where Nb is the neighborhood size in terms of number of individuals, in continuous two-dimensional population (Wright, 1943) , and F0 is the average kinship coefficient between adjacent individuals. In our study, we estimated F0 for the first distance class (1 m distance) as the closest approximation to "adjacent" plants (Vekemans and Hardy, 2004; Oddou-Muratorio and Klein, 2008) and designated it F (1) . The lower and upper bounds for the 95% confidence intervals (CI) of Nb were computed as (F (1) -1) / (b ± 2SEb) , SEb being the st and ard error of the regression slope b (Hardy et al., 2006) . We also calculated the ‘Sp’ statistic, which is the inverse of the neighborhood size Nb under isolation by distance in two-dimensional space (Vekemans and Hardy, 2004) , i.e. the ratio -b/ (1-F (1) ) . This statistic is very useful in the comparison of SGS and gene dispersal across populations and species.
The Nb and Sp are most reliably estimated within a distance between σ (mean gene dispersal distance) and 10-50 σ in two-dimensional space (Rousset, 1997; Vekemans and Hardy, 2004) . Following Volis et al. (2010) we computed the slope b for the full and limited (up to 20m) distance range in each population. From these values we obtained two indirect estimates of neighborhood size and its inverse (Sp) for each population.
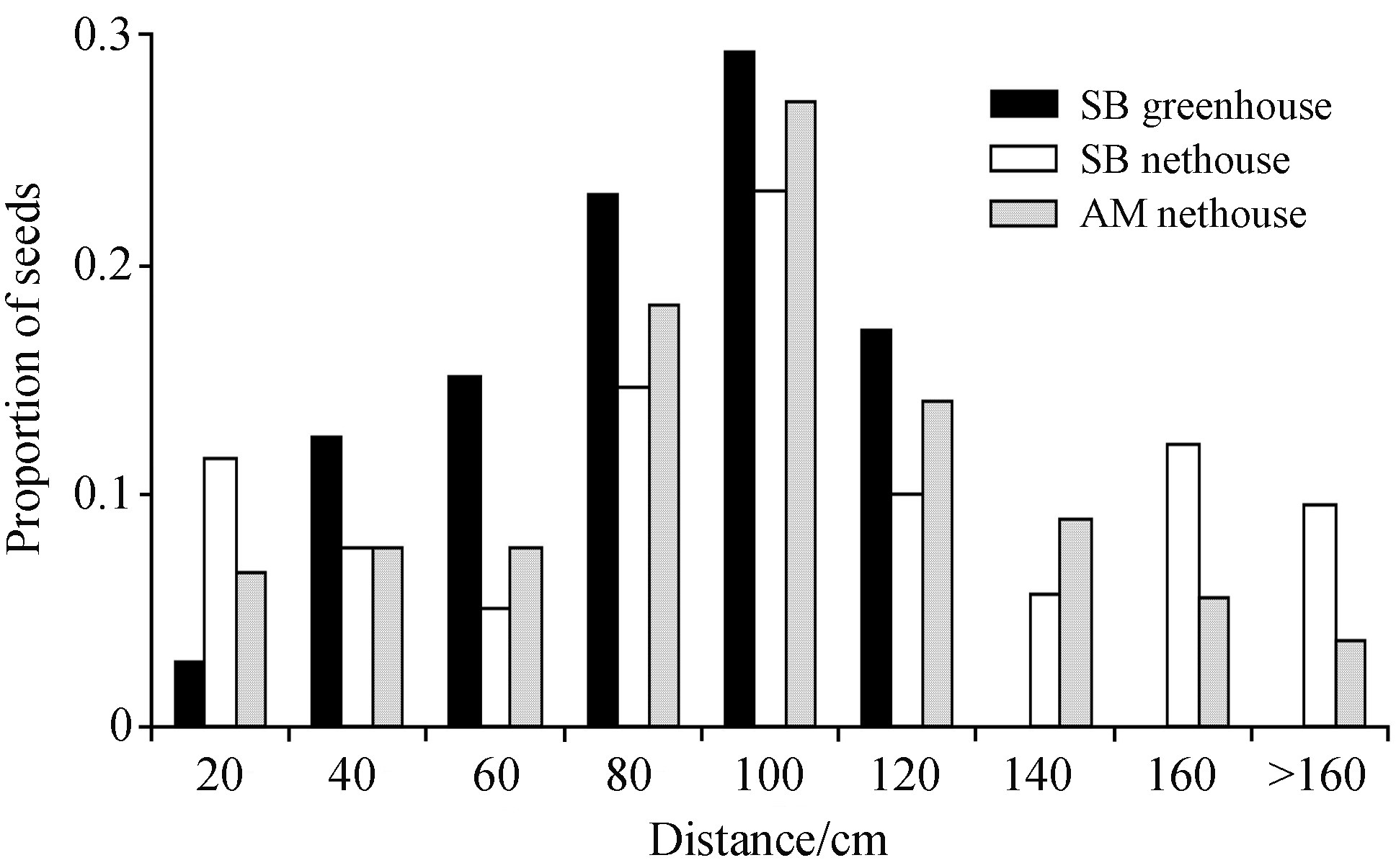
3. ResultsThe histograms for seeds of the three plants indicate that major seed dispersal in A.sterilis is limited to the distance of 1.6m (>90%) and has a sigmoid shape with a mode distance class of 80-100cm from the mother plant (Fig. 2) . This pattern of dispersal was not affected by spikelet size or whether the plants were grown in a greenhouse or a nethouse (60.1±0.9, 48.5±1.0 and 136.2±2.6mg, the SB plants grown in a greenhouse and a nethouse, and the AM plant grown in a nethouse, respectively) . On the other h and , spatial distribution of shattered seeds of the same genotype planted in a nethouse was less leptokurtic than in a greenhouse (37.5% vs. 17.1% of seeds dispersed above 1 m distance) (Fig. 2) . The difference between nethouse and greenhouse was due to wind, present in the former (although nethouse mesh might have some reducing effect on wind velocity, underestimating its effect) and absent in the latter. Thus, wind does contribute to the seed dispersal curve.
|
| Fig.2 Seed dispersal measured as a distance from the mother plant. |
Adult plant density was 10.3 ±1.2, 29.7±6.2, 16.8±3.3 plants per 1 m2 and fecundity was 20.0±1.6, 8.6±0.9, and 6.9±0.8 seeds per plant in years 1996-98 in the SB population. In the BG population, plant density was 117±18.4, 444.8±70.2 and 416.5±67.4 in years 1997-99, and fecundity was 18.3±1.0, 10.5±1.1, and 3.4±0.3 in years 1996, 1997 and 1999.
Genetic diversity statistics for the three populations are given in Table 1. Average number of alleles per locus, A, and expected heterozygosity, He, were similar in all populations, with range 6.8-8.3 and 0.679-0.715, respectively. Similarly, observed heterozygosity, Ho, and inbreeding coefficient FI did not differ among populations, ranging 0.044-0.057 and 0.910-0.931, respectively.
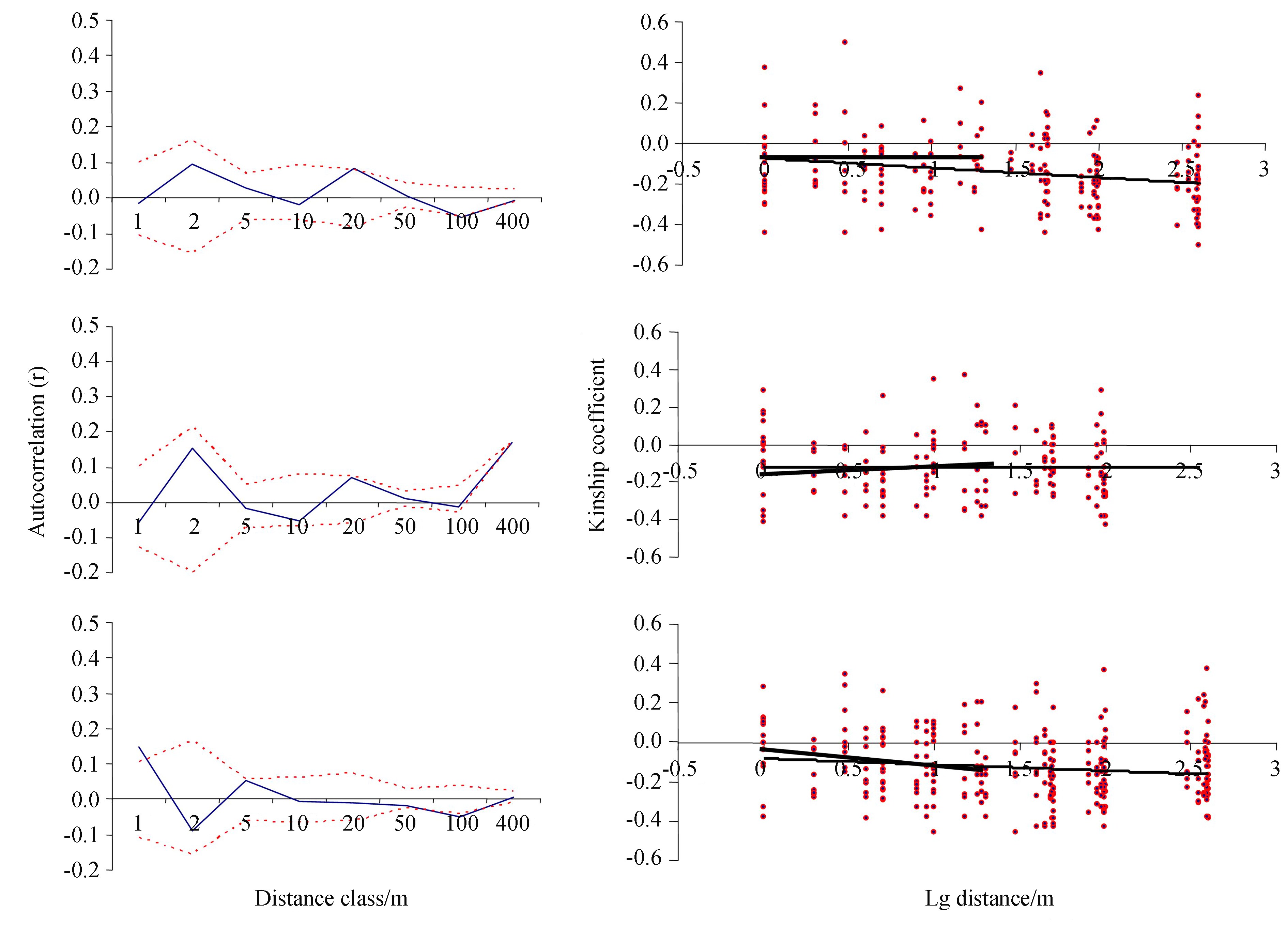
There was a significant autocorrelation for the first distance class of 1 m in the SB population, and no autocorrelation effect for any distance class in the other two populations (Fig. 3) .
|
| Fig.3 Left: The autocorrelogram with 95% confidence interval (dotted lines) for autocorrelation coefficient r (solid line) for populations AM BG and SB, respectively (from top down). Right: The kinship coefficients for each pair of individuals plotted against the logarithm of geographic distance separating them and the estimated regression lines for the distance range 1-400 m and 1-20 m. |
A significant linear decrease of estimated kinship coefficients between pairs of individuals with the logarithm of increasing geographical distance was detected in the SB and AM populations (Fig. 3 and Table 3) . In the SB population the detected pattern was observed for both distance ranges (1-400 and 1-20m) , with regression slope being steeper for the distance range of 1-20m than for 1-400m in the SB population (t=-3.2, P <0.01, t test) (Table 3) .
| Population | Distance range | b ± SE | R2 | p | F0 | Nb(95% CI) Sp | |
| SB | 0-400 | -0.031 ± 0.013 | 0.020 | 0.017 | -0.029 ± 0.043 | 32.2(17.5-196.3) | 0.031 |
| SB | 0-20 | -0.097 ± 0.039 | 0.063 | 0.016 | 10.3(5.7-55.9) | 0.097 | |
| BG | 0-400 | -0.021 ± 0.022 | 0.006 | ns | -0.211 ± 0.071 | ||
| BG | 0-20 | 0.051 ± 0.047 | 0.013 | ns | |||
| AN | 0-400 | -0.047 ± 0.014 | 0.055 | <0.001 | -0.118 ± 0.050 | ||
| AN | 0-20 | 0.004 ± 0.042 | 0.001 | ns |
The average kinship coefficient between individuals 1 m apart was used as the closest approximation to "adjacent" plants. For neither population was the F0 value positive, indicating very low relatedness for even adjacent plants (F0≈0) . Therefore, we used zero value for F0 to calculate the neighborhood size, Nb, for the SB population. Neighborhood size, Nb, estimated for this population was based on regression slopes obtained for two distance ranges, 1-400 and 1-20m, and were 32.2 and 10.3, respectively. Under assumption of F0≈0 Sp becomes equal to the regression slope yielding 0.031 and 0.097 for the SB population (1-400 and 1-20m, respectively) .
4. DiscussionThis study adds to still very limited knowledge of the effects of seed dispersal, breeding system, and species range position on SGS at fine spatial scale.
Seed dispersal typically fits a unimodal distribution, with a peak at or close to the mother plant, and a tail of progressively fewer seeds with increasing distance from the plant (Levin and Kerster, 1974; Willson, 1993; Portnoy and Willson, 1993; Schupp and Fuentes, 1995) . Although many environmental and biotic effects were not accounted for in this study (soil properties, surrounding vegetation, biotic seed dispersers) , the approximated seed dispersal curve in wildoat is bell shaped and indicates that major seed dispersal upon maturation was limited to distances around 1 m. Wind appears to contribute to seed dispersal, making the curve less leptokurtic. Again, estimations on the effect of wind were limited in our experiment because strong winds may be rare events that do not happen every year and throughout A.sterilis distribution. Thus, the actual effect of wind on seed dispersal in this species may be stronger than evident in our experiment. Furthermore, we do not know the importance of occasional long seed dispersal resulting from spikelets entrapped in animals′ fur in this species. Nevertheless, our study shows that in A.sterilis the major factor in seed dispersal is gravity in close (less than 2m) vicinity of the mother plant, with a minor additional effect of wind.
As we showed, A.sterilis is a predominant selfer (FI>0.90) . The number of alleles, as well as expected and observed heterozygosity, did not differ among the three populations, suggesting a common breeding mode and similar extent of population genetic variation. The SGS pattern was detected by spatial autocorrelation analysis only in the most peripheral population, which is located at the arid edge of the species range. Even in this population, the SGS was limited to 1 m distance. In this population the values of regression slope, neighborhood size Nb and Sp statistic indicated higher SGS at the scale of 20m as compared with 400m.
It is interesting to compare the results of this study with methodologically identical study of wild barley (Volis et al., 2010) . Sampling design, population locations and dates of collecting were the same in the two studies. In both wild barely and wild oat species SGS had a very fine spatial scale of about 1 m. However, while strong SGS was detected in all studied populations of wild barley, only one population of wild oat showed SGS, and even that was very weak. These results suggest stronger gene flow in wild oat as compared with wild barley.
Both wild oat and wild barley are predominantly selfing annual grasses with large (0.5-1g) seeds falling at maturity close to the mother plant. However, the two species′ seed dispersal curves (approximated by exponential function for barley and Gaussian function for oat) and a difference in the tail thickness suggest a more important role for wind in oat compared with barley seed dispersal. This is consistent with the morphology of the dispersal units (spikelets) of the two species. The wild barley spikelet has a single long awn, while the A.sterilis spikelet has two short curved awns, making the latter more aerodynamically efficient. In addition, wild oat core populations in mesic Mediterranean climate zone usually have higher plant density than corresponding wild barley populations (up to 445 plants/m2 vs. max density of 195 plants/m2 in wild barley) (Volis et al., 2010 and this study) . These two studies show that predominant selfing and spatially limited seed dispersal lead to the creation of fine-scale SGS, but only if the gene flow and plant densities are not too high.
The three populations did not differ in genetic diversity; thus, lack of genetic structure in two core (Mediterranean) populations appears to be a result of a higher intensity of within-population gene flow in these populations compared with the peripheral (desert) population. This is expected in the species periphery due to lower population density, lower number and higher isolation in patches of suitable environment.
Studies comparing fine-scale SGS across a species range are rare (Gapare and Aitken, 2005; Pandey and Rajora, 2012; Volis et al., 2014) . In our previous study conducted on wild barley, we did not find a clear difference in intensity of SGS between core and peripheral populations. This can be explained by extremely limited seed dispersal in wild barley, almost inevitably creating fine SGS even under high plant density. In wild oat, however, the seed dispersal kernel is longer and the effect of plant density becomes important for creation of SGS. Similarly, a difference in SGS in peripheral versus core populations has been found in two tree species with long seed dispersal kernels. Gapare and Aitken (2005) found r and om distribution of genotypes in the core populations and positive SGS up to 500m in the peripheral populations of Sitka spruce. Pandey and Rajora (2012) detected higher SGS in peripheral (up to 90m) as compared with core populations (up to 15m) in the eastern white cedar.
5. ConclusionsTaken together, the two methodologically identical studies on wild barley (Volis et al., 2010) and wild oat (this study) show that predominant selfing coupled with spatially limited seed dispersal favors creation of fine-scale SGS. However, this fine-scale SGS can be prevented by high plant density.
Our results also support the theoretical prediction that intensity of SGS increases from the species core to periphery as a result of decreased within-population gene flow related to low plant density.
| Born, C., Olivier, J.H., Marie-Héléne, C., et al., 2008. Small-scale spatial genetic structure in the Central African rainforest tree species Aucoumea klaineana: a stepwise approach to infer the impact of limited gene dispersal, population history and habitat fragmentation. Mol. Ecol, 17, 2041-2050. |
| De-Lucas, A.I., González-Martinez, S.C., Vendramin, G.G., et al., 2009. Spatial genetic structure in continuous and fragmented populations of Pinus pinaster Aiton. Mol. Ecol, 18, 4564-4576. |
| Doligez, A., Baril, C., Joly, H.I., et al., 1998. Fine-scale spatial genetic structure with nonuniform distribution of individuals. Genetics, 148, 905-919. |
| Fu, Y.B., Chong, J., Fetch, T., et al., 2007. Microsatellite variation in Avena sterilis oat germplasm. Theor. Appl. Genet, 114, 1029-1038. |
| Gapare, W.J., Aitken, S.N., 2005. Strong spatial genetic structure in peripheral but not core populations of Sitka spruce [Picea sitchensis (Bong.) Carr.]. Mol. Ecol, 14, 2659-2667. |
| Hamrick, J.L., Godt, M.J.W., 1996. Effects of life history traits on genetic diversity in plant species. Philos. Trans. R. Soc. Lond. B-Biol. Sci, 351, 1291-1298. |
| Hardy, O.J., 2003. Estimation of pairwise relatedness between individuals and characterization of isolation-by-distance processes using dominant genetic markers. Mol. Ecol, 12, 1577-1588. |
| Hardy, O.J., Maqqia, L., Bandou, E., et al., 2006. Fine-scale genetic structure and gene dispersal inferences in 10 Neotropical tree species. Mol. Ecol, 15, 559-571. |
| Harlan, R.J., Zohary, D., 1966. Distribution of wild wheats and barley. Science, 153, 1074-1080. |
| Ingvarsson, P.K., 2002. A metapopulation perspective on genetic diversity and differentiation in partially self-fertilizing plants. Evolution, 56, 2368-2373. |
| Jump, A.S., Rico, L., Coll, M., et al., 2012. Wide variation in spatial genetic structure between natural populations of the European beech (Fagus sylvatica) and its implications for SGS comparability. Heredity, 108, 633-639. |
| Levin, D.A., Kerster, H.W., 1974. Gene flow in seed plants. Evol. Biol, 7, 139-220. |
| Loiselle, B.A., Sork, V.L., Nason, J., et al., 1995. Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am. J. Bot, 82, 1420-1425. |
| Meeus, S., Honnay O., Jacquemyn, H., 2013. Differences in fine-scale spatial genetic structure across the distribution range of the distylous forest herb Pulmonaria officinalis (Boraginaceae). Bmc Genetics, 14. |
| Oddou-Muratorio, S., Klein, E.K., 2008. Comparing direct vs. indirect estimates of gene flow within a population of a scattered tree species. Mol. Ecol, 17, 2743-2754. |
| Pandey, M., Rajora, O.P., 2012. Higher fine-scale genetic structure in peripheral than in core populations of a long-lived and mixed-mating conifer-eastern white cedar (Thuja occidentalis L.). BMC Evol. Biol, 12, 48. |
| Peakall, R., Smouse, P.E., 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes, 6, 288-295. |
| Phillips, T.D., Murphy, J.P., Goodman, M.M., 1993. Isozyme variation in germplasm accessions of the wild oat Avena sterilis L. Theor. Appl. Genet, 86, 54-64. |
| Portnoy, S., Willson, M.F., 1993. Seed dispersal curves: behavior of the tails of the distribution. Evol. Ecol, 7, 25-44. |
| Ritland, K., 1996. Estimators for pairwise relatedness and individual inbreeding coefficients. Genet. Res, 67, 175-185. |
| Rogers, S.O., Benedich, A.J., 1985. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol, 5, 69-76. |
| Rousset, F., 1997. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics, 145, 1219-1228. |
| Rousset, F., 2000. Genetic differentiation between individuals. J. Evol. Biol, 13, 58-62. |
| Schupp, E.W., Fuentes, M., 1995. Spatial pattern of seed dispersal and the unification of plant population ecology. Ecoscience, 2, 267-275. |
| Smouse, P.E., Peakall, R., 1999. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity, 82, 561-573. |
| Vekemans, X., Hardy, O.J., 2004. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol, 13, 921-935. |
| Volis, S., 2012. Demographic consequences of delayed germination in two annual grasses from two locations of contrasting aridity. Perspectives in Plant Ecology, Evolut. Syst, 14, 335-340. |
| Volis, S., 2014. Dormancy-related seed positional effect in two populations of an annual grass from locations of contrasting aridity. PLoS ONE 9, e93061. |
| Volis, S., Song, M., Zhang, Y., et al., 2014. Fine-scale spatial genetic structure in emmer wheat and the role of population range position. Evol. Biol, 41, 166-173. |
| Volis, S., Zaretsky, M., Shulqina, I., 2010. Fine-scale spatial genetic structure in a predominantly selfing plant: role of seed and pollen dispersal. Heredity, 105, 384-393. |
| Wight, C.P., Tinker, N.A., Kianian, S.F., et al., 2003. A molecular marker map in ‘Kanota’ x ‘Ogle’ hexaploid oat (Avenas pp.) enhanced by additional markers and a robust framework. Genome, 46, 28-47. |
| Williams, D.A., Wang, Y., Borchetta, M., et al., 2007. Genetic diversity and spatial structure of a keystone species in fragmented pine rockland habitat. Biol. Conserv, 138, 256-268. |
| Willson, M.F., 1993. Dispersal mode, seed shadows, and colonization patterns. Vegetatio, 107/108, 261-280. |
| Wright, S., 1943. Isolation by distance. Genetics, 28, 114-138. |
| Zohary, D., 1983. Wild genetic resources of crops in Israel. Israel J. Bot, 32, 97-127. |



