b. School of Pharmacy, Xi'an Jiaotong University, Xi'an 710061, China;
c. College of Life Science, Shanxi Normal University, Taiyuan, China;
d. Botanic Institute of Barcelona (IBB), CSIC-CMCNB, Barcelona 08038, Catalonia, Spain;
e. Escuela de Ciencias Ambientales, Universidad Espíritu Santo (UEES), Samborondón 091650, Ecuador;
f. Shaanxi Engineering Research Centre for Conservation and Utilization of Botanical Resources, Xi'an Botanical Garden of Shaanxi Province, Xi'an 710061, China;
g. School of Biological Sciences, Guizhou Education University, Guiyang, China
East Asia is widely known to harbor many paleoendemic plant species. Currently, ~600 plant genera are endemic to East Asia, an important fraction of which are relicts of the Arcto-Tertiary flora (Manchester et al., 2009). Several of these relict plant lineages are endangered at present (e.g., Cercidiphyllum, Davidia, Dipteronia, Euptelea, or Tetracentron), with a scattered distribution in mixed mesophytic forests of China (Manchester et al., 2009; Huang et al., 2011, 2012; López-Pujol et al., 2011). The fossil record indicates that some of these relict plant lineages were once widely distributed in Europe or North America (Wen, 1999; Milne and Abbott, 2002; Manchester et al., 2009), although they have since gone extinct in these regions due to unsuitable environmental conditions (Tang et al., 2018). In East Asia, these relict plant lineages survived in long-term stable mountain refugia that provided suitable environmental conditions. Refugia were found in northern Vietnam, and southwestern China, including south-eastern and eastern Yunnan, western and northern Guizhou, eastern Sichuan and Chongqing (Tang et al., 2018).
The extant distribution patterns of ancient relict species may be the result of complex evolutionary events. The demographic history of these relict species has been shown to be affected by mountain uplift and climatic fluctuations (Hampe and Jump, 2011; Hoffmann and Sgrò, 2011). The interaction of these processes may have created new ecological niches, thereby providing opportunities for the divergence of species (Liu et al., 2013). However, the evolutionary history of these paleoendemic genera remains mostly unknown due to the paucity of reliable fossil records and genetic data, although some recent progress has been made by integrating all these data, e.g. Cercidiphyllum (Qi et al., 2012), Cyclocarya (Kou et al., 2016), Euptelea (Cao et al., 2016), Taiwania (Chou et al., 2011), or Tetracentron/Trochodendron (Sun et al., 2014b). Additionally, some of these relict genera have persisted over time with little morphological change, which may hide diversification events (Nagalingum et al., 2011; Cao et al., 2016). Disentangling the evolutionary scenarios that have led to distribution patterns of ancient relict species requires an integrative approach that includes fossil-calibrated phylogenetic analyses, surveys of genetic diversity/phylogeography, and analyses of ecological niche.
Dipteronia Oliv. (Sapindaceae) is an ancient relict genus endemic to China, comprising two extant species, Dipteronia sinensis Oliv. and D. dyeriana Henry. Both species are diploid (2n = 18) broad-leaved deciduous small trees (the former being taller), with bisexual/staminate and wind-pollinated flowers that turn into winged nutlets (those of D. dyeriana being much larger than those of D. sinensis) dispersed by gravity, wind and/or water. Although D. sinensis and D. dyeriana are morphologically similar, they currently occur in different habitats with no-overlapping distribution ranges. According to Flora of China (Xu et al., 2008), D. sinensis has a relatively extensive range in central and south-western China, with most populations distributed in isolated stands at elevations of 1000–2500 m in mountain riparian warm-temperate deciduous forests. Although D. dyeriana also inhabits mountain riparian forests, current populations are restricted to south-eastern Yunnan Province (Xu et al., 2008), with an area of occupancy of less than 300 km2 (Gibbs and Chen, 2009); however, until the late 20th century this narrow endemic also occurred in south-central Yunnan as well as Xingyi County in south-western Guizhou (Sun et al., 2006; Chen et al., 2017). Currently, D. dyeriana is listed as ‘Endangered’ (EN) according to the latest version of the IUCN red list both at the national (Qin et al., 2017) and global (Rivers et al., 2017) level, as well as in the Red List of Maples (Gibbs and Chen, 2009). Dipteronia sinensis is classified as ‘Near Threatened’ (NT) (Gibbs and Chen, 2009), and it has been suggested that D. sinensis might become ‘Vulnerable’ (VU) because of deforestation and poor regeneration. Although both species were initially protected in China, included in the National List of Rare and Endangered Plant Species in 1984 (D. dyeriana listed as ‘second grade’ and D. sinensis as ‘third grade’ nationally protected), only the first was included in the further Catalogue of the National Protected Key Wild Plants (also as ‘second-grade’).
The fossil record indicates that Dipteronia once occurred extensively in North America, beginning in the Paleocene and continuing through the Oligocene, with the greatest number of occurrences in the middle to late Eocene (McClain and Manchester, 2001). In East Asia, the fossil record is less known, but Paleocene fossils, even older than those of North America, are known from the Zeya-Bureya Basin in the Russian Far East (Manchester et al., 2009). Apart from this, the only known Asian fossils are those from Lühe (Yunnan, China), dating from the early Oligocene (Ding et al., 2018). Dipteronia has long been considered as closely related to Acer, being placed within Aceraceae (Hall, 1961), although at present both genera are included within Sapindaceae (APG, 2016). However, due to the lack of sufficient genetic markers, no clear conclusions have been drawn regarding the phylogenetic relationships of Dipteronia and Acer (Yang et al., 2010; Zhou et al., 2016a; Harris et al., 2017). Recent phylogenetic analyses based on phylogenomic datasets have confirmed, nevertheless, the reciprocal monophyly of Dipteronia and Acer, having diverged during the Paleocene (Feng et al., 2019). According to these authors, the divergence of D. sinensis and D. dyeriana can be dated around the Paleocene/Eocene boundary (Feng et al., 2019), which is noticeably older than the dating based on recent phylogeographic studies (Bai et al., 2017).
Over the last few years, several studies have reconstructed the phylogeographic history and genetic differentiation patterns of Dipteronia. These studies indicate that most populations of Dipteronia experienced genetic bottlenecks, with low gene flow detected between D. sinensis and D. dyeriana but also between populations of each species (Yang et al., 2007, 2008; Bai et al., 2017). A recent study characterized the whole genomes of Dipteronia species and inferred their lineage divergence and demographic history, as well as mutation load (Feng et al., 2024). However, many aspects of the evolutionary story of this relict genus remain unknown, particularly (1) the relative roles of ecological differentiation and genetic divergence driving the speciation or lineage diversification of Dipteronia, and (2) how the Cenozoic climate changes and the complex orography of China contributed to the persistence and evolution of this ancient genus. Revealing the long and complex evolutionary history of Dipteronia may also provide further insight into how Arcto-Tertiary relict plants in East Asia survived, evolved, and diversified.
Phylogeographic and genetic diversity surveys, particularly in combination with ecological niche modeling (ENM) and GIS-based methods, have been successfully used to locate glacial refugia and migratory corridors of previous time periods (Chan et al., 2011; Gavin et al., 2014). In this study, we used an integrative approach and extensive sampling to investigate the evolutionary patterns of the two extant Dipteronia species. We hypothesized that (1) D. sinensis and D. dyeriana are genetically and ecologically distinct; (2) populations of D. sinensis will show both genetic and ecological differentiation; (3) late Neogene climatic changes did not strongly affect the distribution of the species given their occurrence in long-term stable refugia; and (4) the range-restricted D. dyeriana would show less genetic diversity than the more widespread D. sinensis. To test these hypotheses, we combined maternally-inherited (three cpDNA regions) and biparentally-inherited markers [two single copy nuclear genes (SCNGs) and 15 expressed sequence tag-simple sequence repeat (EST-SSR) loci] to determine genetic variability, demographic dynamics and diversification patterns of D. sinensis and D. dyeriana. We complemented this genetic analysis with niche comparison analyses on environmental space, ecological niche modeling, and landscape connectivity analysis.
2. Materials and methods 2.1. Population samplingLeaf materials were collected from 40 populations (N = 697) of Dipteronia sinensis and five populations (N = 92) of D. dyeriana, covering the entire geographical range of the genus (Fig. 1A and Table S1). All samples (N = 789) were surveyed for EST-SSR variation, whereas subsets were sequenced at both chloroplast DNA (cpDNA) and nuclear DNA (nDNA) regions (N = 390 and 50 for D. sinensis and D. dyeriana, respectively; Table S1). Aesculus chinensis Bunge was selected as an outgroup. Fresh leaves were collected and dried immediately with silica gel. Voucher specimens were deposited at the Evolutionary Botany Laboratory of Northwest University (NWU) (Xi'an, China).
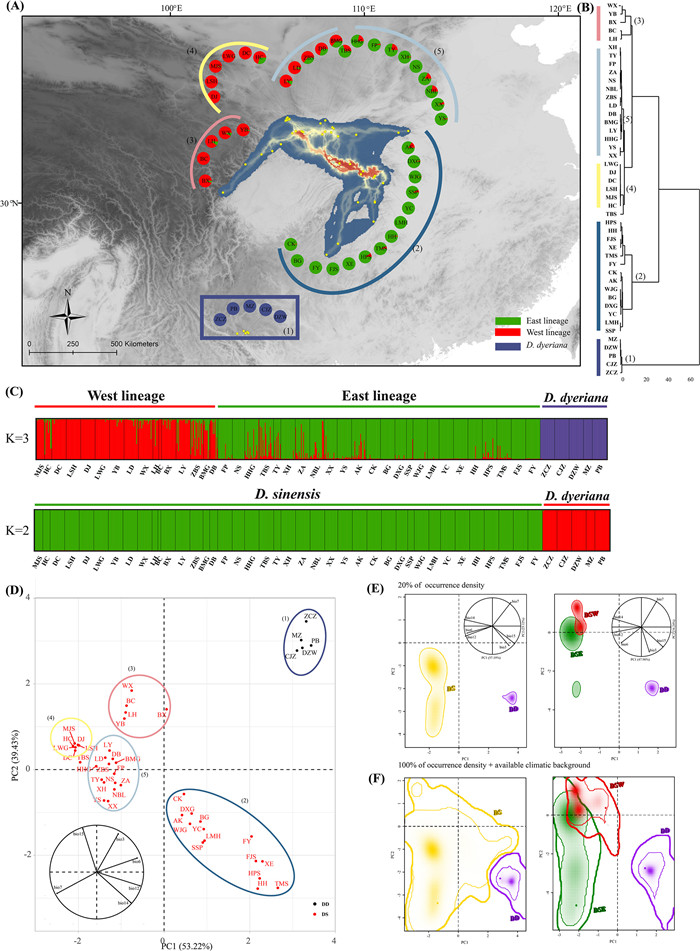
|
| Fig. 1 (A) Geographic distribution of STRUCTURE clusters (K = 3) for Dipteronia, and potential dispersal corridors for D. sinensis at present represented in the background. The color-coded grouping is based to UPGMA clusters (B) UPGMA clustering representation based on the climatic values for the first three axes of the standard principal component analysis (PCA). Population codes are identified in Table S1 (C) Histogram of the STRUCTURE (K = 3, K = 2) assignment test for 45 populations of Dipteronia based on EST-SSRs (D) PCA performed with climatic values for the populations of D. sinensis and D. dyeriana. (E) PCA-env representing global climatic space constructed over all background areas and realized niches of Dipteronia (left, total occurrences dataset, N = 267: DS, 250, DD, 17; right, partial dataset, N = 55: DSE, 23; DSW, 15; DD, 17), plotting a solid line representing 20% of the occurrence density (up), and (F) 100% of occurrence density with a thin line and 100% of available climatic background with a thick line (down). The upper graph includes the contribution and direction of each variable to the two-first components of the PCA-env. |
Total genomic DNA was extracted from dried leaf tissue using the Plant Genomic DNA Extraction Kit (Tiangen, Beijing, China). Three chloroplast DNA markers (trnS-psbZ, psbJ-rpl33, psbA-trnH) and two single copy nuclear genes (SCNGs: DS43372, DS11899) developed in previous studies were sequenced for analyzing a total of 440 individuals representing all sampled populations (Zhou et al., 2016a) (Table S2). Polymerase chain reactions (PCR) were carried out on a SimpliAmpTM Thermal Cycler (Applied Biosystems, Carlsbad, CA, USA) with a volume of 25 μL containing 12.5 μL 2 × Taq PCR Master Mix, 10–30 ng genomic DNA, 1 μL bovine serum albumin (BSA), and 0.3 μM of each primer. The thermo-cycling conditions for cpDNA and SCNGs PCR were as follows: 5 min at 94 ℃, followed by 35 cycles of 50 s at 94 ℃, 40 s annealing at 50–60 ℃ (depending on the specific primers used) and 90 s extension at 72 ℃, with a final extension at 72 ℃ for 7 min. PCR products were checked on a 2% agarose gel and sequenced in both directions with an ABI 3730XL DNA Analyzer (Sangon Biotech Shanghai, China). The raw sequences were manually edited and aligned using BioEdit v.7.2.5 (Hall, 1999). Fifteen specific EST-SSR markers for Dipteronia developed in previous studies were used for genotyping all 789 samples (Zhou et al., 2016b; Table S3). The amplification conditions of EST-SSR were set as follows: 5 min at 94 ℃, followed by 40 cycles of 30 s at 94 ℃, 50 s annealing at 52 ℃, 45 s extension at 72 ℃, with a final extension cycle of 7 min at 72 ℃. PCR products were checked on 2% agarose gel and examined on an ABI 3730XL DNA analyzer (Sangon Biotech Shanghai, China), and the data were scored and compiled using GeneMarker® v.2.2 (SoftGenetics, State College, PA, USA). All polymorphisms detected in the cpDNA and SCNG fragments are shown in Tables S4–S8.
2.3. Genetic and phylogeographic analyses of cpDNA, SCNGs, and EST-SSRsBasic statistics such as haplotype diversity (Hd) and nucleotide diversity (π) for cpDNA were calculated in DnaSP v.5.10.01 (Librado and Rozas, 2009). For the SCNGs, we also used DnaSP to estimate basic genetic parameters such as number of aggregating sites (S), nucleotide polymorphism in terms of θw (Watterson, 1975) and nucleotide diversity at total sites (θwt and πt), silent sites (θwsil and πsil) and nonsynonymous sites (θwa and πa), number of haplotypes (Nh), haplotype diversity (Hd) and minimum number of recombination events (Rm). Network relationships of cpDNA and SCNGs haplotypes were constructed with NETWORK v.4.6 (Bandelt et al., 1999). Geographical distributions of haplotypes were visualized in ArcGIS v.10.2 (ESRI, Redlands, CA, USA). The average gene diversity within populations (HS), total gene diversity (HT) and population differentiation with unordered (GST) and ordered (NST) haplotypes (cpDNA/SCNG) were estimated using PERMUT v.1.0 (Petit et al., 2005), which was also employed to test whether NST is significantly larger than GST (1000 permutations).
For the 15 EST-SSR loci, MICROCHECKER v.2.2.3 (Van Oosterhout et al., 2010) was used to test for the presence of null alleles and genotyping errors. Linkage disequilibrium (LD) between pairs of loci was surveyed using FSTAT v.2.9.3 (Goudet, 2001). Significance levels of LD or departures from Hardy–Weinberg equilibrium (HWE) were corrected by the sequential Bonferroni method (Rice, 1989). Genetic diversity indices (total number of alleles, TA; number of private alleles, PA; expected heterozygosity, HE; observed heterozygosity, HO; inbreeding coefficient, FIS; the percentage of polymorphic loci, PPL) were estimated with FSTAT v.2.9.3 or GENALEX v.6.5 (Peakall and Smouse, 2012) across all loci for all populations. AR (allelic richness) was estimated after rarefaction to a common sample size of eight gene copies in HP-RARE v.1.1 (Kalinowski, 2005). FSTAT was also used to calculate the number of detected alleles (NA), the number of effective alleles (NE), gene diversity (HS), overall gene diversity (HT), and genetic differentiation between populations (FST) per locus and overall loci (Weir and Cockerham, 1984), while the standardized genetic differentiation G’ST (Hedrick, 2005) was calculated with SMOGD (Crawford, 2010).
Genetic structure of Dipteronia populations was surveyed using the Bayesian clustering method implemented in STRUCTURE v.2.3.4 (Pritchard et al., 2000) based on 15 EST-SSR loci. Twenty independent runs were performed for each number of clusters (K) from 1 to 20, with 100,000 iterations as burn-in followed by 500,000 Markov chain Monte Carlo (MCMC) replications. Both the ‘plateau’ ln Pr (X|K) selection method (Pritchard et al., 2000) and the ΔK method (Evanno et al., 2005), which are implemented in STRUCTURE HARVESTER (Earl and vonHoldt, 2012), were used to determine the optimal number of genetic clusters. Subsequently, STRUCTURE analysis was individually run for D. sinensis under the same parameter settings (excepting K = 1–15). Principal coordinate analysis (PCoA) was also conducted on the EST-SSR data using GENALEX. In addition, STRUCTURE analysis was run for all 45 Dipteronia populations with the same settings (excepting K = 1–15) based on the three cpDNA markers and two SCNGs.
Analysis of molecular variance (AMOVA) was conducted in ARLEQUIN v.3.5 (Excoffier and Lischer, 2010) using both Φ-statistics and R-statistics to quantify variation within populations, among populations and among groups of populations, with significance of fixation indices being tested by 1000 permutations. Isolation-by-distance (IBD) was tested through Mantel tests, which check for relationships between genetic and geographic distances of all Dipteronia populations and 40 D. sinensis populations. Mantel tests were conducted for cpDNA and EST-SSRs with 1000 permutations using the ‘vegan’ package in R v.3.6.1 (Dixon, 2003; Core, 2017).
2.4. Divergence time estimationDivergence times among cpDNA haplotypes were estimated with BEAST v.1.8.0 (Drummond and Rambaut, 2007) using Aesculus chinensis as an outgroup. A birth-death process was specified as tree prior, with fossils of Dipteronia (Ding et al., 2018) and the estimated divergence time from a previous study (Feng et al., 2019) used as calibration points. All calibration priors were treated as a lognormal relaxed clock approach. The Dipteronia-A. chinensis crown group was constrained to a mean age of 60 million years ago (Ma) and standard deviation (SD) of 1 Ma, and the Dipteronia crown group was constrained to a mean age of ~32 Ma and SD of 1 Ma. The most appropriate nucleotide substitution model (HKY + G) was selected using Modeltest v.3.7 (Posada and Crandall, 1998). MCMC of BEAST analyses was run for 50,000,000 generations, with sampling every 5000 steps. MCMC samples were checked in Tracer v.1.7 (Rambaut et al., 2018) to ensure the convergence of the chains through sufficient effective sample sizes (ESS) value (> 200). The final consensus tree was generated using TreeAnnotator v.1.8.0; the first 10% of samples were discarded as burn-in (Drummond et al., 2012).
2.5. Historical demographic analysesNeutrality tests (Tajima's D and Fu's Fs) were conducted using ARLEQUIN to test for deviations from neutrality. To further infer population spatial expansions, the mismatch distribution analysis (MDA) was carried out in ARLEQUIN. The sum of squared deviations (SSD) and the Harpending's raggedness index (HRag) between observed and expected mismatch distributions were used as test statistics with 1000 bootstrap replicates. The extended Bayesian skyline plot (EBSP) was implemented in BEAST (Drummond and Rambaut, 2007) to estimate the demographic history of the D. dyeriana populations and the two D. sinensis lineages (detected with STRUCTURE; see Results) by combining cpDNA and SCNG loci. A strict molecular clock assumed a substitution rate of 3.15 × 10−10 for cpDNA and 6.5 × 10−9 for SCNGs. MCMC parameters were set as follows: 200 million generations, sampled every 20,000 generations. The 95% highest posterior density intervals were viewed in Tracer v.1.7 and the resultant extended Bayesian skyline plot was constructed using R script (Rambaut et al., 2018).
2.6. Genetic barriers and gene flow analysesBased on the EST-SSR dataset, Barrier v.2.2 (Manni et al., 2004) was used to identify genetic barriers among the studied populations by means of Monmonier's algorithm. The significance of our calculations was tested by bootstrapping 1000 matrices of Nei's genetic distance DA (Nei et al., 1983) which were previously obtained with Microsatellite Analyzer (MSA) v.4.05 (Dieringer and Schlötterer, 2003). Historical gene flow among three groups (based on STRUCTURE results) was surveyed using MIGRATE v3.13 (Beerli, 2006) with MCMC maximum likelihood (ML) method based on the EST-SSR dataset. We used the Brownian motion mutation model with constant mutation rates for all loci to estimate M (M = m/μ; where m and M are the migration rate and the mutation-scale migration rate, respectively, and μ the mutation rate per generation), with initial parameters based on FST calculations. We used 10 short chains of 10,000 steps and three long chains of 100,000 steps, and genealogies were recorded with a sampling increment of 100 and a burn-in of 10,000. Three independent runs were performed to ensure the convergence of the final results. We also used the ESR-SSR dataset to estimate contemporary migration rates in BAYESASS v.3.0 (Wilson and Rannala, 2003). Most run parameters of BAYESASS analysis were set at default values. Parameter values were estimated after 1,000,000 burn-in steps to allow stabilizing or convergence out of 3,000,000 iterations, sampling every 2000 iterations. Three independent operations were conducted to minimize the convergence problem.
2.7. Tests of population history by ABC (Approximate Bayesian computation) modelingBased on the STRUCTURE results, seven alternative scenarios of population history for three lineages (DSW, DSE and DD) were summarized (Fig S1 and Table S9) and simulated in DIYABC v.2.1 (Cornuet, 2014) based on all 15 EST-SSR loci and the two SCNGs. All parameters were set under the uniform priors, and a goodness-of-fit test was conducted to check the priors of all parameters before implementing the simulation (Table S9). The average generation time for Dipteronia was assumed as 50 years in this study. Each simulation based on the SSR dataset was summarized using the following variables: mean number of alleles, mean genetic diversity for each lineage, mean classification index, FST and shared allele distance between pairs of lineages. Statistics for each simulation with SCNGs were summarized as follows: number of segregating sites, number of haplotypes, mean pairwise differences for each lineage (W) and mean pairwise differences between pairs of lineages (B). To obtain the best model for Dipteronia species, at least 1,000,000 simulated datasets were run for each scenario. To identify the most likely scenario, 1% of the stimulated datasets closest to the observed data for the logistic regression and direct approaches were selected, and relative posterior probability (PP) with 95% confidence intervals (95% CI) was estimated for each scenario. Finally, the most likely scenario was selected according to the parameters’ posterior distributions.
2.8. Occurrence data and climatic variablesCurrent distribution information for the two species of Dipteronia was recovered from specimens deposited in the Chinese Virtual Herbarium platform (http://www.cvh.ac.cn), from National Specimen Information Infrastructure (http://www.nsii.org.cn), from published articles, from our personal contacts, and from the sampling sites of this study. A total of 410 occurrences were found (Table S10; 40 for D. dyeriana and 370 for D. sinensis). A set of 19 bioclimatic variables under current conditions (ca. 1960–1990) were downloaded from the WorldClim database (http://www.worldclim.org/) at 2.5 arc-min (ca. 5 km) resolution. To identify species-specific environmental variables, candidate models were selected after correlation analysis in points generated from background bias file (Fig. S2). Firstly, a Pearson correlation analysis was conducted to exclude models with combinations of highly correlated variables (r ≥ |0.8|). Secondly, we calculated Variance Inflation Factors (VIF) with the vif function to avoid multi-collinearity; only variables with VIF values < 5 were retained. Finally, six climatic variables were selected as data predictors, namely isothermality (bio3), minimum temperature of the coldest month (bio6), temperature annual range (bio7), annual precipitation (bio12), precipitation of the driest month (bio14), and precipitation seasonality (bio15). All statistical analyses were performed in R v.3.6.1 (Core, 2017) in RStudio platform (Racine, 2012).
2.9. Niche comparison analyses on environmental space (E-space)Firstly, a principal component analysis (hereafter standard PCA) was used to examine the climatic variability of the realized niches in the genetically-studied populations (N = 45) across the total climatic space. A simple agglomerative hierarchical clustering method (UPGMA) was conducted based on the means of the principal components (PCs) (with an eigenvalue ≥1) to examine the relationships among realized niches of these populations. The distance matrix was calculated using the squared Euclidian distance employing the dist function and the resulting dendrogram was drawn with the average Hierarchical Clustering Method.
To test for niche divergence/conservatism between the two Dipteronia species and the three inferred lineages [namely, the west (DSW) and east lineages (DSE) of D. sinensis, and D. dyeriana (DD)], we conducted a PCA-env (a PCA calibrated on the entire environmental space of the study background) with the methodological framework developed by Broennimann et al. (2012) and the R script extracted from Herrando-Moraira et al. (2019). The backgrounds for the occurrences of the two species and/or the three lineages (DD, DSE, and DSW) were selected from minimum convex polygons with a buffer size of 0.3°, as proposed by Silva et al. (2016). Values of the six climatic variables were recovered to construct the available environmental space for the two principal axes. We built smoothed density distributions of both species' (and the three Dipteronia lineages') occurrences with a kernel density function to correct the original observed occurrences and then we projected them in the gridded environmental space (500 × 500 grid cells). To compare the two species, we used the dataset composed by the genetically-studied populations plus the two species’ total occurrences (N = 267 after removing duplicate occurrences within each pixel from Table S10). For the PCA-env conducted with the three lineages (DSE, DSW, and DD), however, we used only the genetically-studied populations for D. sinensis, as it is not possible to know which of the occurrences of this species from Table S10 belong to lineage DSE and which to lineage DSW (the sample size was, finally, N = 80).
We further calculated Schoener's D over the E-space between lineages, which is a comparative metric of niche overlap, ranging from 0 (no overlap) to 1 (complete overlap) (Schoener, 1970). In addition, niche equivalency tests (considering only the occurrences) and niche similarity tests (considering both occurrences and background climates) were used to evaluate niche divergence/conservatism (Warren et al., 2008; Broennimann et al., 2012). These tests compare the observed D values (Dobs) with a null distribution of 100 simulated D values (Dsim). A Dobs bigger or smaller than Dsim (P < 0.05) indicates niche conservatism or divergence, respectively. Alternatively, if the value of Dobs falls into the 95% of Dsim values (P > 0.05), we could not reject the hypothesis of niche divergence or conservatism between groups. We ran both analyses in a one-side test, and each one was performed twice with options ‘greater’ or ‘lower’ to evaluate higher or lower niche equivalencies/similarities than randomly expected, respectively, using the ecospat package (Di Cola et al., 2017). Furthermore, we quantified niche dynamics by calculating three indices: unfilling, stability, and expansion, based on the areas of the first two components of PCA-env (Petitpierre et al., 2012). All E-space analyses were performed in R.
2.10. Distribution changes on geographic space (G-space)We assessed shifts in geographic distribution for each species and across different times in geological history, including several stages of the Pleistocene (Last Glacial Maximum or LGM, ~21 ka, Last Interglacial or LIG, ~130 ka, and Marine Isotope Stage 19 or MIS19, ~0.787 Ma), Pliocene (the mid-Pliocene Warm Period or mPWP, ~3.205 Ma, and Marine Isotope Stage M2 or MIS M2, ~3.3 Ma) and present, based on ENMs in G-space using Maxent v.3.4.1 (Phillips et al., 2006). Past climate layers were retrieved from Oscillayers database (Gamisch, 2019). For the G-space analyses, we used the same six uncorrelated climatic variables as for the E-space (bio3, bio6, bio7, bio12, bio14, and bio15). All model sets were built with the following conditions: 100 replicates using the subsample method as resampling strategy, selecting 25% of random occurrence records for model testing, and with the options extrapolate, do clamping. We used two measures of model performance: the area under the receiver operating characteristic (ROC) curve (AUC) and the true skill statistic (TSS). According to Swets (1988), AUC values > 0.9 indicate high accuracy, 0.7–0.9 good accuracy, and < 0.7 low accuracy. Values with a TSS higher than 0.5 are considered as optimal in terms of power prediction (Allouche et al., 2006). In addition, we also estimated the mean habitat suitability (HS) values of the three lineages (DSE, DSW, and DD, N = 45) at the abovementioned six geologic stages. HS was measured as MaxEnt's logistic output, which can be cautiously interpreted as the predicted probability of presence.
2.11. Population connectivityTo explore possible historical dispersal routes of D. sinensis, we conducted a landscape connectivity analysis based on the combination of ENM models and the cpDNA haplotype network (Chan et al., 2011). The ENM was inverted to obtain a friction cost layer (i.e., 1-ENM) with the ‘raster calculator’ tool of ArcGIS. The obtained friction cost layer was used to generate independent least cost path (LCP) rasters for all sampled populations. Then, a population connectivity map was produced by summing the LCP rasters among all populations with shared and sister haplotypes. To reflect landscape heterogeneity in dispersal processes, the LCPs were classified into different default categories and subsequently summed for all pairwise comparisons of the LCPs corridor layers (for details, see Brown, 2014).
3. Results 3.1. cpDNA phylogeography and diversityThe concatenated fragments of cpDNA comprised 1305 bp (psbA-trnH: 561 bp, psbJ-rpl33: 265 bp and trnS-psbZ: 479 bp). A total of 17 cpDNA haplotypes were identified based on variable nucleotide sites (Tables S4–S6). Fourteen haplotypes (H1–H14) were specific to D. sinensis and three (H15–H17) to D. dyeriana; i.e., there were no shared haplotypes between the two Dipteronia species (Fig. S3). Moreover, there were >50 mutational steps separating the specific haplotypes for each species (Fig. S3). The most frequent haplotype (H3) of D. sinensis was mainly found in the populations of eastern Qinling Mountains, and the predominant haplotype for D. dyeriana was H16, suggesting that H3 and H16 might be the ancestral haplotypes in D. sinensis and D. dyeriana, respectively.
At the population level, a rather low haplotype diversity (Hd = 0.117, π = 0.008 × 10−2) was detected for Dipteronia based on the cpDNA variation. Higher means of genetic diversity were found for D. sinensis compared to D. dyeriana (Hd = 0.123 vs. 0.071; π = 0.009 × 10−2 vs. 0.000 × 10−2; Table S1). AMOVA analysis indicated strong cpDNA genetic differentiation across all populations (94.03%). The among-taxa variance component (85.55%) was much higher than the among-populations within taxa (12.53%) and within-populations (1.91%) components (Table 1). In addition, a significant phylogeographic structure was detected at the genus level and for D. sinensis (Dipteronia sp.: NST = 0.940, GST = 0.850, P < 0.05; D. sinensis: NST = 0.913, GST = 0.832, P < 0.05). Mantel's test demonstrated a significant IBD for all Dipteronia populations (r = 0.167, P < 0.05) and 40 populations of D. sinensis (r = 0.305, P<0.05) based on cpDNA.
| Source of variation | d.f. | Sum of squares | Variation components | Percentage of variation | Φ-statistics | |
| cpDNA | All samples | |||||
| Among populations | 44 | 3529.292 | 8.152 | 94.03 | ||
| Within populations | 395 | 204.333 | 0.517 | 5.97 | ΦST = 0.94b | |
| Two taxa | ||||||
| Among taxa | 1 | 2083.933 | 23.124 | 85.55 | ΦCT = 0.86b | |
| Among populations within taxa | 43 | 1445.358 | 3.387 | 12.53 | ΦSC = 0.86b | |
| Within populations | 395 | 204.333 | 0.517 | 1.91 | ΦST = 0.98b | |
| DS11898 | All samples | |||||
| Among populations | 44 | 3350.424 | 3.868 | 88.05 | ||
| Within populations | 835 | 438.272 | 0.524 | 11.95 | ΦST = 0.94b | |
| Two taxa | ||||||
| Among taxa | 1 | 2980.097 | 16.761 | 94.70 | ΦCT = 0.95b | |
| Among populations within taxa | 43 | 370.328 | 0.414 | 2.34 | ΦSC = 0.44b | |
| Within populations | 835 | 438.272 | 0.525 | 2.97 | ΦST = 0.97b | |
| DS43372 | All samples | |||||
| Among populations | 44 | 1926.529 | 2.232 | 93.96 | ||
| Within populations | 835 | 119.733 | 0.144 | 6.04 | ΦST = 0.94b | |
| Two taxa | ||||||
| Among taxa | 1 | 1777.530 | 10.007 | 96.96 | ΦCT = 0.97b | |
| Among populations within taxa | 43 | 148.999 | 148.999 | 1.65 | ΦSC = 0.54b | |
| Within populations | 835 | 119.733 | 119.733 | 1.39 | ΦST = 0.99b | |
| EST-SSR | All samples | |||||
| Among populations | 44 | 2939.860 | 1.841 | 44.10 | ||
| Within populations | 1533 | 3577.911 | 2.334 | 55.90 | RST = 0.44b | |
| Two taxa | ||||||
| Among taxa | 1 | 1413.065 | 4.22910 | 56.30 | RCT = 0.56b | |
| Among populations within taxa | 43 | 1526.795 | 0.94907 | 12.63 | RSC = 0.29b | |
| Within populations | 1533 | 3577.911 | 2.33393 | 31.07 | RST = 0.69b | |
| Three groupsa | ||||||
| Among groups | 2 | 1573.038 | 1.68519 | 34.50 | RCT = 0.35b | |
| Among populations within groups | 42 | 1366.822 | 0.86509 | 17.71 | RSC = 0.27b | |
| Within populations | 1533 | 3577.911 | 2.33393 | 47.79 | RST = 0.52b | |
| All populations of D. sinensis | ||||||
| Among populations | 39 | 1405.839 | 0.96838 | 29.14 | ||
| Within populations | 1354 | 3189.161 | 2.35536 | 70.86 | RST = 0.29b | |
| Two lineages of D. sinensis | ||||||
| Among regions | 1 | 159.974 | 0.19585 | 5.72 | RSC = 0.27b | |
| Among populations within regions | 38 | 1245.865 | 0.87565 | 25.55 | RST = 0.31b | |
| Within populations | 1354 | 3189.161 | 2.35536 | 68.73 | ||
| a Genetic clusters delimited according STRUCTURE under the K = 3 scenario. b P < 0.01, 10,000 permutations. | ||||||
The two SCNGs (DS11898, DS43372) had a length of 574 bp and 548 bp, respectively. The polymorphism of DS11898 yielded 33 haplotypes, with 28 haplotypes (N1–N28) being specific to D. sinensis and five to D. dyeriana (N29–N33) (Table S7 and Fig. S4). The polymorphism of DS43372 yielded 14 haplotypes, with 13 haplotypes (S1–S13) and one haplotype (S14) specific to D. sinensis and D. dyeriana, respectively (Table S8 and Fig. S5). Similar to the cpDNA, no shared haplotypes were observed between the two species, with many mutational steps in between (32 and 20 steps for DS11898 and DS43372, respectively; Figs. S4 and S5).
Across all Dipteronia populations, the genetic diversity at DS11898 was higher than that at DS43372 (Hd: 0.910 vs. 0.645; πt: 1.103 × 10−2 vs. 0.814 × 10−2; πsil: 2.219 × 10−2 vs. 1.620 × 10−2; Table S10). At the species level, genetic diversity was higher in D. sinensis than in D. dyeriana (Table S11), similar to the population level (Table S1). Across all populations, AMOVA analyses revealed that most genetic variation resided among populations (DS11898: 88.05%; DS43372: 93.96%; Table 1). When the species level was introduced, AMOVA analyses detected that 94.70% (DS11898) and 96.69% (DS43372) of the among-taxa variation was much higher than both among-population within taxa (DS11898: 2.34%; DS43372: 1.65%) and within-population variation (DS11898: 2.97%; DS43372: 1.39%). A significant phylogeographic structure was detected at the genus level based on the two SCNGs (DS11898: NST = 0.735, GST = 0.345, P < 0.05; DS43372: NST = 0.711, GST = 0.615, P < 0.05).
3.3. Demographic analysis based on molecular dataNeutrality tests and mismatch distribution analysis (MDA) were conducted based on cpDNA and SCNGs for all populations of Dipteronia and separately for each species (Table S12). For all cases, Tajima's D and Fu's FS were non-significant. Moreover, mismatch distributions of haplotypes for all populations and for D. sinenesis rejected the spatial expansion model for cpDNA (SSD, P < 0.05), while D. dyeriana showed an expansion model (P > 0.05) (Table S12). In addition, the mismatch distributions of all populations and of D. sinensis were bimodal or multimodal at least for one of the markers, supporting range stability (Fig. S6). DS43372 was not used to conduct the neutrality tests or MDA for D. dyeriana because only one haplotype was detected. Similarly, the EBSPs showed that the past populations of D. sinensis and D. dyeriana could be inferred with a model of constant size (Fig. S7), and no recent population expansion/shrinkage was detected in the two lineages of D. sinensis (Fig. S7).
3.4. Nuclear microsatellite diversity and population structureMICRO-CHECKER analyses indicated low occurrence of null alleles from 15 EST-SSR loci, and there was no evidence for LD. When all samples were regarded as one population, significant deviation from HWE due to heterozygote deficiency was not observed after the Bonferroni corrections. Genetic diversity at the locus level is summarized in Table S13. Expected heterozygosity (HE) and allelic richness (AR) were, on average, slightly higher for D. sinensis than for D. dyeriana (HE = 0.298 vs. 0.282; AR = 1.90 vs. 1.82; Table S1). For D. sinensis, the mean values of HE and AR for the West lineage (see the STRUCTURE results) were slightly lower than those for the East lineage (West vs. East, HE: 0.294 vs. 0.298, AR: 1.88 vs. 1.90).
Based on the STRUCTURE analysis conducted for all populations, ΔK indicated an ‘optimal’ value of K = 2, which is not incompatible with the results of the ‘plateau’ method as ln Pr (X|K) increases slowly from K = 2 (Fig. S8A, B). At K = 2, all populations were clearly split into D. sinensis and D. dyeriana lineages (Fig. 1C). At K = 3, D. dyeriana still maintained a pure cluster, whereas D. sinensis was split into two different clusters following a geographic pattern (i.e., a ‘West’ cluster and an ‘East’ cluster could be delimited), with only a few populations showing signs of genetic admixture (Fig. 1C). Populations MJS–DB, mainly located in the western part of Qinling Mountains towards the west, clustered into the West lineage (DSW), and the remaining populations (FP–FY) located in the eastern Qinling Mountains towards the east, clustered into the East lineage (DSE) (Fig. 1A–C). When STRUCTURE was exclusively run for D. sinensis populations, K = 2 was the best clustering scheme according to ΔK (Fig. S8C, D), and was identical to the previous structure based on all Dipteronia populations at K = 3 (data not shown). The PCoA jointly explained 57.12% (PC1: 49.30%, PC2: 7.82%) and 28.55% (PC1: 16.54%, PC2: 12.01%) of the variance for all populations and D. sinensis, respectively (Fig. S9A, B). The PCoA results showed a similar genetic structure to that inferred by the STRUCTURE analyses. STRUCTURE results inferred from the cpDNA markers and the SCNGs also showed an ‘optimal’ value of K = 2, with populations clearly split into D. sinensis and D. dyeriana lineages (Figs. S10, S11, S12A, B and S13A, B). When STRUCTURE was exclusively run for D. sinensis populations, K = 6 and K = 3 were the best clustering schemes for cpDNA markers and SCNGs, respectively (Figs. S12C, D and S13C, D). The clustering scheme agreed with those based on all Dipteronia populations at K = 7 for cpDNA and K = 4 for SCNGs (data not shown).
The EST-SSR-derived AMOVA analyses showed high levels of population genetic differentiation across all populations (FST = 0.44, P < 0.001) (Table 1). When the species level was introduced, the proportion of total variation among taxa (56.30%) was higher than that among populations within taxa (12.63%) and within populations (31.07%; Table 1). In contrast, the percentage of total variation attributed to differences among the groups delimited by STRUCTURE under the K = 3 scenario was lower (34.50%; Table 1) than that within populations (47.79%). For D. sinensis, the proportion of variation among populations was lower than that within populations (29.14% vs. 70.86%). In addition, when the two regions of D. sinensis (lineages DSW and DSE) were considered, the within populations component (68.73%) was much higher than the among-region (5.72%) and the among-population within regions (25.55%) components. EST-SSR markers indicated there was a significant correlation between genetic distances and geographic distances for all Dipteronia populations (r = 0.487, P<0.001) and 40 populations of D. sinensis (r = 0.278, P<0.05).
3.5. Molecular dating based on cpDNA dataThe BEAST-derived cpDNA chronogram generated two well-supported clades (100%), namely D. sinensis and D. dyeriana lineages (Fig. 2). The chronogram indicated that the crown age of Dipteronia was ca. 32.11 Ma (95% highest posterior density, HPD: 30.13–33.99 Ma, posterior probability, PP = 1.00). The coalescent time of all D. sinensis cpDNA haplotypes was estimated as 26.45 Ma (95%, HPD 16.24–33.1 Ma) to 2.32 Ma (95%, HPD: 0–9.22 Ma), and the D. dyeriana haplotypes are predicted to have diverged ~12.71 Ma (95%, HPD: 1.34–27.3 Ma) (Fig. 2).
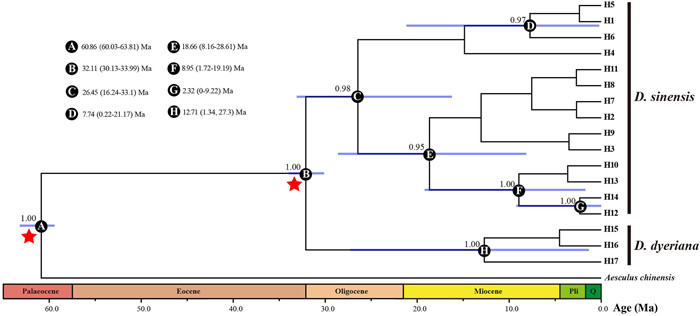
|
| Fig. 2 BEAST-derived chronogram for 17 haplotypes of Dipteronia based on three cpDNA fragments. Blue bars indicate 95% highest posterior densities (HPDs) credibility intervals for node ages (Ma). Posterior probabilities (>0.9) are labelled above nodes. Geological period abbreviations: Pli, Pliocene; Q, Quaternary. Red stars represent calibration points. |
Barrier analysis based on EST-SSR data identified strong genetic barriers between Dipteronia sinensis and D. dyeriana (Fig. S14). Expectedly, much lower levels of historical gene flow were detected between the two Dipteronia species compared to those between the two lineages of D. sinensis (Fig. S15). All rates between the different genetic clusters were more or less symmetric except those detected between D. dyeriana and the eastern lineage of D. sinensis (95% CI: MDD→DSE = 3.79–5.23 vs. MDSE→DD = 0.62–0.92). Multiple runs of BAYESASS yielded very low estimates of contemporary gene flow among the three inferred genetic groups, and the migration rates among these lineages were mostly asymmetrical (Fig. S15).
3.7. Population history inferred from ABC analysesAccording to DIYABC analysis of tree groups based on EST-SSR and SCNGs datasets, scenario 2 had the highest posterior probability (0.7115, 95% CI: 0.6916–0.7313) (Table S14). This scenario described a situation that DSE (East lineage) and DD (Dipteronia dyeriana) simultaneously diverged from the same ancestor, and then DSW (West lineage) diverged from the ancestor of the Eastern lineage (DSE). The mean values for the effective population sizes N1 (DSW), N2 (DSE), N3 (DD) and NA were 3.19 × 105, 3.23 × 104, 1.54 × 104 and 1.94 × 106, respectively. The median values of the divergence time were estimated as t1 = 1.32 × 103 (95% HPD, 2.77 × 102–1.04 × 104) (split of DSW from the DSE) and t2 = 4.51 × 105 (95% HPD, 2.39 × 105–4.97 × 105) (split between DSE and DD). Assuming an average generation time of 50 years for Dipteronia species, the divergence time of t1 and t2 was 6.6 × 104 (95% HPD, 1.39 × 104–5.2 × 104) and 22.55 × 106 (11.9 × 106–24.8 × 106) years ago, respectively (Table S15).
3.8. Niche variation on E-spaceIn the standard PCA analysis for the populations of the two Dipteronia species (N = 45; Fig. 1D), the first three PCs accounted for the 97.47% (PC1 = 53.22%, PC2 = 39.43%, PC3 = 3.71%, Table S16) of the climatic variance. The UPGMA dendrogram (Fig. 1B), derived from PC values of the standard PCA analysis, showed that the range of DSE was climatically most closely related to that of DSW, whereas DD was climatically more distant.
In the PCA-env analyses for the two species (N = 267) and for the three clusters (N = 45), the first two axes explained 82.31% (PC1 = 57.19% and PC2 = 25.12%) and 82.19% (PC1 = 47.96% and PC2 = 34.24%), respectively. In the two PCA-env analyses, the first component (PC1) was mainly explained by annual precipitation, whereas the second was principally loaded by precipitation of the driest month (Fig. S16A, B). Niche plots displaying 20% and 100% of occurrences density showed no overlap between Dipteronia sinensis and D. dyeriana (Fig. 1E and F). However, some overlap in climatic space was detected between DSE and DSW, with niche breath of DSE somewhat larger than that of DSW (Fig. 1E and F). In the two PCA-env plots, the range of D. dyeriana showed very limited niche breadth while being completely differentiated from D. sinensis (Fig. 1E and F).
The overlap D values between the two Dipteronia sinensis lineages (DSE and DSW) was low (D = 0.15). However, D. sinensis and D. dyeriana ranges were completely differentiated (D = 0). In addition, niche equivalency and niche similarity tests did not reveal significant niche conservatism or divergence patterns between DSE, DSW, and DD (Table S17). In agreement with D values, both D. sinensis and D. dyeriana niches were well differentiated in terms of niche dynamics (unfilling = 1, expansion = 1, and stability = 0). The values of niche dynamics for DSE and DSW were also in line with the low but detectable degree of overlap between the two lineages of D. sinensis; DSE ‘expanded’ the niche of DSW by 82%, whereas DSW represented an ‘expansion’ of 57% of the niche of DSE (Table S17).
3.9. ENM models on G-spaceAll ENM models showed moderately high model performance and prediction power (AUC values ≥ 0.947; TSS values ≥ 0.792) (Table S18). The predicted distribution ranges of the two Dipteronia species were generally similar to their current distributions (Fig. 3A). Remarkably, this relict genus showed relatively similar potential distribution ranges during the mid-Pliocene (MIS M2, ca. 3.3 Ma, and mPWP, ca. 3.2 Ma) and the Pleistocene (MIS19, ca. 0.787 Ma; LIG, ca. 130 ka, and LGM, ca. 21 ka) (Fig. 3). Dipteronia sinensis showed a potentially continuous range mostly located on the Qinling-Daba Mountains (hereafter ‘Qinling-Bashan’) since the mid-Pliocene, with few exceptions (at the mPWP there was some range contraction in Wu and Wuling Mountains, whereas during the LGM new potential areas would have appeared in eastern China except for a range contraction in Longmen Shan; Fig. 3). Although the potential distribution range of D. dyeriana experienced relatively large changes (with the maximum expansion and contraction at the LGM and mPWP, respectively), it was always located in south-eastern Yunnan (Fig. 3).
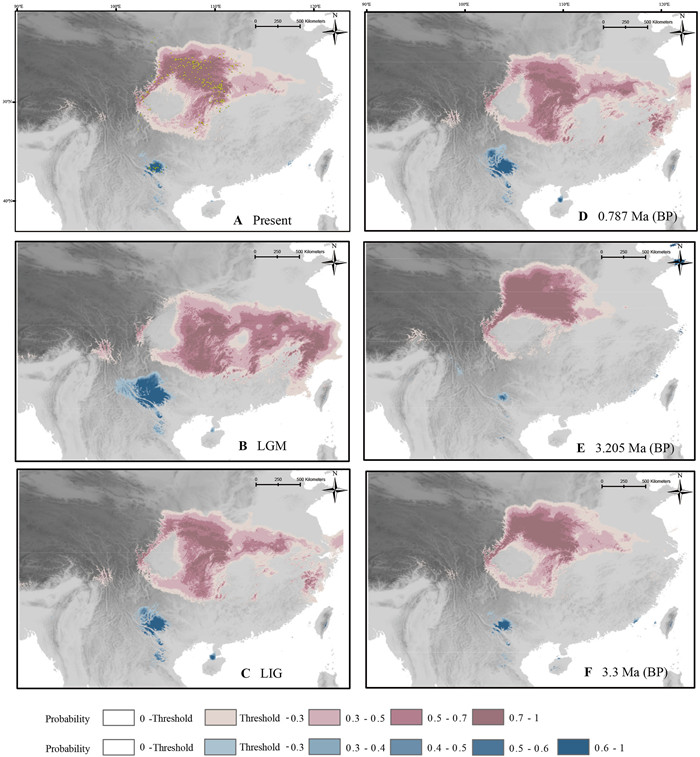
|
| Fig. 3 Potential distribution of two Dipteronia species at present, during the Pleistocene (ca. 0.787 Ma; LIG, ca. 130 ka and LGM, ca. 21ka) and during the mid-Pliocene (MIS M2, ca. 3.3 Ma and mPWP, ca. 3.205 Ma), respectively. |
Our models predict that habitat suitability (HS) was maintained throughout time in Dipteronia (Fig. 4), although the HS (with values generally ranging from 0.6 to 0.8) indicate that DSE was likely more stable than was DSW (value of HS: 0.2–1). This higher variability in HS for DSW was likely partly due to the loss of a large part of the potential range of this lineage during the LGM (around Longmen Shan; Fig. 3B). The HS of D. dyeriana was predicted to have remained very high with the exception of mPWP (3.2 Ma) (Fig. 4), when the range of the species significantly shrank (Fig. 3E).
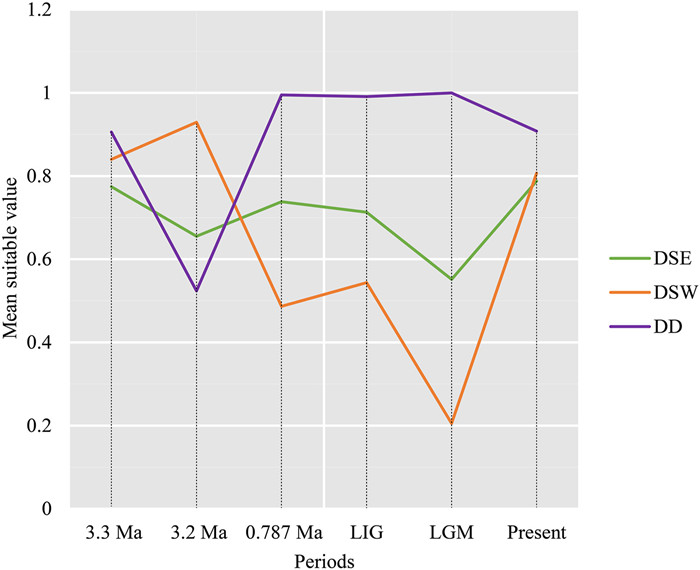
|
| Fig. 4 The mean probability of habitat suitability values for three genetic lineages of Dipteronia (DSE, DSW, and DD; N = 45: DSE, 24; DSW, 16; DD, 5) at present, during the Pleistocene (MIS19, ca. 0.787 Ma; LIG, ca. 130 ka, and LGM, ca. 21 ka) and during the mid-Pliocene (MIS M2, ca. 3.3 Ma, and mPWP, ca. 3.205 Ma), respectively. |
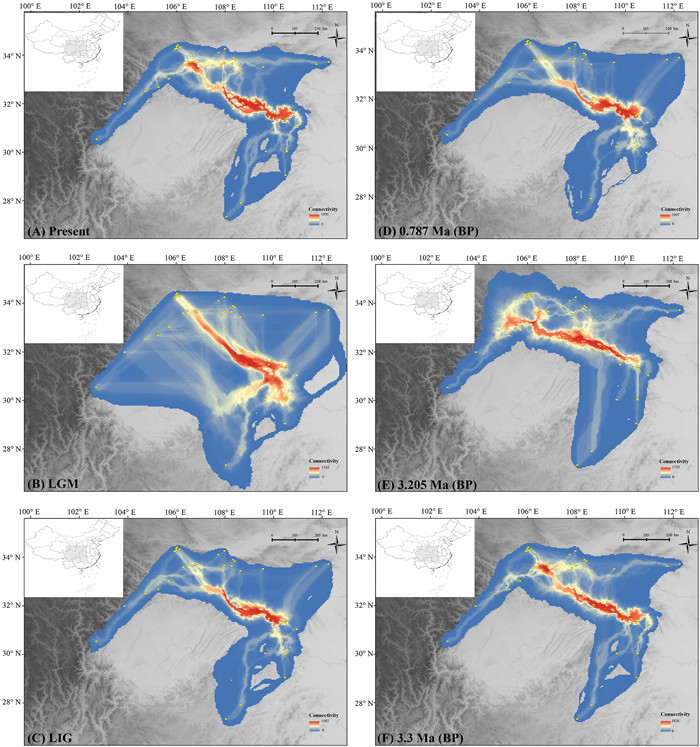
Maps obtained after landscape connectivity analysis showed a permanent east–west oriented dispersal corridor at least since the mid-Pliocene (the areas with the hottest color), which largely coincides with Qinling-Bashan, and slightly extending to the Wu Mountains (Fig. 5).
|
| Fig. 5 cpDNA dispersal corridor of Dipteronia sinensis at present, during the Pleistocene (ca. 0.787 Ma; LIG, ca. 130 ka and LGM, ca. 21ka) and during the mid-Pliocene (MIS M2, ca. 3.3 Ma and mPWP, ca. 3.205 Ma), respectively. |
In recent decades, studies on species delimitation, traditionally based on morphology, have incorporated both genetic and ecological data. Today, the usefulness of this integrative approach is indisputable, with many studies showing high congruence between the different lines of evidence, e.g. in Orinus (Su et al., 2015) or Cynara (Massó et al., 2018). Although the two extant species of Dipteronia are morphologically very similar (they are sometimes treated as subspecies in old works; Renner et al., 2008), modern treatments recognize the two taxa as separate species (Ying et al., 1993; Xu et al., 2008), mostly based on differences in the panicles and the fruits. However, such lack of conspicuous morphological differences must be contextualized within the morphological stasis of many paleoendemic lineages (Milne and Abbott, 2002), including Dipteronia (Ding et al., 2018).
Certainly, our genetic results based on EST-SSRs, cpDNA, and SCNGs consistently indicate that the two Dipteronia species represent highly divergent lineages: (1) K = 2 is by far the best clustering scheme in the STRUCTURE analyses, with each species having its own cluster (Fig. 1, S8 and S10–S13); (2) in the PCoA the populations belonging to each species are clustered together and the two clusters are clearly differentiated (Fig. S9); (3) the first genetic barrier detected with Monmonier's algorithm (B = 1) separates the two species with a maximum bootstrap value (Fig. S14); (4) the among-taxa component of the AMOVA is, by far, the largest (cpDNA: ΦCT = 0.86; DS11898: ΦCT = 0.95; DS43372: ΦCT = 0.97; EST-SSR: RCT = 0.56; Table 1); (5) no shared haplotypes and many mutational steps have been detected between the two species based on both cpDNA and SCNGs (Figs. S3~S5). Additionally, the cpDNA chronogram also indicates that the two Dipteronia species diverged during the Oligocene (32.11 Ma), which is older than the divergence time estimated from the DIYABC analyses (22.55 Ma). The discrepancy of divergence time would be mainly caused by the rapid mutation rate of SSR and SCNGs datasets (Selkoe and Toonen, 2006). Regardless, both cpDNA phylogeny and DIYABC analyses indicted that D. sinensis and D. dyeriana have experienced long independent evolutionary histories [as also found by Feng et al. (2024), who placed their divergence somewhat earlier—to the Paleocene/Eocene boundary].
The Cenozoic global climate changes promoted the diversification of plant lineages but also the extinction of many others (Latham and Ricklefs, 1993; Collinson, 2000; Jaramillo et al., 2006), with a central role played by the three climatic “aberrations” (or profound abnormalities of 103–105 years that occurred around the Paleocene/Eocene, Eocene/Oligocene, and Oligocene/Miocene boundaries; Zachos et al., 2001; Pälike et al., 2006). Our molecular dating indicates that the extant Dipteronia species diverged during the early Oligocene (~32.11 Ma; Fig. 2), i.e., just after the Eocene/Oligocene boundary aberration, which took place around 33.5 Ma (Eldrett et al., 2009). In addition to promoting speciation, the global cooling and increased seasonality and/or aridification that characterized the Eocene–Oligocene transition (Zanazzi et al., 2007; Eldrett et al., 2009; Xiao et al., 2010; Zhang et al., 2012; Sun et al., 2014a) might have also driven the populations of Dipteronia in the Northern Hemisphere to regional extinction. The most recent fossils of Dipteronia from North America are those from the Bridge Creek flora of Oregon, dated to about 32 Ma (McClain and Manchester, 2001). Notably, other plants today endemic to East Asia were also extirpated from North America just before or during the Oligocene, including Craigia, Davidia, Diplopanax, Hovenia, and Keteleeria (Manchester et al., 2009). In contrast, the fossils of D. brownii recently found in Yunnan dating from ca. 32 Ma (Ding et al., 2018), together with the current ranges of D. sinensis and D. dyeriana, clearly suggest that this genus persisted to the present —as many other relict lineages did—in the long-term stable refugia recently identified by Tang et al. (2018) in central and south-western China. As expected, both MDA and EBSPs showed no recent expansion/contraction signals for two D. sinensis lineages and D. dyeriana, indicating stable population sizes for Dipteronia species.
In addition to genetic and morphological evidence, the climatic niche assessment of Dipteronia also indicates that the two extant taxa (D. sinensis and D. dyeriana) can be delimited as independent species. Standard PCA and UPGMA both show clear ecological divergence between the two Dipteronia species, and E-space analyses based on the Broennimann method indicate no climatic niche overlap (Fig. 1B–D and E). For closely related species (e.g., sister species such as in the present study) with partially overlapping or adjacent ranges, ecological divergence would likely be important in facilitating the process of speciation (Nagy, 1997; Nosil, 2008; Nosil et al., 2009; Anacker and Strauss, 2014; Li et al., 2014; Zhou et al., 2014). Indeed, ecological differentiation would provide preconditions for the divergence of different species/lineages following the initial spatial isolation, and may have resulted in some degree of differential adaptation to their corresponding environmental conditions (Liu et al., 2013). This seems to be the case of D. sinensis/dyeriana, since Feng et al. (2024) identified species-specific genes under positive selection likely involved in adaptation (e.g. related to cold stress). Studies have shown that local adaptation generates morphological differences in plant species (Ellis and Weis, 2006; Li et al., 2014; Chapman et al., 2016). According to Flora of China (Xu et al., 2008), there are some differences in plant height, in inflorescence pubescence, and in fruit size between the two Dipteronia species, which suggest that ecological factors might have played an important role in their speciation by divergent selection. However, more functional genomic data are needed to elucidate the details of how these morphological differences are related to abiotic factors (e.g. Chapman et al., 2016).
4.2. Mountains of central and south-western China: refugium and dispersal corridor for DipteroniaAlthough the dating analyses indicated that the onset of diversification of Dipteronia sinensis lineages began during the late Oligocene (around 26 Ma), most intraspecific diversification events took place after the middle Miocene, and the same is applicable for D. dyeriana (Fig. 2). The Late Oligocene Warming Event is characterized by a climatic amelioration that extended until the end of the Mid-Miocene Climatic Optimum (ca. 15 Ma) (Zachos et al., 2001), which would have enabled the existence of suitable habitats for the thermophilic Arcto-Tertiary taxa in central and south-western regions of China, promoting the abundance of relict plant species (Ding et al., 2018). Most branching events in Dipteronia occurred after the Mid-Miocene Climatic Optimum, when the climate became progressively colder (Zachos et al., 2001) but also more seasonal (especially evident with the onset of the East Asian monsoons during the Miocene; An et al., 2001; Liu et al., 2017). The onset and the intensification episodes of the East Asian monsoons are intrinsically linked with the phased uplift of the Qinghai-Tibet Plateau (QTP), with peaks of both phenomena at approximately 15 Ma, 10–8 Ma, and 3.6–2.6 Ma (Wan et al., 2007). The QTP uplift since the Miocene induced the uplift and other geomorphological changes in the adjacent mountain ranges and plateaus of central and south-western China, including the Yunnan-Guizhou Plateau and the Qinling Mountains (Shi et al., 2019). These dramatic climatic and geological changes may have fostered inter/intraspecific differentiation of relict plant lineages distributed in these regions apart from Dipteronia; examples include Cyclocarya paliurus (intraspecific divergence starting about 16.7 Ma; Kou et al., 2016), Davidia involucrata (4.8 Ma; Ma et al., 2016), Euptelea pleiosperma (3.96–3.64 Ma; Cao et al., 2016, 2020), Taxus wallichiana (4.2 Ma; Liu et al., 2009, 2013), Tetracentron sinense (9.6 Ma; Sun et al., 2014b), and Urophysa (inter/intraspecific divergence; 8.9 Ma; Xie et al., 2017).
Mountains have often served as refugia for many species distributed in the temperate and tropical regions of the world, because of their very diverse topography, which offers a wide array of climatic and edaphic conditions, even in the harshest periods (e.g., glacial periods of the Pleistocene) (Tian et al., 2018; Suissa et al., 2021). The distribution ranges of the two Dipteronia species fall partially (D. sinensis) or totally (D. dyeriana) within the major long-term refugia for relict species of East Asia (Tang et al., 2018). Mountainous regions of central and south-western China (and contiguous regions of Vietnam and Myanmar) may have been refugia for paleo-species, as these areas would have maintained stable climatic conditions favorable for their growth. Accordingly, ENMs indicate that the mountains where the two Dipteronia species are dwelling would have been suitable since mid-Pliocene with very few exceptions (Fig. 3), which can also be seen from the graph of habitat suitability (HS) (Fig. 4). The topographically diverse landscapes of Qinling-Bashan likely provided suitable climatic conditions for D. sinensis across the six time-periods considered. In contrast, the Longmen Mountains, which sustain most of the populations of D. sinensis located east to 106°E, were not suitable during the LGM (Fig. 3B), thus leading to fewer HS values for the west lineage (DSW) compared to the east one (DSE) at the LGM (Fig. 4). Consistent with these findings, DSW showed lower genetic diversity and allelic richness than DSE (HE: 0.294 vs. 0.320; Table S1). In addition, the loss of genetic diversity is even more evident when the values for the populations exclusively occurring in Longmen Mountains are averaged (HE = 0.205, considering BC, BX, LH, WX, and YB populations). Negative effects of the harsh conditions of the LGM on levels of genetic diversity are well known (Gavin et al., 2014; Chung et al., 2017). Even a positive correlation between the expected heterozygosity (HE) and environmental LGM habitat suitability (HS) has been reported (Chung et al., 2018). The other part of the distribution range that has been not suitable for one of the six-time periods considered in addition to Longmen Mountains, Wuling Mountains (and partially Wu Mountains; Fig. 4), also shows levels of genetic diversity below the average for D. sinensis (HE = 0.298): HE = 0.258, considering populations from both Wu (HH, HPS) and Wuling (FJS, FY, TMS) Mountains; HE = 0.229, only considering populations from Wuling Mountains.
In addition to serving as late Neogene refugia, mountains have often acted as dispersal corridors for plants, allowing for range expansions/contractions but also population connectivity, thus ensuring species integrity (Luebert and Weigend, 2014; Chung et al., 2017; Yu et al., 2017). Recent studies on central and southern China indicate that mountain ranges serve as dispersal corridors for plants, e.g., Nanling Mountains (Tian et al., 2018) and some small ranges located in the Poyang Lake Basin (Fan et al., 2017). Our genetic plus ecological data clearly suggest that Daba Shan-west Qinling Mountains is (and it has been at least since mid-Miocene) a dispersal corridor for D. sinensis, with a slight extension to the Wu Mountains (Fig. 5). Such a dispersal corridor would have contributed to the relatively low divergence among populations (FST = 0.29 based on EST-SSR) in D. sinensis and the very small AMOVA's among-lineage (DSW vs. DSE) component (below 6%; Table 1). Although STRUCTURE analysis detected two genetic lineages that match with an east–west divide, the existence of a contact zone with a clear pattern of admixture and the lack of a clear genetic separation of the two lineages in the PCoA (Fig. S9) supports a corridor that would have allowed gene exchange through time (Fig. S15). Consistent with this theory, the genetic barriers do not match with the two genetic lineages depicted by STRUCTURE (Fig. S14). Despite the cpDNA haplotype diversification starting from late Oligocene, species integrity has been maintained, as no obvious genetic boundaries were observed between the two D. sinensis lineages with STRUCTURE (and the same occurred with SCNGs; Figs. S10 and S11).
The lack of sharp genetic boundaries between the two Dipteronia sinensis lineages is mirrored by the absence of clear ecological boundaries: the standard PCA, the UPGMA and the PCA-env show that there is some niche overlap between the two lineages. In the UPGMA, the populations within the admixture area as indicated by STRUCTURE (basically the populations that are approximately located at 33.5–34.5°N and 106–108.5°E) are included in the same cluster (cluster no. 5, in light blue in Fig. 1); moreover, the cluster that is sister to the rest (no. 2, deep blue) matches quite well with BARRIER analysis, which might indicate that a second incipient divide—delineated by the Han River valley that separates Qinling and Daba Mountains before they meet at about 106°E—could be drawn. Such slight differentiation is also apparent in the graphical output of the PCA-env with 20% of the occurrence density (Fig. 1E). Ecological factors have been shown to promote and maintain intraspecific differentiation in other plant species of subtropical China such as Taxus wallichiana (Liu et al., 2013) and Pinus yunnanensis (Wang et al., 2013). Although ecological divergence has not resulted in the emergence of new species, our results suggest that D. sinensis is in an incipient stage of ecological speciation.
Although we have been able to detect incipient events of both genetic and ecological differentiation within Dipteronia sinensis, the occurrence of a dispersal corridor along the Qinling-Bashan would have ensured both genetic and ecological integrity. Compared with the role as refugium, which is widely recognized for both plants and animals (Fang et al., 2013; Qu et al., 2014; Yuan et al., 2014), the role of Qinling-Bashan as a corridor has only been reported in animals (e.g., for Sinopotamon acutum see Fang et al., 2015; for Rhinopithecus roxellana see Li et al., 2018). We believe that this corridor may have played a major role not only for D. sinensis but also for other plant and animal species native to China given that, because its east–west orientation, it would have enabled the connection between the Hengduan Mountains (the richest biodiversity hotspot in the whole East Asia; Myers et al., 2000) and Wuling Mountains (the main mountain range with a north–south orientation in central China). The connection of these three main orographic units, as shown by the ENMs of our study (Fig. 3), have likely contributed to continental migrations of the regional flora and fauna.
Regarding Dipteronia dyeriana, ENMs indicate that populations may have persisted in south-eastern Yunnan since mid-Pliocene, although experiencing large range contractions and expansions. The greatest expansion of the distribution range of D. dyeriana occurred during the LGM. Although this may appear rather surprising for a species that inhabits the southernmost part of the subtropical belt of China (i.e., almost falling within the tropical belt), D. dyeriana is usually found at high elevations (above 2000 m). Similar to D. dyeriana, several montane (e.g., Pinus kwangtungensis; Tian et al., 2010) as well as many alpine plants [e.g. Picea likiangensis (Li et al., 2013); Rosa sericea complex (Gao et al., 2015)] native to China likely experienced range expansions during the LGM, one of the coldest time-periods of the Pleistocene. As reported for P. kwangtungensis, populations of D. dyeriana likely expanded to nearby lowlands during the glacial periods.
4.3. Levels of genetic diversity of Dipteronia sinensis vs. D. dyeriana and recommendations for conservationThe narrow endemic Dipteronia dyeriana show lower levels of intrapopulation genetic diversity than its widespread congener D. sinensis with all the markers used: cpDNA (Hd = 0.071 vs. 0.123; π = 0.000 × 10−2 vs. 0.009 × 10−2), two SCNGs (Hd = 0.000/0.455 vs. 0.279/0.609; π = 0.000/0.104 × 10−2 vs. 0.058/0.195 × 10−2), and EST-SSR (HE = 0.282 vs. 0.298). This is an expected result as range size is one of the main determinants of genetic variability within populations in plant species (Hamrick et al., 1990; Nybom, 2004). Other factors are demographic stability and effective population sizes (Ne); so, large fluctuations on Ne or a dynamic of recurrent extinction/recolonization may have lowered values of genetic diversity (Nunney, 2020). Although both species of Dipteronia may have suffered important fluctuations of their range since the mid-Miocene (Fig. 3), those of D. dyeriana were probably more dramatic. For example, the latter nearly went extinct during the mPWP (ca. 3.205 Ma; Fig. 3E). In addition, D. dyeriana likely suffered considerable losses to its distribution range during modern times (e.g., south-central Yunnan and Xingyi county in SW Guizhou), as a consequence of habitat fragmentation (Sun et al., 2006; Chen et al., 2017). Similarly, a recent population genomics study confirmed that D. dyeriana has lower genome-wide nucleotide diversity than the more widespread D. sinensis (Feng et al., 2024).
Although the two species are threatened (Dipteronia dyeriana) or nearly threatened (D. sinensis) and D. dyeriana is protected at the national level, to our knowledge routine conservation practices for this endangered genus have yet to be implemented, apart from the generic protection provided by nature reserves to the individuals that grow within them. To save costs, both in situ and ex situ conservation measures should focus on those populations with the highest levels of genetic diversity and/or unique alleles/haplotypes. Examples of populations with high priority include (1) LWG, which shows the highest levels of intrapopulation genetic diversity with EST-SSR and among the highest with cpDNA and SCNGs, and harbors a private haplotype (H5); (2) YS, with the highest number of private SSR alleles (three) and a private cpDNA haplotype (H11); or (3) FJS, with the highest number of private cpDNA haplotypes (two; H12 and H13) and one private SSR allele. In contrast, populations with low levels of genetic diversity (e.g., DB or XE) should be subject to restoration activities such as reinforcements, although from genetically close individuals to avoid the breakdown of the co-adapted gene complexes and outbreeding depression (Fenster and Dudash, 1994); thus, it is not avisable to mix (1) DSE and DSW or (2) populations located north and south of the Han River.
5. ConclusionsOur integrative approach has unraveled the patterns of genetic variability and genetic and ecological divergence of the paleo-endemic genus Dipteronia. The ancestors of the two Dipteronia species likely persisted in mountain refugia in central and south-western China and evolved independently, at least since the beginning of the Oligocene. The long-term spatial isolation likely promoted both ecological and genetic divergence, leading to the two current widely-recognized species. At the same time, the climatic (global cooling) and geological changes (uplift of Yunnan-Guizhou Plateau and Qinling Mountains) that occurred during the last 15 Ma might have fostered intraspecific differentiation. In the more widespread D. sinensis there is incipient ecological and genetic divergence driven by an east–west divide (western part of Qinling Mountains towards the west–eastern Qinling Mountains towards the east) but also a north–south divide (delineated by the Han River valley). The occurrence of long-term stable refugia but, particularly, of a dispersal corridor along Daba Shan-west Qinling would have ensured the genetic and ecological integrity of D. sinensis. As expected, the threatened narrow endemic D. dyeriana shows lower levels of intrapopulation genetic diversity than its more widespread congener.
AcknowledgementsSpecial thanks to Cindy Q. Tang for providing information on the occurrence of Dipteronia. This study was co-supported by the National Natural Science Foundation of China (Grant No. 31470311) and the Ph.D. Programs Foundation of the Ministry of Education of China (Grant No.20136101130001).
CRediT authorship contribution statement
Tao Zhou: Writing – original draft, Software, Methodology, Investigation, Data curation. Xiaodan Chen: Software, Methodology, Formal analysis. Jordi López-Pujol: Writing – review & editing, Software, Methodology, Data curation. Guoqing Bai: Methodology, Investigation. Sonia Herrando-Moraira: Software, Methodology. Neus Nualart: Software, Methodology. Xiao Zhang: Software, Investigation. Yuemei Zhao: Investigation, Data curation. Guifang Zhao: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors have no competing interests to declare.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.04.008.
Allouche, O., Tsoar, A., Kadmon, R., 2006. Assessing the accuracy of species distribution models: prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol., 43: 1223-1232. DOI:10.1111/j.1365-2664.2006.01214.x |
An, Z., Kutzbach, J.E., Prell, W.L., et al., 2001. Evolution of Asian monsoons and phased uplift of the Himalaya-Tibetan plateau since Late Miocene times. Nature, 411: 62-66. DOI:10.4319/lo.2001.46.1.0062 |
Anacker, B.L., Strauss, S.Y., 2014. The geography and ecology of plant speciation: range overlap and niche divergence in sister species. Proc. Roy. Soc. B-Biol. Sci., 281: 20132980. DOI:10.1098/rspb.2013.2980 |
APG [The Angiosperm Phylogeny Group], 2016. An update of the Angiosperm phylogeny group classification for the orders and families of flowering plants: APG Ⅳ. Bot. J. Linn. Soc., 181: 1-20. DOI:10.1111/boj.12385 |
Bai, G., Zhou, T., Zhang, X., et al., 2017. Genetic differentiation and population genetic structure of the Chinese endemic Dipteronia Oliv. revealed by cpDNA and AFLP data. Forests, 8: 424. DOI:10.3390/f8110424 |
Bandelt, H.J., Forster, P., Röhl, A., 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol., 16: 37-48. DOI:10.1093/oxfordjournals.molbev.a026036 |
Beerli, P., 2006. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics, 22: 341-345. DOI:10.1093/bioinformatics/bti803 |
Broennimann, O., Fitzpatrick, M.C., Pearman, P.B., et al., 2012. Measuring ecological niche overlap from occurrence and spatial environmental data. Global Ecol. Biogeogr., 21: 481-497. DOI:10.1111/j.1466-8238.2011.00698.x |
Brown, J.L., 2014. SDMtoolbox: a python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods Ecol. Evol., 5: 694-700. DOI:10.1111/2041-210X.12200 |
Cao, Y.N., Comes, H.P., Sakaguchi, S., et al., 2016. Evolution of East Asia’s Arcto-tertiary relict Euptelea (Eupteleaceae) shaped by late Neogene vicariance and Quaternary climate change. BMC Evol. Biol., 16: 66. DOI:10.1186/s12862-016-0636-x |
Cao, Y.N., Zhu, S.S., Chen, J., et al., 2020. Genomic insights into historical population dynamics, local adaptation, and climate change vulnerability of the East Asian Tertiary relict Euptelea (Eupteleaceae). Evol. Appl., 13: 2038-2055. DOI:10.1111/eva.12960 |
Chan, L.M., Brown, J.L., Yoder, A.D., 2011. Integrating statistical genetic and geospatial methods brings new power to phylogeography. Mol. Phylogenet. Evol., 59: 523-537. DOI:10.1016/j.ympev.2011.01.020 |
Chapman, M.A., Hiscock, S.J., Filatov, D.A., 2016. The genomic bases of morphological divergence and reproductive isolation driven by ecological speciation in Senecio (Asteraceae). J. Evol. Biol., 29: 98-113. DOI:10.1111/jeb.12765 |
Chen, C., Lu, R.S., Zhu, S.S., et al., 2017. Population structure and historical demography of Dipteronia dyeriana (Sapindaceae), an extremely narrow palaeoendemic plant from China: implications for conservation in a biodiversity hot spot. Heredity, 119: 95-106. DOI:10.1038/hdy.2017.19 |
Chou, Y.W., Thomas, P.I., Ge, X.J., et al., 2011. Refugia and phylogeography of Taiwania in East Asia. J. Biogeogr., 38: 1992-2005. DOI:10.1111/j.1365-2699.2011.02537.x |
Chung, M.Y., López-Pujol, J., Chung, M.G., 2017. The role of the Baekdudaegan (Korean Peninsula) as a major glacial refugium for plant species: a priority for conservation. Biol. Conserv., 206: 236-248. DOI:10.1016/j.biocon.2016.11.040 |
Chung, M.Y., Vu, S.H., López-Pujol, J., et al., 2018. Comparison of genetic variation between northern and southern populations of Lilium cernuum (Liliaceae): implications for Pleistocene refugia. PLoS One, 13: e0190520. DOI:10.1371/journal.pone.0190520 |
Collinson, M.E., 2000. Cenozoic evolution of modern plant communities and vegetation. In: Rawson, P.F., Culver, S.J. (Eds.), Biotic Response to Global Change: The Last 145 Million Years. Cambridge University Press, Cambridge, pp. 223-243.
|
Core, T.R., 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org.
|
Cornuet, J.M., 2014. DIYABC v2.0: a software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics, 30: 1187-1189. DOI:10.1093/bioinformatics/btt763 |
Crawford, N.G., 2010. smogd: software for the measurement of genetic diversity. Mol. Ecol. Resour., 10: 556-557. DOI:10.1111/j.1755-0998.2009.02801.x |
Di Cola, V., Broennimann, O., Petitpierre, B., et al., 2017. ecospat: an R package to support spatial analyses and modeling of species niches and distributions. Ecography, 40: 774-787. DOI:10.1111/ecog.02671 |
Dieringer, D., Schlötterer, C., 2003. Microsatellite Analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Resour., 3: 167-169. |
Ding, W.N., Huang, J., Su, T., et al., 2018. An early Oligocene occurrence of the palaeoendemic genus Dipteronia (Sapindaceae) from Southwest China. Rev. Palaeobot. Palynol., 249: 16-23. DOI:10.1016/j.revpalbo.2017.11.002 |
Dixon, P., 2003. VEGAN, a package of R functions for community ecology. J. Veg. Sci., 14: 927-930. DOI:10.1111/j.1654-1103.2003.tb02228.x |
Drummond, A.J., Rambaut, A., 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol., 7: 214. DOI:10.1186/1471-2148-7-214 |
Drummond, A.J., Suchard, M.A., Xie, D., et al., 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol., 29: 1969-1973. DOI:10.1093/molbev/mss075 |
Earl, D.A., vonHoldt, B.M., 2012. Structure HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour., 4: 359-361. DOI:10.1007/s12686-011-9548-7 |
Eldrett, J.S., Greenwood, D.R., Harding, I.C., et al., 2009. Increased seasonality through the Eocene to Oligocene transition in northern high latitudes. Nature, 459: 969-973. DOI:10.1038/nature08069 |
Ellis, A.G., Weis, A.E., 2006. Coexistence and differentiation of ‘flowering stones’: the role of local adaptation to soil microenvironment. J. Ecol., 94: 322-335. DOI:10.1111/j.1365-2745.2005.01074.x |
Evanno, G., Regnaut, S., Goudet, J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol., 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Excoffier, L., Lischer, H.E., 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour., 10: 564-567. DOI:10.1111/j.1755-0998.2010.02847.x |
Fan, D., Sun, Z., Li, B., et al., 2017. Dispersal corridors for plant species in the Poyang Lake Basin of southeast China identified by integration of phylogeographic and geospatial data. Ecol. Evol., 7: 5140-5148. DOI:10.1002/ece3.2999 |
Fang, F., Ji, Y., Zhao, Q., et al., 2015. Phylogeography of the Chinese endemic freshwater crab Sinopotamon acutum (Brachyura, Potamidae). Zoo. Scr., 44: 653-666. DOI:10.1111/zsc.12131 |
Fang, F., Sun, H., Zhao, Q., et al., 2013. Patterns of diversity, areas of endemism, and multiple glacial refuges for freshwater crabs of the genus Sinopotamon in China (Decapoda: Brachyura: Potamidae). PLoS One, 8: e53143. DOI:10.1371/journal.pone.0053143 |
Feng, Y., Comes, H.P., Chen, J., et al., 2024. Genome sequences and population genomics provide insights into the demographic history, inbreeding, and mutation load of two ‘living fossil’ tree species of Dipteronia. Plant J., 117: 177-192. DOI:10.1111/tpj.16486 |
Feng, Y., Comes, H.P., Zhou, X.-P., et al., 2019. Phylogenomics recovers monophyly and early Tertiary diversification of Dipteronia (Sapindaceae). Mol. Phylogenet. Evol., 130: 9-17. DOI:10.1016/j.ympev.2018.09.012 |
Fenster, C.B., Dudash, M.R., 1994. Genetic considerations for plant population restoration and conservation. In: Whelan, C.J., Bowles, M.L. (Eds.), Restoration of Endangered Species: Conceptual Issues, Planning and Implementation. Cambridge University Press, Cambridge, pp. 34-62.
|
Gamisch, A., 2019. Oscillayers: a dataset for the study of climatic oscillations over Plio-Pleistocene time-scales at high spatial-temporal resolution. Global Ecol. Biogeogr., 28: 1552-1560. DOI:10.1111/geb.12979 |
Gao, Y.D., Zhang, Y., Gao, X.F., et al., 2015. Pleistocene glaciations, demographic expansion and subsequent isolation promoted morphological heterogeneity: a phylogeographic study of the alpine Rosa sericea complex (Rosaceae). Sci. Rep., 5: 11698. DOI:10.1038/srep11698 |
Gavin, D.G., Fitzpatrick, M.C., Gugger, P.F., et al., 2014. Climate refugia: joint inference from fossil records, species distribution models and phylogeography. New Phytol., 204: 37-54. DOI:10.1111/nph.12929 |
Gibbs, D., Chen, Y., 2009. The Red List of Maples. Botanic Gardens Conservation International, Richmond.
|
Goudet, J., 2001. FSTAT; a program to estimate and test gene diversities and fixation indices version 2.9. 3. http://www.unil.ch/izea/softwares/fstat.html.
|
Hall, B.A., 1961. The floral anatomy of Dipteronia. Am. J. Bot., 48: 918-924. DOI:10.1002/j.1537-2197.1961.tb11731.x |
Hall, T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser., 41: 95-98. |
Hampe, A., Jump, A.S., 2011. Climate relicts: past, present, future. Annu. Rev. Ecol. Evol. Syst., 42: 313-333. DOI:10.1146/annurev-ecolsys-102710-145015 |
Hamrick, J.L., Godt, M.J.W., 1990. Allozyme diversity in plant species. In: Brown, A.H.D., Clegg, M.T., Kahler, A.L., et al. (Eds.), Plant Population Genetics, Breeding, and Genetic Resources. Sinauer Associates, Sunderland, pp. 43-63.
|
Harris, A., Frawley, E., Wen, J., 2017. The utility of single-copy nuclear genes for phylogenetic resolution of Acer and Dipteronia (Acereae, Sapindaceae). Ann. Bot. Fenn., 54: 209-222, 214. DOI:10.5735/085.054.0603 |
Hedrick, P.W., 2005. A standardized genetic differentiation measure. Evolution, 59: 1633-1638. |
Herrando-Moraira, S., Nualart, N., Herrando-Moraira, A., et al., 2019. Climatic niche characteristics of native and invasive Lilium lancifolium. Sci. Rep., 9: 14334. DOI:10.1038/s41598-019-50762-4 |
Hoffmann, A.A., Sgrò, C.M., 2011. Climate change and evolutionary adaptation. Nature, 470: 479-485. DOI:10.1038/nature09670 |
Huang, J.H., Chen, J.H., Ying, J.S., et al., 2011. Features and distribution patterns of Chinese endemic seed plant species. J. Syst. Evol., 49: 81-94. DOI:10.1111/j.1759-6831.2011.00119.x |
Huang, J., Chen, B., Liu, C., et al., 2012. Identifying hotspots of endemic woody seed plant diversity in China. Divers. Distrib., 18: 673-688. DOI:10.1111/j.1472-4642.2011.00845.x |
Jaramillo, C., Rueda, M.J., Mora, G., 2006. Cenozoic plant diversity in the Neotropics. Science, 311: 1893-1896. DOI:10.1126/science.1121380 |
Kalinowski, S.T., 2005. Hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Resour., 5: 187-189. DOI:10.1111/j.1471-8286.2004.00845.x |
Kou, Y., Cheng, S., Tian, S., et al., 2016. The antiquity of Cyclocarya paliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late Early Miocene. J. Biogeogr., 43: 351-360. DOI:10.1111/jbi.12635 |
Latham, R.E., Ricklefs, R.E., 1993. Continental comparisons of temperate-zone tree species diversity. In: Ricklefs, R.E., Schluter, D. (Eds.), Species Diversity in Ecological Communities. University of Chicago Press, Chicago, pp. 294-314.
|
Li, J., Li, D., Xue, Y., et al., 2018. Identifying potential refugia and corridors under climate change: a case study of endangered Sichuan golden monkey (Rhinopithecus roxellana) in Qinling Mountains, China. Am. J. Primatol., 80: e22929. DOI:10.1002/ajp.22929 |
Li, L., Abbott, R.J., Liu, B., et al., 2013. Pliocene intraspecific divergence and Plio-Pleistocene range expansions within Picea likiangensis (Lijiang spruce), a dominant forest tree of the Qinghai-Tibet Plateau. Mol. Ecol., 22: 5237-5255. DOI:10.1111/mec.12466 |
Li, L., Wang, H., Pang, D., et al., 2014. Phenotypic and genetic evidence for ecological speciation of Aquilegia japonica and A. oxysepala. New Phytol., 204: 1028-1040. DOI:10.1111/nph.12967 |
Librado, P., Rozas, J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451-1452. DOI:10.1093/bioinformatics/btp187 |
Liu, J., Möller, M., Provan, J., et al., 2013. Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytol., 199: 1093-1108. DOI:10.1111/nph.12336 |
Liu, X., Dong, B., Yin, Z.Y., et al., 2017. Continental drift and plateau uplift control origination and evolution of Asian and Australian monsoons. Sci. Rep., 7: 40344. DOI:10.1038/srep40344 |
Liu, Y., Wang, Y., Huang, H., 2009. Species-level phylogeographical history of Myricaria plants in the mountain ranges of western China and the origin of M. laxiflora in the Three Gorges mountain region. Mol. Ecol., 18: 2700-2712. DOI:10.1111/j.1365-294X.2009.04214.x |
López-Pujol, J., Zhang, F.M., Sun, H.Q., et al., 2011. Mountains of southern China as “plant museums” and “plant cradles”: evolutionary and conservation insights. Mt. Res. Dev., 31: 261-269, 269. DOI:10.1659/MRD-JOURNAL-D-11-00058.1 |
Luebert, F., Weigend, M., 2014. Phylogenetic insights into Andean plant diversification. Front. Ecol. Evol., 2: 27. |
Ma, Q., Du, Y., Chen, N., et al., 2016. Phylogeography of Davidia involucrata (Davidiaceae) inferred from cpDNA haplotypes and nSSR data. Syst. Bot., 40: 769-810. |
Manchester, S.R., Chen, Z.D., Lu, A.M., et al., 2009. Eastern Asian endemic seed plant genera and their paleogeographic history throughout the Northern Hemisphere. J. Syst. Evol., 47: 1-42. DOI:10.1111/j.1759-6831.2009.00001.x |
Manni, F., Guérard, E., Heyer, E., 2004. Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using Monmonier's algorithm. Hum. Biol., 76: 173-190. DOI:10.1353/hub.2004.0034 |
Massó, S., López-Pujol, J., Vilatersana, R., 2018. Reinterpretation of an endangered taxon based on integrative taxonomy: the case of Cynara baetica (Compositae). PLoS One, 13: e0207094. DOI:10.1371/journal.pone.0207094 |
McClain, A.M., Manchester, S.R., 2001. Dipteronia (Sapindaceae) from the Tertiary of North America and implications for the phytogeographic history of the Aceroideae. Am. J. Bot., 88: 1316-1325. DOI:10.2307/3558343 |
Milne, R.I., Abbott, R.J., 2002. The origin and evolution of Tertiary relict floras. Adv. Bot. Res., 38: 281-314. |
Myers, N., Mittermeier, R.A., Mittermeier, C.G., et al., 2000. Biodiversity hotspots for conservation priorities. Nature, 403: 853-858. DOI:10.1038/35002501 |
Nagalingum, N.S., Marshall, C.R., Quental, T.B., et al., 2011. Recent synchronous radiation of a living fossil. Science, 334: 796-799. DOI:10.1126/science.1209926 |
Nagy, E.S., 1997. Selection for native characters in hybrids between two locally adapted plant subspecies. Evolution, 51: 1469-1480. DOI:10.2307/2411199 |
Nei, M., Tajima, F., Tateno, Y., 1983. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol., 19: 153-170. DOI:10.1007/BF02300753 |
Nosil, P., 2008. Speciation with gene flow could be common. Mol. Ecol., 17: 2103-2106. DOI:10.1111/j.1365-294X.2008.03715.x |
Nosil, P., Harmon, L.J., Seehausen, O., 2009. Ecological explanations for (incomplete) speciation. Trends Ecol. Evol., 24: 145-156. DOI:10.1016/j.tree.2008.10.011 |
Nunney, L., 2020. The limits to knowledge in conservation genetics: the value of effective population size. In: Clegg, M., Hecht, M., MacIntyre, R. (Eds.), Evolutionary Biology. Springer, New York, pp. 179-194.
|
Nybom, H., 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol., 13: 1143-1155. DOI:10.1111/j.1365-294X.2004.02141.x |
Pälike, H., Norris, R.D., Herrle, J.O., et al., 2006. The heartbeat of the Oligocene climate system. Science, 314: 1894-1898. DOI:10.1126/science.1133822 |
Peakall, R., Smouse, P.E., 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics, 28: 2537-2539. DOI:10.1093/bioinformatics/bts460 |
Petit, R., Duminil, J., Fineschi, S., et al., 2005. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol., 14: 689-701. DOI:10.1111/j.1365-294x.2004.02410.x |
Petitpierre, B., Kueffer, C., Broennimann, O., et al., 2012. Climatic niche shifts are rare among terrestrial plant invaders. Science, 335: 1344-1348. DOI:10.1126/science.1215933 |
Phillips, S.J., Anderson, R.P., Schapire, R.E., 2006. Maximum entropy modeling of species geographic distributions. Ecol. Model., 190: 231-259. DOI:10.1504/IJGENVI.2006.010156 |
Posada, D., Crandall, K.A., 1998. Modeltest: testing the model of DNA substitution. Bioinformatics, 14: 817-818. DOI:10.1093/bioinformatics/14.9.817 |
Pritchard, J.K., Stephens, M., Donnelly, P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. DOI:10.1093/genetics/155.2.945 |
Qi, X.S., Chen, C., Comes, H.P., et al., 2012. Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae). New Phytol., 196: 617-630. DOI:10.1111/j.1469-8137.2012.04242.x |
Qin, H., Yang, Y., Dong, S., et al., 2017. Threatened species list of China's higher plants. Biodivers. Sci., 25: 696-744. DOI:10.17520/biods.2017144 |
Qu, Y., Ericson, P.G.P., Quan, Q., et al., 2014. Long-term isolation and stability explain high genetic diversity in the Eastern Himalaya. Mol. Ecol., 23: 705-720. DOI:10.1111/mec.12619 |
Racine, J.S., 2012. RStudio: a platform-independent IDE for R and Sweave. J. Appl. Econom., 27: 167-172. DOI:10.1002/jae.1278 |
Rambaut, A., Drummond, A.J., Xie, D., et al., 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Sys. Biol., 67: 901-904. DOI:10.1093/sysbio/syy032 |
Renner, S.S., Grimm, G.W., Schneeweiss, G.M., et al., 2008. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: implications for North American/Asian disjunctions. Syst. Biol., 57: 795-808. DOI:10.1080/10635150802422282 |
Rice, W.R., 1989. Analyzing tables of statistical tests. Evolution, 43: 223-225. DOI:10.2307/2409177 |
Rivers, M.C., Barstow, M., Crowley, D., 2017. Dipteronia dyeriana. The IUCN Red List of Threatened Species 2017: e.T32340A2815531. https://www.iucnredlist.org/species/32340/2815531.
|
Schoener, T.W., 1970. Nonsynchronous spatial overlap of lizards in patchy habitats. Ecology, 51: 408-418. DOI:10.2307/1935376 |
Selkoe, K.A., Toonen, R.J., 2006. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol. Lett., 9: 615-629. DOI:10.1111/j.1461-0248.2006.00889.x |
Shi, X., Yang, Z., Dong, Y., et al., 2019. Tectonic uplift of the northern Qinling Mountains (Central China) during the late Cenozoic: evidence from DEM-based geomorphological analysis. J. Asian Earth Sci., 184: 104005. DOI:10.1016/j.jseaes.2019.104005 |
Silva, D.P., Vilela, B., Buzatto, B.A., et al., 2016. Contextualized niche shifts upon independent invasions by the dung beetle Onthophagus taurus. Biol. Invasions, 18: 3137-3148. DOI:10.1007/s10530-016-1204-4 |
Su, X., Wu, G., Li, L., et al., 2015. Species delimitation in plants using the Qinghai–Tibet Plateau endemic Orinus (Poaceae: Tridentinae) as an example. Ann. Bot., 116: 35-48. DOI:10.1093/aob/mcv062 |
Suissa, J.S., Sundue, M.A., Testo, W.L., 2021. Mountains, climate and niche heterogeneity explain global patterns of fern diversity. J. Biogeogr., 48: 1296-1308. DOI:10.1111/jbi.14076 |
Sun, J., Ni, X., Bi, S., et al., 2014a. Synchronous turnover of flora, fauna and climate at the Eocene-Oligocene Boundary in Asia. Sci. Rep., 4: 7463. DOI:10.1038/srep07463 |
Sun, W., Zhang, G., Ouyang, Z., 2006. Community characteristics and conservation strategies of a rare species, Dipteronia dyeriana (Aceraceae). Acta Bot. Yunnan., 28: 54-58. DOI:10.1007/s11458-005-0010-9 |
Sun, Y., Moore, M.J., Yue, L., et al., 2014b. Chloroplast phylogeography of the East Asian Arcto-Tertiary relict Tetracentron sinense (Trochodendraceae). J. Biogeogr., 41: 1721-1732. DOI:10.1111/jbi.12323 |
Swets, J., 1988. Measuring the accuracy of diagnostic systems. Science, 240: 1285-1293. DOI:10.1126/science.3287615 |
Tang, C.Q., Matsui, T., Ohashi, H., et al., 2018. Identifying long-term stable refugia for relict plant species in East Asia. Nat. Commun., 9: 4488. DOI:10.1038/s41467-018-06837-3 |
Tian, S., Kou, Y., Zhang, Z., et al., 2018. Phylogeography of Eomecon chionantha in subtropical China: the dual roles of the Nanling Mountains as a glacial refugium and a dispersal corridor. BMC Evol. Biol., 18: 20. DOI:10.1186/s12862-017-1093-x |
Tian, S., López-Pujol, J., Wang, H.-W., et al., 2010. Molecular evidence for glacial expansion and interglacial retreat during Quaternary climatic changes in a montane temperate pine (Pinus kwangtungensis Chun ex Tsiang) in southern China. Plant Syst. Evol., 284: 219-229. DOI:10.1007/s00606-009-0246-9 |
Van Oosterhout, C., Hutchinson, W.F., Wills, D.P.M., et al., 2010. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Resour., 4: 535-538. |
Wan, S., Li, A., Clift, P.D., et al., 2007. Development of the East asian monsoon: mineralogical and sedimentologic records in the northern south China sea since 20 Ma. Palaeogeogr. Palaeoclimatol. Palaeoecol., 254: 561-582. DOI:10.1016/j.palaeo.2007.07.009 |
Wang, B., Mao, J.F., Zhao, W., et al., 2013. Impact of geography and climate on the genetic differentiation of the subtropical pine Pinus yunnanensis. PLoS One, 8: e67345. DOI:10.1371/journal.pone.0067345 |
Warren, D.L., Glor, R.E., Turelli, M., 2008. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution, 62: 2868-2883. DOI:10.1111/j.1558-5646.2008.00482.x |
Watterson, G.A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol., 7: 256-276. DOI:10.1016/0040-5809(75)90020-9 |
Weir, B.S., Cockerham, C.C., 1984. Estimating F-statistics for the analysis of population structure. Evolution, 38: 1358-1370. |
Wen, J., 1999. Evolution of Eastern Asian and Eastern North American disjunct distributions in flowering plants. Annu. Rev. Ecol. Syst., 30: 421-455. DOI:10.1146/annurev.ecolsys.30.1.421 |
Wilson, G.A., Rannala, B., 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics, 163: 1177-1191. DOI:10.1093/genetics/163.3.1177 |
Xiao, G., Abels, H.A., Yao, Z., et al., 2010. Asian aridification linked to the first step of the Eocene-Oligocene Climate Transition (EOT) in obliquity-dominated terrestrial records in Xining Basin, China. J. Earth Sci., 21: 219-220. DOI:10.1007/s12583-010-0216-8 |
Xie, D., Li, M., Tan, J., et al., 2017. Phylogeography and genetic effects of habitat fragmentation on endemic Urophysa (Ranunculaceae) in Yungui Plateau and adjacent regions. PLoS One, 12: e0186378. DOI:10.1371/journal.pone.0186378 |
Xu, T.Z., Chen, Y.S., de Jong, P.C., et al., 2008. Aceraceae. In: Wu, Z., Raven, P.H., Hong, D. (Eds.), Flora of China. Botanical Garden Press, St. Louis, Science Press, Beijing & Missouri, pp. 515-553.
|
Yang, J., Li, S., Sun, G., et al., 2008. Population structure and genetic variation in the genus Dipteronia Oliv. (Aceraceae) endemic to China as revealed by cpSSR analysis. Plant Syst. Evol., 272: 97-106. DOI:10.1007/s00606-007-0641-z |
Yang, J., Qian, Z.Q., Liu, Z.L., et al., 2007. Genetic diversity and geographical differentiation of Dipteronia Oliv. (Aceraceae) endemic to China as revealed by AFLP analysis. Biochem. Syst. Ecol., 35: 593-599. |
Yang, J., Wang, X.M., Li, S., et al., 2010. What is the phylogenetic placement of Dipteronia dyerana Henry? An example of plant species placement based on nucleotide sequences. Plant Biosyst., 144: 634-643. DOI:10.1080/11263504.2010.490032 |
Ying, T.S., Zhang, Y.L., Boufford, D.E., 1993. The Endemic Genera of Seed Plants of China. Beijing: Science Press.
|
Yu, H., Zhang, Y., Wang, Z., et al., 2017. Diverse range dynamics and dispersal routes of plants on the Tibetan Plateau during the late Quaternary. PLoS One, 12: e0177101. DOI:10.1371/journal.pone.0177101 |
Yuan, S., Huang, M., Wang, X.S., et al., 2014. Centers of endemism and diversity patterns for typhlocybine leafhoppers (Hemiptera: Cicadellidae: Typhlocybinae) in China. Insect Sci., 21: 523-536. DOI:10.1111/1744-7917.12040 |
Zachos, J., Pagani, M., Sloan, L., et al., 2001. Trends, rhythms, and aberrations in global Climate 65 Ma to present. Science, 292: 686-693. |
Zanazzi, A., Kohn, M.J., MacFadden, B.J., et al., 2007. Large temperature drop across the Eocene–Oligocene transition in central North America. Nature, 445: 639-642. DOI:10.1038/nature05551 |
Zhang, R., Kravchinsky, V.A., Yue, L., 2012. Link between global cooling and mammalian transformation across the Eocene-Oligocene boundary in the continental interior of Asia. Int. J. Earth Sci., 101: 2193-2200. DOI:10.1007/s00531-012-0776-1 |
Zhou, T., Chen, C., Wei, Y., et al., 2016a. Comparative transcriptome and chloroplast genome analyses of two related Dipteronia Species. Front. Plant Sci., 7: 1512. |
Zhou, T., Li, Z., Bai, G., et al., 2016b. Transcriptome sequencing and development of genic SSR markers of an endangered Chinese endemic genus Dipteronia Oliver (Aceraceae). Molecules, 21: 166. DOI:10.3390/molecules21030166 |
Zhou, Y., Zhang, L., Liu, J., et al., 2014. Climatic adaptation and ecological divergence between two closely related pine species in Southeast China. Mol. Ecol., 23: 3504-3522. DOI:10.1111/mec.12830 |



