b. University of Chinese Academy of Sciences, Beijing 100049, China;
c. Guangxi Key Laboratory of Conservation and Restoration Ecology in Karst Terrain, Guangxi Institute of Botany, Guangxi Zhang Autonomous Region and the Chinese Academy of Sciences, Guilin 541006, China;
d. Institute of Molecular Plant Sciences, University of Edinburgh, Daniel Rutherford Building Max Born Crescent, The King's Buildings, Edinburgh EH9 3BF, UK;
e. Royal Botanic Garden Edinburgh, 20a Inverleith Row, Edinburgh EH3 5LR, UK;
f. State Key Laboratory of Plant Diversity and Specialty Crops, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou 510650, China
The limestone karst formations in southwestern China, characterized by unprecedented edaphic and topographic heterogeneity, host a global biodiversity hotspot with a diverse array of unique and endemic species (Wang et al., 2014; Hao et al., 2015; Pu et al., 2017). The soils of the area are characterized by shallow profiles, limited moisture and nitrogen, while the sedimentary rock outcrops are mainly composed of calcium carbonate, which exerts significant evolutionary pressure on karst plant populations (Zhang et al., 2011; Kang et al., 2015; Wu et al., 2022). The complex karst topography functions as a terrestrial archipelago with small and isolated populations in cave or cave-like microhabitats (Sarthou et al., 2001; Clements et al., 2006; Tseng et al., 2019). Small and isolated populations often experience reduced genetic diversity, increased inbreeding, and increased genetic load, which can lead to reduced fitness and an increased risk of extinction (Spielman et al., 2004; Jangjoo et al., 2016; Ma et al., 2021a; Rodger et al., 2021).
Recent advances in sequencing technologies and bioinformatic tools have revolutionized the field of conservation genetics, providing unprecedented opportunities for comprehensive genome-wide analysis of nucleotide variation (Davey et al., 2011; Funk et al., 2012; Bertorelle et al., 2022; Paez et al., 2022; Robinson et al., 2023). We can now investigate genetic structure, divergence trajectories, historical population fluctuations, and the impact of inbreeding at the genomic scale. Additionally, these advances allow us to assess the risk of extinction due to deleterious mutations in small populations (Liu et al., 2020; Hohenlohe et al., 2021). These breakthroughs in genomic research have already shed light on the genetics of threatened species, and provided invaluable insights into the conservation needs of several endangered animals and plants, such as Rhododendron (Ma et al., 2021a), Acer yangbiense (Ma et al., 2021b), pangolins (Hu et al., 2020; Wang et al., 2022), killer whales (Kardos et al., 2023), and scimitar-horned oryx (Humble et al., 2023). Despite significant advances in genomic research on endangered animal species, only a few plants have been subjects of conservation genomic studies (Yang et al., 2018; Li et al., 2020; Feng et al., 2024; Tao et al., 2024), and there is currently no knowledge of genome-wide inbreeding and mutation load in karst plants. Understanding genome-wide diversity, inbreeding, and the burden of accumulated deleterious mutations (i.e., mutation load) in small populations is essential for predicting and increasing population persistence and resilience.
Begonia L. (Begoniaceae, Cucurbitales), one of the largest angiosperm genera, thrives in tropical forests and has significant economic value as an ornamental plant (Goodall-Copestake et al., 2010). The limestone karst landscapes in southwestern China and northern Vietnam support a remarkable diversity of Begonia species (Peng et al., 2013), which thrive in moist, shaded areas such as karst walls, cave entrances, and watersheds. Previous studies on Begonia have primarily focused on resource surveys, morphological anatomy, cytology, and phylogenetic relationships (Brennan et al., 2012; Moonlight et al., 2017; Emelianova and Kidner, 2022; Oginuma and Peng, 2022; Tsai et al., 2022). Although a few studies have used microsatellite markers or chloroplast DNA to investigate population genetic structure in several Begonia species (Goodall-Copestake et al., 2010; Twyford et al., 2013; Chung et al., 2014; Xiao et al., 2023), genome-wide population genetic studies have yet to be reported in this mega-diverse genus. The recent release of a high-quality nuclear genome of Begonia masoniana facilitates population and conservation genetic studies in this group (Li et al., 2022).
In this study, we focus on the Begonia masoniana complex, which consists of four closely related species (B. liuyanii, B. longgangensis, B. masoniana and B. variegata). B. liuyanii, B. longgangensis and B. masoniana are endemic to southwestern Guangxi, near the border with northern Vietnam (Fig. 1a), whereas B. variegata is endemic to northern Vietnam. The four species of the B. masoniana complex are morphologically distinct (Peng et al., 2005, 2013). Geographically, these species are distributed in similar habitat with small and scattered ranges in limestone karst landscapes. Due to habitat destruction caused by human activities, B. liuyanii, B. longgangensis and B. masoniana are currently classified as vulnerable (VU) species in China (Qin et al., 2017). Understanding patterns of genetic diversity, levels of inbreeding, and genetic load will help guide future conservation efforts.
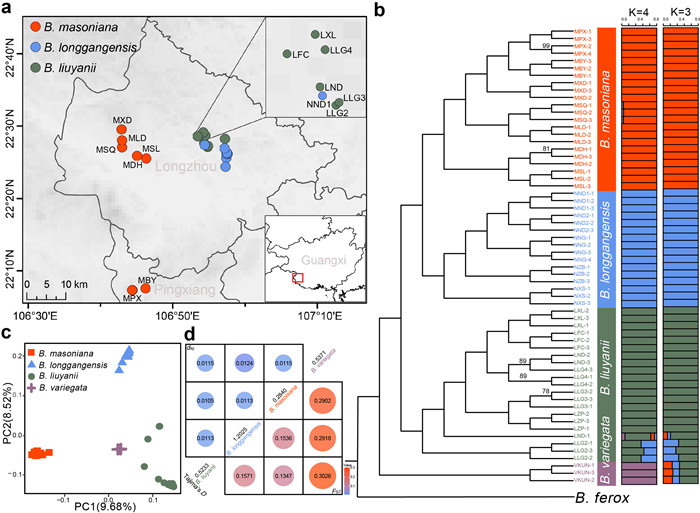
|
| Fig. 1 Geographic location, phylogenetic relationships, population structure, and genomic divergence in the Begonia masoniana complex. (a) Geographic locations of sampling sites. (b) Maximum-likelihood (ML) nuclear phylogeny and population structure. Except for the explicitly labeled branches, all branches in the tree have bootstrap values of 100. (c) Principal component analysis (PCA). (d) The values in the upper left corner represent dxy, while the values in the lower right corner represent FST. The diagonal values correspond to Tajima's D. |
Here we investigate genomic divergence and mutation load in the Begonia masoniana complex using whole-genome re-sequencing data. We aimed to (1) clarify population structure and phylogenetic relationships, with a particular focus on understanding the contribution of gene flow, especially in species pairs with sympatric distributions; (2) infer demographic history, including population divergence, fluctuations in effective population size (Ne), and their potential correlations with climatic fluctuations; and (3) assess levels of inbreeding and mutation load accumulation in these species. Our results have important implications for the development of more targeted and effective conservation strategies for small and isolated populations in karst regions.
2. Materials and methods 2.1. Population sampling, genome sequencing and variant callingWe collected 71 samples from different locations in the karst landscapes of southern China (Fig. 1a). These samples included 62 individuals of Begonia liuyanii (21), B. longgangensis (16), B. masoniana (22) and B. variegata (3) (Table S1). Nine individuals of B. ferox were used as the outgroup for phylogenetic analyses. The distribution ranges of these species were highly restricted, with B. liuyanii and B. longgangensis displaying geographic sympatry, while individuals of B. masoniana are allopatric to the other species (Fig. 1a). The three individuals of B. variegata were collected from northern Vietnam and cultivated at Kunming Institute of Botany, Chinese Academy of Sciences, however, the specific location of these samples is unknown.
Genomic DNA was extracted from dried leaves using a customized cetyltrimethylammonium bromide (CTAB) method (Doyle, 1987). Each sample was re-sequenced on Illumina Novaseq 6000 PE150 platform using paired-end sequencing libraries. Quality control was performed using FastQC v.0.11.8 followed by trimming of raw reads using TRIMMOMATIC v.0.39 (Bolger et al., 2014; Chen et al., 2018). Clean reads were aligned to the Begonia masoniana reference genome using the BWA-MEM algorithm within BWA v.0.7.15 (Li, 2013). Then, SAMTOOLS v.0.1.19 was used to convert sequence alignment files in sequence alignment map (SAM) format into binary alignment map (BAM) format and sort them by chromosome number, and calculated depth and mapping rate of each sequence (Li et al., 2009). Picard v.2.20.2 (DePristo et al., 2011) was then used to mark and remove PCR duplicates. The resulted BAM files constitute Dataset 1 for this research. Single nucleotide polymorphisms (SNPs) and genotypes were called using the Genome Analysis Toolkit v.4.1.2.0 (GATK) (McKenna et al., 2010). Only bases with a quality score of ≥ 30 were included and all indels and variants containing more than two alleles were excluded. To minimize the influence of mapping bias, we further discarded the following sites: (i) sites with anomalous depth (mean depth < 5 or > 100), and (ii) sites with more than 30% missing data (genotyped in fewer than 70% of all individuals). A total of 102,645,147 sites were finally obtained (Dataset 2). SNPs with a minor allele frequency < 0.05 were then also removed using VCFTOOLS v.0.1.13 (Danecek et al., 2011), leaving 2,711,146 SNPs (Dataset 3) for downstream analysis. Next, a moderate linkage disequilibrium (LD) pruning was performed using PLINK v.1.90, removing SNPs within a window of 50 SNPs with r2 > 0.5, resulting in 574,987 SNPs (Dataset 4).
2.2. Population genetic structure and genetic parametersWe used a Python script (https://github.com/edgardomortiz/vcf2phylip) to convert the SNPs from Dataset 3 into concatenated sequences for all individuals. Then, we constructed a maximum likelihood (ML) phylogenetic tree using IQ-TREE v.1.6.12 based on these concatenated sequences, using Begonia ferox as an outgroup (Nguyen et al., 2015). Bootstrap values were calculated from 1000 repetitions and subsequently visualized with FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). Next, the identity-by-state (IBS) kinship matrix between individuals was analyzed based on Dataset 2 using VCF2PCACluster v.1.1.40 from github (https://github.com/hewm2008/VCF2PCACluster).
Subsequent analyses were based on Dataset 4. For population structure analysis, we used ADMIXTURE v.1.3.0 (Alexander and Lange, 2011). For each K value ranging from 1 to 10, we conducted 10 replicates and determined the optimal K based on the cross-validation error. To confirm the result, we performed principal component analysis (PCA) on all individuals using PLINK v.1.90 to examine genetic structure and relatedness. In addition, we used the R package ggplot2 to plot the results (Villanueva and Chen, 2019).
We also calculated the unbiased nucleotide diversity (π), absolute divergence (dxy) and fixation statistics (FST) by PIXY v.1.2.6 (Korunes and Samuk, 2021) using Dataset 2 through 200-kb windows. The values of Tajima's D were then calculated using VCFTOOLS v.0.1.13. Whole genome heterozygosity (He) represents the proportion of heterozygous sites in the whole genome, excluding any gaps, and we used ANGSD v.1.9 software to calculate the He of each individual (Korneliussen et al., 2014).
2.3. Inferring demographic historyTo reconstruct the demographic history using the pairwise sequential Markovian coalescent (PSMC) model (Li and Durbin, 2011), we selected the individual with the highest sequencing depth and three additional individuals from different populations each of the species (Figs. 2a and S3). Parameter -p (which specifies the atomic time intervals) was set as “4 + 25*2 + 4 + 6” and other parameters were set as default. The generation time was set at 2 years, and the mutation rate (μ) was set to 6.5 × 10-9 per site per year, following the estimates in rice (Molina et al., 2011). A bootstrapping approach with 100 replicates was performed to evaluate the variation in the inferred Ne trajectories (Fig. 2a).
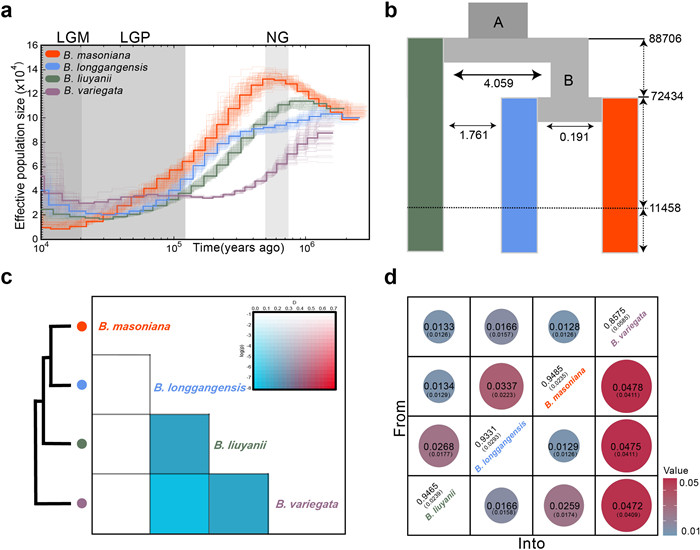
|
| Fig. 2 Demographic history and gene flow in the Begonia masoniana complex. (a) Chronological variation of effective population size (Ne) in four species inferred by pairwise sequentially Markovian coalescent (PSMC) model, where the 100 thin curves represent the estimates for 100 sequences randomly re-sampled from the original sequence. The three shaded rectangles represent three glaciations. (b) Best model of dadi. A represents the common ancestor of the three species, while B represents the common ancestor of B. longangensis and B. masoniana. Solid bidirectional arrows and numerical values represent the strength of genetic exchange between species, while dashed arrows represent time. Dashed lines represent the onset of species isolation when they entered refugia. (c) D-statistic test of Dsuite detected three distinct migration events. (d) Estimated migration rates (posterior mean) of four species based on BayesAss3 SNPs (BA3-SNPs), from populations on the y-axis to populations on the x-axis. Parentheses indicate the standard deviations of the marginal posterior distribution for each estimate. |
In addition, we aimed to confirm the specific demographic history using dadi based on site frequency spectrum (SFS) calculations. However, for an accurate estimation of population history based on SFS calculation, it is advisable to have a sample size more than 10 individuals (Gutenkunst et al., 2009; Beichman et al., 2017). Therefore, B. variegata was excluded from this analysis due to small sample size (N = 3). We used the Python script easySFS.py (https://github.com/isaacovercast/easySFS) to estimate the folded SFS based on LD-pruned SNPs data without missing sites (Gutenkunst et al., 2009). We then analyzed the SFSs of the three species using the diffusion approximation method of dadi and fitted 20 demographic models (Fig. S4), following the demographic modeling workflow (dadipipeline) described by Portik et al. (2017). First, we performed a comprehensive optimization process (dadi_Run_Optimizations.py), which included model fitting with specific configurations for a given number of replicates. Next, we used the parameters derived from the best-performing repetition to initiate a subsequent round of model fitting with updated configurations. This optimization process consisted of four rounds, with each round consisting of 10, 20, 30, and 40 repetitions, respectively. The four-round optimization process contributed significantly to reducing discrepancies in the likelihood values obtained at the end of the runs. Finally, to establish confidence intervals (CIs) for the parameters derived from the demographic models with the highest likelihood, we used a procedure that involved the generation of 100 bootstrap replicates of the spectra.
2.4. Estimating gene flow and inbreedingTo identify historical introgression events, Patterson's D-statistic (Durand et al., 2011) was performed on the whole genome of the four species using Dtrios implemented in Dsuite v.0.5 r48 (Malinsky et al., 2021). The D-statistic is derived from the assessment of the proportional occurrence of two different topological configurations in variable genetic sites, referred to as “ABBAs” and “BABAs”. In summary, in the context of a given relationship, represented as (((P1, P2) P3) P4), the genetic alleles hosted by P4 (the outgroup) are designated as the ancestral state A, while the altered state is denoted as B (Durand et al., 2011). To estimate contemporary migration rates, we used the Bayesian MCMC method of BayesAss3-SNPs (BA3-SNPs) (Mussmann et al., 2019; Wilson and Rannala, 2003). Delta values (-m, -a, and -f) were each set to 0.5, and we ran the analysis for 10,000,000 iterations with a burn-in of 1,000,000 steps, and 1000 sampling intervals. These analyses were performed on Dataset 3.
Individual-based FIS values were calculated with Dataset 2 by using VCFTOOLS v.0.1.13. Population scale inbreeding coefficients were calculated using PLINK v.1.90 for SNPs across different lineages. To further investigate inbreeding, we used PLINK v.1.90 to identify runs of homozygosity (ROH), retaining those longer than 100 kb (Yang et al., 2018; Ma et al., 2021a). The inbreeding coefficient based on ROH (FROH) was calculated for each individual as the total length of all its ROH divided by the total length of the autosomal genome (herein 799,826,415 bp).
2.5. Characterization of deleterious mutations and gene annotationsTo estimate the genetic load in four species of the Begonia masoniana complex, we annotated SNPs across the entire genome using SNPEFF v.4.3t (Cingolani et al., 2012). Individuals of B. ferox were used as an outgroup to determine the ancestral state for each SNP. The ancestral state was defined as the nucleotide that was homozygous in the outgroup individuals and matched at least one of the alleles within the ingroup (Chen et al., 2020b). We then classified each species within the ingroup into heterozygous mutation sites and homozygous mutation sites based on the ancestral and derived states from the outgroup. We followed the Grantham score (GS) rating criteria (Grantham, 1974), which quantifies the occurrence of three distinct mutation types: synonymous mutations, tolerated mutations, and deleterious mutations. Sites with scores ranging from 5 to 150 were designated as “tolerated” mutation sites, while SNPs with a GS ≥ 150 were categorized as “deleterious mutation” sites (Li et al., 1984). However, mutations that significantly affect the transcription of the entire gene are considered highly deleterious, and from this point on, we referred to them as loss-of-function (LOF) mutations. We then calculated the ratios of three types of harmful mutations to synonymous mutations and all derived sites, respectively.
Finally, we characterized the detected LOF mutations by gene ontology (GO) analysis and Kyoto encyclopedia of genes and genomes (KEGG) according to the gene annotation results of the previous study (Li et al., 2022) using TBtools (Chen et al., 2020a). Analyses were performed for our selected candidate genes, focusing on those genes associated with statistically significant terms (P < 0.05).
3. Results 3.1. Sampling and sequencingRaw reads were trimmed and mapped onto the Begonia masoniana reference genome, resulting in a mean depth of 15.90× (Table S1) and generating 71 BAM files (e.g., Dataset 1). Then SNPs were called using HaplotypeCaller in GATK and filtered using stringent criteria (hard filter in GATK and VCFTOOLS), which finally generated Dataset 2 (102,645,147 sites). After using maf 0.05 and mac 1, we retained 2,711,146 high quality nuclear SNPs as Dataset 3, which were further reduced to 574,987 SNPs through LD pruning (Dataset 4).
3.2. Population genetic structure and differentiationAdmixture identified K = 3 as the optimal number of clusters (Fig. S1). At K = 3, the populations of Begonia longgangensis and B. masoniana formed into two distinct clusters, while the third cluster includes both B. liuyanii and B. variegata, with individuals of B. variegata showing substantial admixture (Fig. 1b). However, when K = 4, four distinct clusters were observed for the corresponding four species (Fig. S1). Nevertheless, we observed genetic admixture in several individuals, which may be indicative of potential hybridization between species (Fig. 1b). The inferred phylogenetic relationships based on nuclear SNPs revealed four genetic clusters with strong support (Fig. 1b), which is consistent with the pattern identified by structure analysis. Such a genetic pattern was also supported by PCA analysis (Figs. 1c and S2).
Pairwise estimates of genetic differentiation between species ranged from 0.1347 (Begonia masoniana vs B. liuyanii) to 0.3026 (B. variegata vs B. liuyanii) for FST, and from 0.0113 (B. longgangensis vs B. liuyanii/B. masoniana) to 0.0124 (B. longgangensis vs B. variegata) for dxy (Fig. 1d). These results suggest pronounced genomic divergence among these species. In addition, intraspecific differentiation (FST) is higher in B. masoniana (0.1437) compared to B. liuyanii (0.0946) and B. longgangensis (0.0827) (Table S3).
3.3. Demographic history and gene flowThe PSMC analysis suggested that all four species had experienced a severe bottleneck in response to global temperature fluctuations throughout a series of glacial and interglacial periods (Figs. 2a and S3). Specifically, Begonia liuyanii and B. masoniana exhibited a significant expansion prior to Naynayxungla glaciation (NG, 0.5–0.72 million years ago [Mya]) (Zheng et al., 2001), followed by a sharp decline in Ne until the end of the last glacial maximum (LGM, 10–20 thousand years ago [kya]) (Bai et al., 2018), with the most substantial decline occurring in B. masoniana (Figs. 2a and S3). Additionally, the positive values of Tajima's D in each group may also indicate a population bottleneck in their evolutionary history (Fig. 1d and Table S2). This long-term bottleneck corresponded with global temperature fluctuations in the karst region (Zachos et al., 2008). Moreover, around ten thousand years ago, the effective population size (Ne) of B. masoniana reached its minimum among the four species, with B. masoniana having a Ne less than twenty thousand (Figs. 2a and S3).
We used coalescent-based simulations in dadi to estimate gene flow and divergence events amongst the three species (Begonia liuyanii, B. longgangensis, B. masoniana) (Tables S4 and S5). The best fitted model fit the actual data relatively well (Figs. 2b and S5). These results suggest that B. liuyanii diverged from ancestor A approximately 88.7 kya (95% CIs: 66.1–11.1 kya) with high gene flow with ancestor B. About 72.4 kya (95% CIs: 57.2–87.6 kya), B. longgangensis and B. masoniana also diverged, with only B. longgangensis maintaining a more pronounced historical gene flow with B. liuyanii. Then about 11,000 years ago, three species became isolated from each other (Fig. 2b). We also found that the Ne of B. masoniana was the largest during the early stages of divergence, around 70,000 years ago (Fig. 2b; Table S5), a pattern also observed in the PSMC analysis (Fig. 2a).
Gene flow is present among four closely distributed lineages, albeit with low contemporary migration rates (Fig. 2c and d). Dsuite detected three instances of gene flow (Fig. 2c and Table S6). Notably, gene flow between Begonia liuyanii and B. longgangensis reinforces the evidence for genetic admixture in the LLG2 population observed in the admixture analysis, and the IBS analysis also confirms the close genetic relationship between LLG2 of B. liuyanii and B. longgangensis (Figs. 1b and S6; Table S7). Moreover, the BA3-SNP analysis reveals diverse and asymmetric contemporary migration rates among the four species, although they all remain relatively low (Fig. 2d).
3.4. Inbreeding and genetic diversityAmongst the four species, we observed the highest levels of inbreeding in Begonia masoniana (Fig. 3 and Table S8). The FIS for B. masoniana is 0.212, while for the other species it is less than 0.1 (Table S8). Similar trends were also observed in FROH values, which were found to be 0.116 for B. liuyanii, 0.106 for B. longgangensis, 0.263 for B. masoniana, and 0.030 for B. variegata, respectively (Fig. 3a; Table S8). Additionally, it is evident that individuals of B. masoniana have accumulated a greater number of long ROH segments. As a result, B. masoniana exhibits the lowest heterozygosity among the four species (mean heterozygosity rate = 0.01). However, all of the four species show a similar level of nucleotide diversity (π ranging from 0.005 to 0.007) (Fig. 3b and c; Tables S2 and S8).
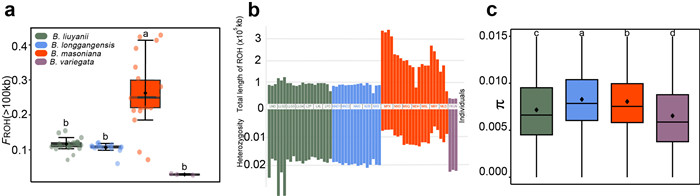
|
| Fig. 3 Inbreeding and genetic diversity in the Begonia masoniana complex. (a) Fraction of long run of homozygosity (ROH) greater than 100 kb in the genome of each population. (b) Comparison of total long ROH fragments and heterozygosity for each individual. (c) Nucleotide diversity within each population. The line in the middle of the box represents the median values, the diamond-shaped black dots inside the boxes represent the mean values. Statistical significance (P < 0.05) is indicated by letters above the plots. Different letters represent significant differences between groups. |
The accumulation of genetic load varies among species, with Begonia masoniana notably carrying a relatively higher homozygous genetic load (Fig. 4 and Table S9). When compared with synonymous mutations, tolerated mutations predominate in both homozygous and heterozygous states across all species, with a lower occurrence of strongly deleterious mutations and a relatively small proportion of LOF mutations (Fig. 4a and b). Moreover, in B. masoniana, the average proportion of observed missense homozygosity is higher than in the other three species for three types of missense mutations, while its missense heterozygous sites were notably the least frequent when compared with the other three species (Fig. 4a and b; Table S9).
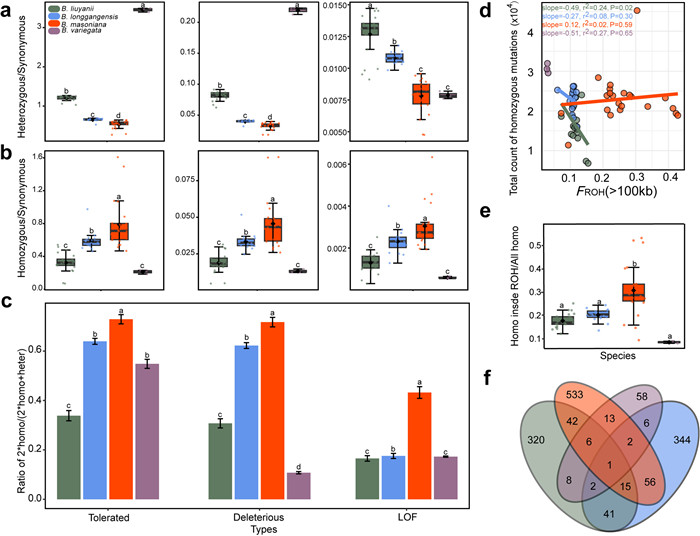
|
| Fig. 4 Mutation load and its correlation with inbreeding for the Begonia masoniana complex. (a) Ratio of heterozygous to synonymous sites for tolerated, deleterious, and loss-of-function (LOF) mutations. (b) Ratio of homozygous to synonymous sites for tolerated, deleterious, and LOF mutations. (c) The proportion of homozygous mutations in the coding region of derived mutations. Error bars represent the standard error. (d) Correlation between genome-wide FROH and total count of homozygous deleterious sites. (e) Proportion of homozygous mutations in the run of homozygosity (ROH) segment larger than 100 kb accounted for all homozygous mutations. (f) Venn diagram showing private or shared homozygous LOF mutations among species. The line in the middle of the box represents the median values, the diamond-shaped black dots inside the boxes represent the mean values. Statistical significance (P < 0.05) is indicated by letters above the plots. Different letters represent significant differences between groups. |
We further assessed mutation load by comparing the proportions of homozygous sites to all derived sites for three mutation types. We found that Begonia masoniana showed average proportions exceeding 0.6 for tolerated and deleterious types, with LOF mutations being more than twice as prevalent as those in other species (Fig. 4c).
Furthermore, we found a positive correlation between FROH levels and the total count of genome-wide heterozygous deleterious mutations in all species except Begonia masoniana (Fig. S7). In contrast, when assessing the association between FROH and the total number of homozygous deleterious mutations, only B. masoniana had a slight positive correlation without statistical significance (P = 0.59) (Fig. 4d). In addition, B. masoniana showed the highest proportion of homozygous deleterious mutations within the ROH segments compared to the other three species (Fig. 4e).
3.6. Functional enrichment of genes with loss-of-function mutationsA total of 435 genes in Begonia liuyanii, 467 genes in B. longgangensis, 668 genes in B. masoniana, and 96 genes in B. variegata were identified as carrying homozygous LOF mutations, in some cases the same genes with LOF mutations were found in multiple species. B. masoniana not only had the largest number of homozygous LOF mutant genes, but also had 533 unique homozygous LOF mutant genes (Fig. 4f and Table S10). In B. masoniana, homozygous LOF mutant genes were enriched in “oxidoreductase activity” (GO:0016627, P < 0.05) and pathways related to leaf, phyllome, shoot system, tissue and plant organ development (Fig. S8 and Table S11). GO pathway analysis identified “ATP-dependent activity (GO:0140657, P < 0.05)” as significantly enriched in homozygous LOF mutant genes in B. liuyanii, B. masoniana, and B. longgangensis. Moreover, the results indicated that these LOF mutation genes were significantly associated with phosphate metabolism and DNA repair in the four species (Figs. S8 and S9; Tables S11 and S12).
4. Discussion 4.1. Population structure of the Begonia masoniana complexIncreased geographic distance leads to decreased gene flow and consequently increased genetic differentiation between populations due to increased genetic drift (Bohonak, 2002). Karst landscapes in southern China, characterized by ubiquitous local-scale soil type mosaics, harbor a majority of species that are narrow endemics. Previous studies suggested that species divergence of karst plants has been influenced by uniquely heterogeneous habitats (Wang et al., 2017a; Zhu et al., 2022). Recent population genetic studies in Begonia species have revealed significant population structure due to geographic isolation (Matolweni et al., 2000; Forrest et al., 2005; Hughes and Hollingsworth, 2008; Thomas et al., 2012; Chung et al., 2014; Twyford et al., 2014). However, no study to date has attempted to assess whole-genome differentiation in this mega-diverse genus, nor has it been linked to edaphic or landscape factors.
Our admixture analysis, together with PCA and phylogenetic tree construction, revealed that the Begonia masoniana complex exhibits four distinct genetic clusters (Fig. 1). Furthermore, despite the close phylogenetic relationship between B. masoniana and B. longgangensis (Fig. 1b), they exhibited minimal gene flow (Fig. 2b and c) and no recent geographic overlap (Fig. 1a). It appears that the significant genetic structure among species can be attributed to the distinctive karst topography, leading to species isolation and subsequent speciation. These findings are consistent with previous observations documenting strong reproductive barriers among certain Begonia species in nature (Twyford et al., 2015).
We found genomic admixture between the close neighbors Begonia liuyanii and B. longgangensis (Fig. 1b), likely due to their close geographic proximity (Fig. 1a). Additional tests confirmed the occurrence of gene flow between these two species (Fig. 2). IBS analysis suggests a relatively close genetic relationship between the LLG2 population of B. liuyanii and B. longgangensis, further indicating hybridization between these species (Fig. S6 and Table S7). Although hybridization has been widely observed in Begonia species, it has been suggested that the majority of hybrid individuals belong to the F1 generation (Li et al., 2015; Tian et al., 2017, 2018). Therefore, despite the high genetic differentiation caused by karst habitat heterogeneity, natural hybridization can still occur between species in sympatry and parapatry.
4.2. Demographic history of the Begonia masoniana complex following climate-induced divergence and isolationOur reconstructed demographic history suggests a consistent decline in population size after NG for the four species. Remarkably, the population size of Begonia masoniana appeared not to stabilize during interglacial periods, but to continue to decline throughout the LGM and Holocene (Figs. 2a and S3). This pattern likely reflects the influence of Quaternary glaciations on the B. masoniana complex. Previous studies suggested that historical climate temperature fluctuations may have influenced the speciation of karst plants and imposed constraints on their distribution (Kong et al., 2017; Ke et al., 2022). We used dadi to infer an optimal model of speciation events. However, previous studies have shown that a substantial sample size (N > 10) is essential for a reliable analysis of the SFS-based modeling (Gutenkunst et al., 2009; Beichman et al., 2017). Due to the unknown sampling locations and a small sample size (N = 3) for B. variegata, our analysis focused solely on the other three species. Notably, these divergence events are consistent with the timing of the last glacial period (LGP, 11.7–115 kya) (Chen et al., 2011). Furthermore, B. longgangensis and B. liuyanii are characterized by higher gene exchange compared to other species pairs. In addition, dadi analysis revealed that these three species continued to be isolated until approximately 11,000 years ago (Fig. 2b), which ultimately strengthened the formation of distinct new species.
Another notable finding is that after differentiation, Begonia masoniana virtually ceased gene flow with other species (Fig. 2b). The species was also observed to have a higher level of inbreeding than the other species, which may underlie the lower heterozygosity and more extended ROH in B. masoniana (Fig. 3). These results indicate that B. masoniana, which experienced the highest level of isolation and had the smallest recent Ne (Figs. 2a and S3), also suffered the most severe inbreeding. Small and isolated populations are at higher risk of experiencing inbreeding depression due to the increased likelihood of mating between closely related individuals (Bortoluzzi et al., 2020). It is reasonable to speculate that these patterns could be due to the prolonged isolation experienced by B. masoniana, potentially leading to increased inbreeding and the accumulation of deleterious mutations. These findings highlight the impact of Quaternary glacial climate fluctuations on the genetic differentiation and population structure of karst endemics.
Previous studies have found that endangered species with small populations have high levels of inbreeding, resulting in reduced Ne and low genetic diversity (Willi et al., 2006; Provan and Bennett, 2008; Jangjoo et al., 2016). However, contrary to our expectations, despite the more pronounced isolation and lower Ne in Begonia masoniana (Fig. 1, Fig. 2a), the genetic diversity of this species is similar to that of the other species (Fig. 3c). We speculate that the high genetic diversity may have resulted from large ancestral populations of B. masoniana accumulating a high level of genetic polymorphisms prior to Naynayxungla glaciation (Figs. 2a and S3). Relatively high genetic diversity has been reported for many endangered plants with small populations, such as Rhododendron meddianum (Zhang et al., 2021), Cupressus chengiana (Li et al., 2020), Tricyrtis ishiiana (Setoguchi et al., 2010), and Rhododendron protistum var. gigantum (Wu et al., 2014). Therefore, assessing the conservation status of species requires multifaceted approaches that consider not only genetic diversity within populations, but also demographic history.
4.3. The accumulation of deleterious mutations due to geographic isolationSmall and isolated populations that have experienced long periods of decline are particularly vulnerable to the influence of genetic drift (LaBar and Adami, 2017; Rodger et al., 2021; Humble et al., 2023). This, in turn, compromises the efficiency of purifying selection to purge deleterious variants (Whitlock, 2000; Wang et al., 2021). Consequently, these populations are susceptible to the accumulation of deleterious alleles, i.e., mutation load, which can lead to species extinction, also known as “mutation meltdown” (Poon and Otto, 2000; Charlesworth, 2009; Agrawal and Whitlock, 2012; Robinson et al., 2019, 2023).
In our study, we observed the accumulation of deleterious mutations in the Begonia masoniana complex, particularly in the case of B. masoniana. We found that compared to all synonymous mutations or to all derived alleles, B. masoniana had the highest accumulated proportion of three types of homozygous mutations (Fig. 4b and c). Under evolutionary constraints, homozygous derived allele mutations represent realized genetic load and are predicted to have fitness consequences (Marsden et al., 2016; Feng et al., 2019; van der Valk et al., 2019). This suggests that B. masoniana may face a higher risk of extinction compared to other species. Notably, our analysis shows a positive correlation between the accumulation of deleterious type homozygous mutations and FROH in B. masoniana (Fig. 4d), although the correlation is not statistically significant. In addition, we found that the proportion of deleterious homozygous mutations within ROH segments in B. masoniana is the highest among all four species (Fig. 4e). This suggests that an increased inbreeding within this species may contribute to the accumulation of genetic load, potentially affecting the fitness and long-term survival of the population. Empirical and simulation studies have shown that long-term small effective population sizes (Ne) could be conducive to the reduction of deleterious variation through the purging of strongly deleterious mutations, which could help the long-term survival of endangered species (Yang et al., 2018; Feng et al., 2019; van der Valk et al., 2019; Khan et al., 2021), but this may not hold true for B. masoniana in karst landscapes. However, the genetic differentiation (FST) within B. masoniana is higher than that of B. liuyanii and B. longgangensis (Table S3). It seems that B. masoniana populations may have experienced recent population size decline, which led to an increase of genetic drift and decrease of efficacy for purging deleterious mutations.
4.4. The functional enrichment of loss-of-function genes and conservation implicationsWe analyzed the genes affected by the homozygous putative deleterious LOF mutations in the four species and found that a number of GO and KEGG categories were involved in processes related to DNA repair and phosphate metabolism (Figs. S8 and S9; Tables S11 and S12). Notably, there is the marked enrichment of nitrogen metabolism pathways (KEGG, 00910) in Begonia longgangensis (Fig. S9 and Table S12), which may be intricately linked to the distinctive N, P-deficient karst soil environment (Wang et al., 2017b). These findings suggest a potential adaptive evolutionary strategy adopted by the B. masoniana complex in response to the unique karst soil conditions. In B. masoniana, we also observed a significant enrichment of LOF genes associated with oxidative reduction processes (GO, GO0016627). Moreover, we found that genes associated with GO pathways related to leaf, phyllome, shoot system, tissue and plant organ development were highly enriched in the LOF mutant genes (Fig. S8 and Table S11). This observation suggests a higher burden of deleterious mutations compared to other species, posing a significant threat to its long-term survival. Taken together with previous analyses (Grossen et al., 2020; Ma et al., 2021b; Wang et al., 2021), we propose that B. masoniana may be at higher risk of extinction due to low Ne, a higher genetic load, and prolonged isolation. Therefore, conservation efforts should prioritize B. masoniana. Introducing hybridization via exogenous breeding programs could mitigate the genetic erosion resulting from recent demographic events.
However, although hybridization between Begonia liuyanii and B. longgangensis can mitigate the accumulation of homozygous deleterious mutations, both species still carry a substantial burden of heterozygous deleterious mutations (Fig. 4b). In the event of further isolation, their genetic integrity could be rapidly compromised. The fragile karst ecosystem in Guangxi has witnessed an increasing influx of human activities, leading to a degree of anthropogenic interference with karst flora in recent years (Chen and Xu, 2023). Therefore, it is imperative to protect the ecological habitats of these species and mitigate the risk of anthropogenic habitat degradation. The results of this study highlight the importance of not relying solely on genetic diversity when assessing the conservation status of these plants, but also considering factors such as population size and accumulation of deleterious mutations for a comprehensive evaluation of conservation strategies.
AcknowledgmentWe are grateful to Ms. Jifeng Long from Nonggang National Nature Reserve, Guangxi Zhuang Autonomous Region and Ms. Jingxiu Li from Kunming Institute of Botany, the Chinese Academy of Sciences for their kind help on sample collecting. This work was supported by Key-Area Research and Development Program of Guangdong Province (Grant No. 2022B1111230001) and National Natural Science Foundation of China (31860048).
Data availability statement
The genome re-sequencing data has been deposited to SRA database at NCBI under BioProject PRJNA1043235.
CRediT authorship contribution statement
Yiqing Chen: Writing – original draft, Formal analysis, Data curation. Lina Dong: Resources, Investigation, Funding acquisition. Huiqin Yi: Methodology. Catherine Kidner: Writing – review & editing. Ming Kang: Writing – review & editing, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.04.001.
Agrawal, A.F., Whitlock, M.C., 2012. Mutation load: the fitness of individuals in populations where deleterious alleles are abundant. Annu. Rev. Ecol. Evol. Syst., 43: 115-135. DOI:10.1146/annurev-ecolsys-110411-160257 |
Alexander, D.H., Lange, K., 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics, 12: 1-6. DOI:10.1186/1471-2105-12-1 |
Bai, W.N., Yan, P.C., Zhang, B.W., et al., 2018. Demographically idiosyncratic responses to climate change and rapid Pleistocene diversification of the walnut genus Juglans (Juglandaceae) revealed by whole-genome sequences. New Phytol., 217: 1726-1736. DOI:10.1111/nph.14917 |
Beichman, A.C., Phung, T.N., Lohmueller, K.E., 2017. Comparison of single genome and allele frequency data reveals discordant demographic histories. G3-Genes Genom. Genet., 7: 3605-3620. DOI:10.1534/g3.117.300259 |
Bertorelle, G., Raffini, F., Bosse, M., et al., 2022. Genetic load: genomic estimates and applications in non-model animals. Nat. Rev. Genet., 23: 492-503. DOI:10.1038/s41576-022-00448-x |
Bohonak, A.J., 2002. IBD (Isolation by Distance): a program for analyses of isolation by distance. J. Hered., 93: 153-154. DOI:10.1093/jhered/93.2.153 |
Bolger, A.M., Lohse, M., Usadel, B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30: 2114-2120. DOI:10.1093/bioinformatics/btu170 |
Bortoluzzi, C., Bosse, M., Derks, M.F.L., et al., 2020. The type of bottleneck matters: insights into the deleterious variation landscape of small managed populations. Evol. Appl., 13: 330-341. DOI:10.1111/eva.12872 |
Brennan, A.C., Bridgett, S., Shaukat Ali, M., et al., 2012. Genomic resources for evolutionary studies in the large, diverse, tropical genus, Begonia. Trop. Plant Biol., 5: 261-276. DOI:10.1007/s12042-012-9109-6 |
Charlesworth, B., 2009. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet., 10: 195-205. DOI:10.1038/nrg2526 |
Chen, C., Xu, Y., 2023. Spatial heterogeneity of human activities and its driving factors in karst areas of Southwest China over the past 20 years. Front. Environ. Sci., 11: 1225888. DOI:10.3389/fenvs.2023.1225888 |
Chen, D., Kang, H., Liu, C., 2011. An overview on the potential Quaternary glacial refugia of plants in China mainland. Bull. Bot. Res., 31: 623-632. |
Chen, S., Zhou, Y., Chen, Y., et al., 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34: i884-i890. DOI:10.1093/bioinformatics/bty560 |
Chen, C., Chen, H., Zhang, Y., et al., 2020a. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant, 13: 1194-1202. DOI:10.1016/j.molp.2020.06.009 |
Chen, J., Glemin, S., Lascoux, M., 2020b. From drift to draft: how much do beneficial mutations actually contribute to predictions of Ohta's slightly deleterious model of molecular evolution?. Genetics, 214: 1005-1018. DOI:10.1534/genetics.119.302869 |
Chung, K.F., Leong, W.C., Rubite, R.R., et al., 2014. Phylogenetic analyses of Begonia sect. Coelocentrum and allied limestone species of China shed light on the evolution of Sino-Vietnamese karst flora. Bot. Stud., 55: 1. DOI:10.1186/1999-3110-55-1 |
Cingolani, P., Platts, A., Wang le, L., et al., 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin), 6: 80-92. DOI:10.4161/fly.19695 |
Clements, R., Sodhi, N.S., Schilthuizen, M., et al., 2006. Limestone karsts of Southeast Asia: imperiled arks of biodiversity. BioScience, 56: 733-742. DOI:10.1641/0006-3568(2006)56[733:LKOSAI]2.0.CO;2 |
Danecek, P., Auton, A., Abecasis, G., et al., 2011. The variant call format and VCFtools. Bioinformatics, 27: 2156-2158. DOI:10.1093/bioinformatics/btr330 |
Davey, J.W., Hohenlohe, P.A., Etter, P.D., et al., 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet., 12: 499-510. DOI:10.1038/nrg3012 |
DePristo, M.A., Banks, E., Poplin, R., et al., 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43: 491-498. DOI:10.1038/ng.806 |
Doyle, J., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull., 19: 11-13. |
Durand, E.Y., Patterson, N., Reich, D., et al., 2011. Testing for ancient admixture between closely related populations. Mol. Biol. Evol., 28: 2239-2252. DOI:10.1093/molbev/msr048 |
Emelianova, K., Kidner, C., 2022. Comparative transcriptome analysis of two closely related Begonia species reveals divergent patterns in key light-regulated pathways. Edinb. J. Bot., 79: 1-18. DOI:10.24823/ejb.2022.398 |
Feng, S., Fang, Q., Barnett, R., et al., 2019. The genomic footprints of the fall and recovery of the crested ibis. Curr. Biol., 29: 340-349.e7. DOI:10.1016/j.cub.2018.12.008 |
Feng, Y., Comes, H.P., Chen, J., et al., 2024. Genome sequences and population genomics provide insights into the demographic history, inbreeding, and mutation load of two ‘living fossil’ tree species of Dipteronia. Plant J., 117: 177-192. DOI:10.1111/tpj.16486 |
Forrest, L.L., Hughes, M., Hollingsworth, P.M., 2005. A phylogeny of Begonia using nuclear ribosomal sequence data and morphological characters. Syst. Bot., 30: 671-682. DOI:10.1600/0363644054782297 |
Funk, W.C., McKay, J.K., Hohenlohe, P.A., et al., 2012. Harnessing genomics for delineating conservation units. Trends Ecol. Evol., 27: 489-496. DOI:10.1016/j.tree.2012.05.012 |
Goodall-Copestake, W.P., Pérez-Espona, S., Harris, D.J., et al., 2010. The early evolution of the mega-diverse genus Begonia (Begoniaceae) inferred from organelle DNA phylogenies. Biol. J. Linn. Soc., 101: 243-250. DOI:10.1111/j.1095-8312.2010.01489.x |
Grantham, R., 1974. Amino acid difference formula to help explain protein evolution. Science, 185: 862-864. DOI:10.1126/science.185.4154.862 |
Grossen, C., Guillaume, F., Keller, L.F., et al., 2020. Purging of highly deleterious mutations through severe bottlenecks in Alpine ibex. Nat. Commun., 11: 1001. DOI:10.1038/s41467-020-14803-1 |
Gutenkunst, R.N., Hernandez, R.D., Williamson, S.H., et al., 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genetics, 5: e1000695. DOI:10.1371/journal.pgen.1000695 |
Hao, Z., Kuang, Y., Kang, M., 2015. Untangling the influence of phylogeny, soil and climate on leaf element concentrations in a biodiversity hotspot. Funct. Ecol., 29: 165-176. DOI:10.1111/1365-2435.12344 |
Hohenlohe, P.A., Funk, W.C., Rajora, O.P., 2021. Population genomics for wildlife conservation and management. Mol. Ecol., 30: 62-82. DOI:10.1111/mec.15720 |
Hu, J.Y., Hao, Z.Q., Frantz, L., et al., 2020. Genomic consequences of population decline in critically endangered pangolins and their demographic histories. Natl. Sci. Rev., 7: 798-814. DOI:10.1093/nsr/nwaa031 |
Hughes, M., Hollingsworth, P.M., 2008. Population genetic divergence corresponds with species-level biodiversity patterns in the large genus Begonia. Mol. Ecol., 17: 2643-2651. DOI:10.1111/j.1365-294X.2008.03788.x |
Humble, E., Stoffel, M.A., Dicks, K., et al., 2023. Conservation management strategy impacts inbreeding and mutation load in scimitar-horned oryx. Proc. Natl. Acad. Sci. U.S.A., 120: e2210756120. DOI:10.1073/pnas.2210756120 |
Jangjoo, M., Matter, S.F., Roland, J., et al., 2016. Connectivity rescues genetic diversity after a demographic bottleneck in a butterfly population network. Proc. Natl. Acad. Sci. U.S.A., 113: 10914-10919. DOI:10.1073/pnas.1600865113 |
Kang, M., Wang, J., Huang, H., 2015. Nitrogen limitation as a driver of genome size evolution in a group of karst plants. Sci. Rep., 5: 11636. DOI:10.1038/srep11636 |
Kardos, M., Zhang, Y., Parsons, K.M., et al., 2023. Inbreeding depression explains killer whale population dynamics. Nat. Ecol. Evol., 7: 675-686. DOI:10.1038/s41559-023-01995-0 |
Ke, F., Vasseur, L., Yi, H., et al., 2022. Gene flow, linked selection, and divergent sorting of ancient polymorphism shape genomic divergence landscape in a group of edaphic specialists. Mol. Ecol., 31: 104-118. DOI:10.1111/mec.16226 |
Khan, A., Patel, K., Shukla, H., et al., 2021. Genomic evidence for inbreeding depression and purging of deleterious genetic variation in Indian tigers. Proc. Natl. Acad. Sci. U.S.A., 118: e2023018118. DOI:10.1073/pnas.2023018118 |
Kong, H., Condamine, F.L., Harris, A.J., et al., 2017. Both temperature fluctuations and East Asian monsoons have driven plant diversification in the karst ecosystems from southern China. Mol. Ecol., 26: 6414-6429. DOI:10.1111/mec.14367 |
Korneliussen, T.S., Albrechtsen, A., Nielsen, R., 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics, 15: 356. DOI:10.1186/s12859-014-0356-4 |
Korunes, K.L., Samuk, K., 2021. pixy: unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol. Ecol. Resour., 21: 1359-1368. DOI:10.1111/1755-0998.13326 |
LaBar, T., Adami, C., 2017. Evolution of drift robustness in small populations. Nat. Commun., 8: 1012. DOI:10.1038/s41467-017-01003-7 |
Li, H., 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA–MEM. arXiv: Genomics.
|
Li, H., Durbin, R., 2011. Inference of human population history from individual whole-genome sequences. Nature, 475: 493-496. DOI:10.1038/nature10231 |
Li, W.H., Wu, C.I., Luo, C.C., 1984. Nonrandomness of point mutation as reflected in nucleotide substitutions in pseudogenes and its evolutionary implications. J. Mol. Evol., 21: 58-71. DOI:10.1007/BF02100628 |
Li, H., Handsaker, B., Wysoker, A., et al., 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25: 2078-2079. DOI:10.1093/bioinformatics/btp352 |
Li, C., Tian, D., Li, X., et al., 2015. Morphological and molecular identification of natural hybridization between Begonia hemsleyana and B. macrotoma. Sci. Hortic., 192: 357-360. DOI:10.1016/j.scienta.2015.06.031 |
Li, J., Milne, R.I., Ru, D., et al., 2020. Allopatric divergence and hybridization within Cupressus chengiana (Cupressaceae), a threatened conifer in the northern Hengduan Mountains of western China. Mol. Ecol., 29: 1250-1266. DOI:10.1111/mec.15407 |
Li, L., Chen, X., Fang, D., et al., 2022. Genomes shed light on the evolution of Begonia, a mega-diverse genus. New Phytol., 234: 295-310. DOI:10.1111/nph.17949 |
Liu, D., Zhang, L., Wang, J., et al., 2020. Conservation genomics of a threatened Rhododendron: contrasting patterns of population structure revealed from neutral and selected SNPs. Front. Genet., 11: 757. DOI:10.1201/9781351187435-95 |
Ma, H., Liu, Y., Liu, D., et al., 2021a. Chromosome-level genome assembly and population genetic analysis of a critically endangered rhododendron provide insights into its conservation. Plant J., 107: 1533-1545. DOI:10.1111/tpj.15399 |
Ma, Y., Liu, D., Wariss, H.M., et al., 2021b. Demographic history and identification of threats revealed by population genomic analysis provide insights into conservation for an endangered maple. Mol. Ecol., 31: 767-779. |
Malinsky, M., Matschiner, M., Svardal, H., 2021. Dsuite-Fast D-statistics and related admixture evidence from VCF files. Mol. Ecol. Resour., 21: 584-595. DOI:10.1111/1755-0998.13265 |
Marsden, C.D., Ortega-Del Vecchyo, D., O'Brien, D.P., et al., 2016. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci. U.S.A., 113: 152-157. DOI:10.1073/pnas.1512501113 |
Matolweni, L.O., Balkwill, K., McLellan, T., 2000. Genetic diversity and gene flow in the morphologically variable, rare endemics Begonia dregei and Begonia homonyma (Begoniaceae). Am. J. Bot., 87: 431-439. DOI:10.2307/2656639 |
McKenna, A., Hanna, M., Banks, E., et al., 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20: 1297-1303. DOI:10.1101/gr.107524.110 |
Molina, J., Sikora, M., Garud, N., et al., 2011. Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl. Acad. Sci. U.S.A., 108: 8351-8356. DOI:10.1073/pnas.1104686108 |
Moonlight, P.W., Reynel, C., Tebbitt, M., 2017. Begonia elachista Moonlight & Tebbitt sp. nov., an enigmatic new species and a new section of Begonia (Begoniaceae) from Peru. Eur. J. Taxon., 281: 1-13. DOI:10.11646/phytotaxa.307.1.1 |
Mussmann, S.M., Douglas, M.R., Chafin, T.K., et al., 2019. BA3-SNPs: contemporary migration reconfigured in BayesAss for next-generation sequence data. Methods Ecol. Evol., 10: 1808-1813. DOI:10.1111/2041-210x.13252 |
Nguyen, L.T., Schmidt, H.A., von Haeseler, A., et al., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol., 32: 268-274. DOI:10.1093/molbev/msu300 |
Oginuma, K., Peng, C.I., 2022. Karyomorphology of Taiwanese Begonia (Begoniaceae): taxonomic implications. J. Plant Res., 115: 225-235. |
Paez, S., Kraus, R.H.S., Shapiro, B., et al., 2022. Reference genomes for conservation. Science, 377: 364-366. DOI:10.1126/science.abm8127 |
Peng, C.I., Ku, S.M., Leong, W.C., 2005. Begonia liuyanii (sect. Coelocentrum, Begoniaceae), a new species from limestone areas in Guangxi, China. Bot. Bull. Acad. Sinica, 46: 245-245. |
Peng, C.I., Yang, H.A., Kono, Y., et al., 2013. Novelties in Begonia sect. Coelocentrum: B. longgangensis and B. ferox from limestone areas in Guangxi, China. Bot. Stud., 54: 44. DOI:10.1186/1999-3110-54-44 |
Poon, A., Otto, S.P., 2000. Compensating for our load of mutations: freezing the meltdown of small populations. Evolution, 54: 1467-1479. |
Portik, D.M., Leache, A.D., Rivera, D., et al., 2017. Evaluating mechanisms of diversification in a Guineo-Congolian tropical forest frog using demographic model selection. Mol. Ecol., 26: 5245-5263. DOI:10.1111/mec.14266 |
Provan, J., Bennett, K.D., 2008. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol., 23: 564-571. DOI:10.1016/j.tree.2008.06.010 |
Pu, G., Lv, Y., Xu, G., et al., 2017. Research progress on karst Tiankeng ecosystems. Bot. Rev., 83: 5-37. DOI:10.1007/s12229-017-9179-0 |
Qin, H.N., Yang, Y., Dong, S.Y., et al., 2017. Threatened species list of China's higher plants. Biodiv. Sci., 25: 696-744. DOI:10.17520/biods.2017144 |
Robinson, J.A., Räikkönen, J., Vucetich, L.M., et al., 2019. Genomic signatures of extensive inbreeding in Isle Royale wolves, a population on the threshold of extinction. Sci. Adv., 5: eaau0757. DOI:10.1126/sciadv.aau0757 |
Robinson, J., Kyriazis, C.C., Yuan, S.C., et al., 2023. Deleterious variation in natural populations and implications for conservation genetics. Annu. Rev. Anim. Biosci., 11: 93-114. DOI:10.1146/annurev-animal-080522-093311 |
Rodger, Y.S., Pavlova, A., Sinclair, S., et al., 2021. Evolutionary history and genetic connectivity across highly fragmented populations of an endangered daisy. Heredity, 126: 846-858. DOI:10.1038/s41437-021-00413-0 |
Sarthou, C., Samadi, S., Boisselier-Dubayle, M.-C., 2001. Genetic structure of the saxicole Pitcairnia geyskesii (Bromeliaceae) on inselbergs in French Guiana. Am. J. Bot., 88: 861-868. DOI:10.2307/2657038 |
Setoguchi, H., Mitsui, Y., Ikeda, H., et al., 2010. Genetic structure of the critically endangered plant Tricyrtis ishiiana (Convallariaceae) in relict populations of Japan. Conserv. Genet., 12: 491-501. |
Spielman, D., Brook, B.W., Frankham, R., 2004. Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. U.S.A., 101: 15261-15264. DOI:10.1073/pnas.0403809101 |
Tao, T., Milne, R.I., Li, J., et al., 2024. Conservation genomic investigation of an endangered conifer, Thuja sutchuenensis, reveals low genetic diversity but also low genetic load. Plant Divers., 46: 78-90. DOI:10.1016/j.pld.2023.06.005 |
Thomas, D.C., Hughes, M., Phutthai, T., et al., 2012. West to east dispersal and subsequent rapid diversification of the mega-diverse genus Begonia (Begoniaceae) in the Malesian archipelago. J. Biogeogr., 39: 98-113. DOI:10.1111/j.1365-2699.2011.02596.x |
Tian, D., Li, C., Xiao, Y., et al., 2017. Occurrence and characteristics of natural hybridization in Begonia in China. Biodiv. Sci., 25: 654-674. DOI:10.17520/biods.2017050 |
Tian, D., Xiao, Y., Tong, Y., et al., 2018. Diversity and conservation of Chinese wild begonias. Plant Divers., 40: 75-90. DOI:10.1016/j.pld.2018.06.002 |
Tsai, M.Y., Kuan, C., Guo, Z.L., et al., 2022. Stomatal clustering in Begonia improves water use efficiency by modulating stomatal movement and leaf structure. Plant Environ. Interact., 3: 141-154. DOI:10.1002/pei3.10086 |
Tseng, Y.H., Huang, H.Y., Xu, W.B., et al., 2019. Phylogeography of Begonia luzhaiensis suggests both natural and anthropogenic causes for the marked population genetic structure. Bot. Stud., 60: 20. DOI:10.1186/s40529-019-0267-9 |
Twyford, A.D., Ennos, R.A., Kidner, C.A., 2013. Development and characterization of microsatellite markers for Central American Begonia sect. Gireoudia (Begoniaceae). Appl. Plant Sci., 1: 1200499. DOI:10.3732/apps.1200499 |
Twyford, A.D., Kidner, C.A., Ennos, R.A., 2014. Genetic differentiation and species cohesion in two widespread Central American Begonia species. Heredity, 112: 382-390. DOI:10.1038/hdy.2013.116 |
Twyford, A.D., Kidner, C.A., Ennos, R.A., 2015. Maintenance of species boundaries in a Neotropical radiation of Begonia. Mol. Ecol., 24: 4982-4993. DOI:10.1111/mec.13355 |
van der Valk, T., Diez-Del-Molino, D., Marques-Bonet, T., et al., 2019. Historical genomes reveal the genomic consequences of recent population decline in eastern gorillas. Curr. Biol., 29: 165-170.e6. DOI:10.1016/j.cub.2018.11.055 |
Villanueva, R.A.M., Chen, Z.J., 2019. ggplot2: Elegant graphics for data analysis. Taylor & Francis, Beijing.
|
Wang, J., Kang, M., Huang, H., 2014. Long-distance pollen dispersal ensures genetic connectivity of the low-density tree species, Eurycorymbus cavaleriei, in a fragmented karst forest landscape. Conserv. Genet., 15: 1163-1172. DOI:10.1007/s10592-014-0608-x |
Wang, J., Ai, B., Kong, H., et al., 2017a. Speciation history of a species complex of Primulina eburnea (Gesneriaceae) from limestone karsts of southern China, a biodiversity hot spot. Evol. Appl., 10: 919-934. DOI:10.1111/eva.12495 |
Wang, J., Feng, C., Jiao, T., et al., 2017b. Genomic signature of adaptive divergence despite strong nonadaptive forces on edaphic islands: a case study of Primulina juliae. Genome Biol. Evol., 9: 3495-3508. DOI:10.1093/gbe/evx263 |
Wang, P., Burley, J.T., Liu, Y., et al., 2021. Genomic consequences of long-term population decline in brown eared pheasant. Mol. Biol. Evol., 38: 263-273. DOI:10.1093/molbev/msaa213 |
Wang, Q., Lan, T., Li, H., et al., 2022. Whole-genome resequencing of Chinese pangolins reveals a population structure and provides insights into their conservation. Commun. Biol., 5: 821. DOI:10.1038/s42003-022-03757-3 |
Whitlock, M.C., 2000. Fixation of new alleles and the extinction of small populations: drift load, beneficial alleles, and sexual selection. Evolution, 54: 1855-1861. |
Willi, Y., Van Buskirk, J., Hoffmann, A.A., 2006. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst., 37: 433-458. DOI:10.1146/annurev.ecolsys.37.091305.110145 |
Wilson, G.A., Rannala, B., 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics, 163: 1177-1191. DOI:10.1093/genetics/163.3.1177 |
Wu, F.Q., Shen, S.K., Zhang, X.J., et al., 2014. Genetic diversity and population structure of an extremely endangered species: the world's largest Rhododendron. AoB Plants, 7: plu082. |
Wu, G.L., Ye, Q., Liu, H., et al., 2022. The evolutionary rate of leaf osmotic strength drives diversification of Primulina species in karst regions. J. Syst. Evol., 51: 843-851. DOI:10.3390/antibiotics11070843 |
Xiao, Y., Li, X.J., Jiang, X.L., et al., 2023. Spatial genetic patterns and distribution dynamics of Begonia grandis (Begoniaceae), a widespread herbaceous species in China. Front. Plant Sci., 14: 1178245. DOI:10.3389/fpls.2023.1178245 |
Yang, Y., Ma, T., Wang, Z., et al., 2018. Genomic effects of population collapse in a critically endangered ironwood tree Ostrya rehderiana. Nat. Commun., 9: 5449. DOI:10.1038/s41467-018-07913-4 |
Zachos, J.C., Dickens, G.R., Zeebe, R.E., 2008. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature, 451: 279-283. DOI:10.1038/nature06588 |
Zhang, J.G., Chen, H.S., Su, Y.R., et al., 2011. Spatial variability and patterns of surface soil moisture in a field plot of karst area in southwest China. Plant Soil Environ., 57: 409-417. DOI:10.17221/374/2010-PSE |
Zhang, X.J., Liu, X.F., Liu, D.T., et al., 2021. Genetic diversity and structure of Rhododendron meddianum, a plant species with extremely small populations. Plant Divers., 43: 472-479. DOI:10.1016/j.pld.2021.05.005 |
Zheng, B., Xu, Q., Shen, Y., 2001. The relationship between climate change and Quaternary glacial cycles on the Qinghai–Tibetan Plateau: review and speculation. Quat. Int., 97–98: 93-101. |
Zhu, X., Liang, H., Jiang, H., et al., 2022. Phylogeographic structure of Heteroplexis (Asteraceae), an endangered endemic genus in the limestone karst regions of southern China. Front. Plant Sci., 13: 999964. DOI:10.3389/fpls.2022.999964 |



