b. CAS Key Laboratory of Tropical Forest Ecology, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla 666303, China;
c. Xi'an Center for Disease Control and Prevention, Xi'an 710068, China
Genome-scale data, while promising for illuminating phylogenetic relationships, frequently pose a conundrum by yielding conflicting topologies and highly variable gene tree distributions (Pease et al., 2016). This complexity likely arises from the reticulate evolution observed in many taxa, where genetic information exchange occurs through diverse biological processes, deviating from a strictly bifurcating mode (Gernandt et al., 2018). In the context of reticulate evolution, phylogenetic inference often reveals discordances between gene trees and species trees, a phenomenon termed phylogenomic conflict (Smith et al., 2020). This conflict is principally driven by population processes such as incomplete lineage sorting (ILS) and interspecific gene flow (introgression) (Hibbins and Hahn 2022). The challenge intensifies as interspecific gene flow generates genomic patterns similar to those arising from ILS, making precise quantification and phylogenetic inference formidable (Pease et al., 2016).
Recent advances in sequencing technologies and the emergence of various methods, notably in phylogenomic, have significantly improved our ability to discern and quantify ILS and gene flow (Cai et al., 2021). The inherent flexibility of tree-based methods allows for the construction of evolutionary relationships based on the analysis of orthologous genes (Cheon et al., 2020), easily obtained from transcriptome data. Transcriptomes are cost-effective and suitable for sequencing without prior knowledge of the target genome's size and complexity, particularly valuable when working with non-model organisms (One Thousand Plant Transcriptomes Initiative, 2019). These advancements also empower the assessment of the influence of recombination rate variation on introgression and ILS (Feng et al., 2023), providing valuable perspectives on the relevance of genes in deciphering the evolution of life.
Within the realm of spruce taxa, the phenomena of introgression and ILS have frequently captured attention due to substantial effective population sizes, limited interspecific divergence, and extended generation times (Feng et al., 2019). Understanding the characteristics of Picea species holds paramount importance for advancing our knowledge of the taxonomy, ecology, and environmental applications of spruce taxa. The Picea likiangensis species complex (PLSC) is of particular interest, displaying reticulate relationships and recent divergence events. It is proposed that geological and climatic changes during the Pliocene and Pleistocene epochs facilitated the incomplete divergence of lineages within the PLSC across the Qinghai-Tibet Plateau (Li et al., 2013). Subsequent studies illuminated the penetrable boundaries between taxa within the PLSC and related species (Sun et al., 2018; Ru et al., 2018). The PLSC has been identified as a contributor to the origin of multiple species including Picea purpurea and Picea brachytyla (Ru et al., 2018; Wang et al., 2023). P. purpurea has notably influenced the evolution of P. likiangensis var. rubescens, while Picea spinulosa has been closely clustered with P. likiangensis var. linzhiensis. However, the lack of related taxa may undervalue ILS and mislead our understanding of divergence between the PLSC lineages. Moreover, it is still unclear whether homologous recombination, which plays decisive roles in generating the distribution of phylogenetic signals (Rivas-González et al., 2023), influence the genomic patterns of both ILS and introgression within spruce and even the reconstruction of evolutionary history.
To address these gaps, our study collected 279 samples from six spruce species, which together represent the extended PLSC (e-PLSC) as a representative model system. These species, including Picea spinulosa (45), P. purpurea (48), P. likiangensis var. likiangensis (49), P. likiangensis var. linzhiensis (57), P. likiangensis var. rubescens (70) and P. farreri (10) (Fig. 1A; Tables S1 and S2), formed a cohesive cluster and were identified as the sister lineage to Picea wilsonii (Feng et al., 2019). Our goal is to investigate the relative impacts of introgression and ILS on diversification within the e-PLSC, and how recombination rates influence them, offering new insights into the complex evolutionary history of these spruce species.
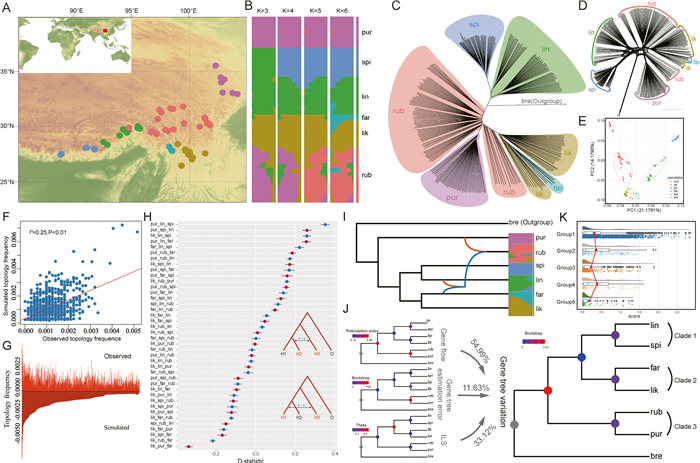
|
| Fig. 1 A: The distribution of the sampled trees within the e-PLSC. Red, yellow, green, light-blue, blue and purple circles indicate locations from where Picea likiangensis var. rubescens (rub), P. likiangensis var. likiangensis (lik), P. likiangensis var. linzhiensis (lin), P. farreri (far), P. spinulosa (spi), P. purpurea (pur) samples were collected for RNA-seq. B: Bar-plots indicative of assignment probabilities from ADMIXTURE analysis of 289 transcriptomes. The best number of clusters (K) is 6, suggested by cross-validation analysis in ADMIXTURE. C: The NJ tree. Red, yellow, green, light-blue, blue and purple indicate species of P. likiangensis var. rubescens (rub), P. likiangensis var. likiangensis (lik), P. likiangensis var. linzhiensis (lin), P. farreri (far), P. spinulosa (spi), P. purpurea (pur), respectively. D: SplitsTree networks for the concatenated nuclear datasets of P. likiangensis var. rubescens (rub), P. likiangensis var. likiangensis (lik), Picea farreri (far), P. likiangensis var. linzhiensis (lin), P. spinulosa (spi), P. purpurea (pur) and Picea breweriana (bre). E: Principal component analysis (PCA) plots showing the first two principal components. F: Comparison of simulated frequencies based on coalescent probabilities of gene topologies of different classes with the corresponding observed frequencies. G: Observed and simulated topology frequency. H: The results of the ABBA-BABA test. The X-axis represents the D-statistic. I: The likely hybrid events among taxa, inferred by PhyloNet. J: Relative contributions of ILS, estimation error, and gene flow for taxa within the e-PLSC. K: Recombination rates for all genes, including the density distribution, boxplots and abnormal values, and the estimated values for each gene represented by points, for each of three groups respectively. Each point represents the recombination rate of a gene, and the red line in the box is the average recombination rate in that group. Group 1, Group 2, Group 3, Group 4 and Group 5 represent the distribution of recombination rates of all genes, genes in the top ten percent of D-statistic scores (calculated using pur_lin_spi as P1, P2, P3), genes with D-statistic scores close to zero (calculated using pur_lin_spi as P1, P2, P3), genes with the top 50 ILS scores, genes with the bottom 20 ILS scores, respectively. The abnormal values are represented by diamonds. |
In addition to the 279 samples of the e-PLSC, we also collected 2 samples of Picea breweriana to serve as an outgroup. The average depth of the entire transcriptome was determined to be 51.81 ×, with an average mapping rate of 73.94% and an average transcriptome coverage of 72.98% (Table S2). The generated VCF file which stored the mutation information contained 239,730 SNPs after strict quality filtering. Seven high-quality transcriptomes were successfully assembled, yielding a total of 95,212 contigs for P. likiangensis var. rubescens, 226,501 for P. likiangensis var. likiangensis, 129,898 for P. likiangensis var. linzhiensis, 115,956 for P. farreri, 114,980 for P. spinulosa, 76,820 for P. purpurea and 67,811 for P. breweriana. The N50 values for these assemblies ranged from 980 to 1882 and BUSCO values ranged from 82.0 to 87.5, indicating high quality and completeness transcriptomes (Table S3). OrthoMCL identified a total of 7203 ortholog genes across the seven species.
Our extensive analyses of transcriptome-based SNP variation, including phylogenetic analyses, principal component analysis, and genetic clustering (Figs. 1B–E, S1 and S2), have robustly highlighted the genetic distinctiveness of the e-PLSC. This finding aligns cohesively with previous studies (Ru et al., 2018; Sun et al., 2018; Wang et al., 2023). An intriguing revelation from our results is the extensive incongruity observed between the chloroplast and nuclear trees, based on expanded sampling (Figs. 1C and D and S1). The plastid genomes of a small number of samples are classified into branches of non-local groups, which contradicts the expected pedigree information (Fig. S1). This incongruence may suggest a complex network of evolutionary relationships within this group. Introgression and ILS have emerged as plausible explanations for this incongruence (Garg and Chattopadhyay, 2021), with their prevalence among the six taxa indicating a substantial role in shaping the genomic landscape. Additionally, these results strongly suggests that P. likiangensis var. rubescens may have undergone hybridization events with P. purpurea, P. likiangensis var. likiangensis, and P. likiangensis var. linzhiensis. Further exploration of the specific mechanisms and genomic regions underlying these patterns will provide valuable insights into the evolutionary dynamics and genetic interactions within this group of spruce species.
In addition, we simulated 20,000 gene trees under the multi-species coalescent model on the basis of the ASTRAL tree inferred from the 7203 ortholog genes obtained from OrthoMCL. We found considerable agreement between simulated and empirical gene trees (overall correlation coefficient, Spearman's ρ = 0.25, P < 0.01; Fig. 1F and G), suggesting that the multi-species coalescent model is quite a good fit to our data, and ILS can well explain the conflicting topologies in the e-PLSC. Our further investigation leveraged D-statistics, PhyloNet analysis, QuIBL, and methods from Cai et al. (2021), accounting for ILS to detect and quantify introgression, consistently shedding lights on the extent of introgression and ILS among clades (Fig. 1H–J and Table S4). Of particular note is our robust substantiation of the intricate nature of the P. likiangensis var. rubescens component, a phenomenon previously suggested and corroborated through our ADMIXTURE and PhyloNet analyses (Fig. 1B and I; Wang et al., 2023). The presence of a complex genetic component, combined with polyphyly of P. likiangensis var. rubescens and the fact that P. purpurea is nested within the P. likiangensis var. rubescens clade in the phylogeny (Fig. 1B–D), strongly suggests the existence of a genetic transitional zone along the distribution of P. likiangensis var. rubescens. The existence of this genetic transitional zone may have played a crucial role in the expansion of P. likiangensis var. rubescens, as suggested by the observation of negative Tajima's D value (Fig. S3 and Table S5). These findings suggest that introgression has played a role in shaping the genetic makeup of these spruce populations. Some of the genes that show high levels of introgression are involved in abiotic stress responses (Figs. S4–S7 and Tables S6–S9), may be associated with the sharing of genetic traits and may contribute to the formation of homoplasy and reticulation.
Our research strongly indicates that models positing a strictly bifurcating tree can be supplanted by network models, which hypothesize reticulation events to account for most of gene tree discordances (Wen et al., 2018). Using phylogenetic network methods and visualizing the outcomes using tools like Densitree and Dendroscope, we obtained deeper insights into the relationships between the three clades within the e-PLSC, revealing the reticulated evolutionary patterns (Figs. 1I and J and S8). Furthermore, our investigation reveals a possible correlation between the extent of interspecific gene flow and geographical distance (Figs. 1H and S12). For instance, we observed more significant gene exchange between P. spinulosa and P. likiangensis var. linzhiensis than P. spinulosa and P. purpurea (Fig. 1H), potentially due to wind-mediated pollen transport facilitated by the limited geographic distance between P. spinulosa and P. likiangensis var. linzhiensis (Fig. 1A). Intriguingly, P. spinulosa exhibited a larger genetic distance compared to P. farreri (Figs. S3 and S9 and Table S10) when compared with other species within the e-PLSC, possibly attributed to a combination of lineage sorting and limited introgression. P. purpurea appears to undergo introgression more frequently than P. spinulosa, while the latter exhibits a higher prevalence of ILS (Fig. 1H–J). Furthermore, our analysis reveals that Clade 1 and Clade 2 display greater levels of ILS, whereas between Clade 1 + Clade 2 and Clade 3, interspecific gene flow is more pronounced (Fig. 1J). Within Clade 3, interspecific gene flow and estimation errors contribute significantly. In aggregate, it becomes apparent that interspecific gene flow has a more substantial impact than ILS (54.99% vs. 33.12%), aligning with the findings of our QuIBL, and ILS simulation analyses (Fig. 1H–J and Table S4). Importantly, our results unequivocally demonstrate substantial introgression and ILS within the e-PLSC. This extensive gene exchange highlights the critical role of hybridization in shaping the genetic diversity of these spruce species. It's worth noting that this gene flow persists even in the presence of marked genetic differentiation among these species, underscoring the dynamic nature of species boundaries in this complex system.
A striking finding in our study is the correlation between regions with high recombination rates and elevated levels of introgression and ILS (Fig. 1K). This observation indicates that recombination may play a significant role in shaping the phylogenetic patterns within the e-PLSC. Interestingly, genomic regions with lower recombination rates exhibit decreased levels of ILS and introgression (Fig. 1K), thereby demonstrating higher accuracy in capturing species relationships. This highlights the influence of recombination rates on the accuracy of phylogenetic inferences, consistent with previous studies (Feng et al., 2023), and underscores the importance of considering these recombination rates in future phylogenetic research. Furthermore, it's worth noting that these genes in genomic regions with high recombination rates may be subject to natural selection, adding an additional layer of complexity to their role in the evolutionary dynamics of the e-PLSC and likely contributing to the increased genetic diversity and the potential for adaptive evolution within these populations. For instance, genes associated with high introgression and ILS levels display relevance to adaptation in spruces. Further gene ontology (GO) enrichment showed that genes in genomic regions with high recombination rates were associated with abiotic stresses, response to cold and hypoxia, and response to UV light (Figs. S4–S7, S10 and S11; Tables S6–S9, S11 and S12), suggesting that recombinants from introgression might have facilitate the adaptive evolution of the e-PLSC. Introgression introduces adaptive variation from closely related species, and influences phenotypes (Pardo-Diaz et al., 2012), while recombination may disrupt linkage and generate new combinations of selected alleles (Schumer et al., 2018). On the other hand, natural selection favors advantageous traits, and accelerates the accumulation and fixation of beneficial mutations. Therefore, within the e-PLSC, the frequent gene flow and environmental selection are plausible drivers to the increased levels of introgression and ILS in regions with elevated recombination rates.
In conclusion, our comprehensive analysis of spruce transcriptomes has unveiled the pervasive occurrence of introgression and ILS within the e-PLSC, weaving a complex network of evolutionary trajectories. Our investigation highlights the substantial incongruence between chloroplast and nuclear trees, primarily attributed to ILS and gene flow, with interspecific gene flow (54.99%) exerting a greater influence than ILS (33.12%) on the conflicting topological structures within the e-PLSC. Upon closer examination of the gene tree topologies, we have found an intriguing synchronization: regions characterized by high levels of introgression and ILS often coincide with elevated rates of recombination, possibly affected by natural selection. Conversely, regions with low recombination rates appear to be more conducive to accurate reconstruction of the tree of life. Furthermore, this study provides valuable resources and insights for future studies on the molecular characterization and functional genomics of spruce.
AcknowledgementsThis study was supported by the National Natural Science Foundation of China (grant no. 32001085, 31971392, 31960319).
Data availability
All sequencing reads that support the findings of this study have been deposited in the NCBI Sequence Read Archive under project number PRJNA1069340. All the assembled transcriptomes and supplementary materials that support the findings of this study are available from the Dryad Digital Repository: https://datadryad.org/stash/share/5OKHQkKtdWFGrqJ4J3wyY1iU15kGhl2KKZ41pcPdcLw.
CRediT authorship contribution statement
Hui Zhu: Writing – original draft, Visualization, Software, Investigation, Formal analysis, Data curation. Weixiao Lei: Visualization, Software, Resources, Formal analysis, Data curation. Qing Lai: Software, Resources, Formal analysis. Yongshuai Sun: Writing – review & editing, Validation, Supervision, Project administration, Investigation, Funding acquisition, Conceptualization. Dafu Ru: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare no competing interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.04.004.
Cai, L., Xi, Z., Lemmon, E.M., et al., 2021. The perfect storm: gene tree estimation error, incomplete lineage sorting, and ancient gene flow explain the most recalcitrant ancient angiosperm clade. Malpighiales. Syst. Biol., 70: 491-507. DOI:10.1093/sysbio/syaa083 |
Cheon, S., Zhang, J., Park, C., 2020. Is phylotranscriptomics as reliable as phylogenomics?. Mol. Biol. Evol., 37: 3672-3683. DOI:10.1093/molbev/msaa181 |
Feng, C., Wang, J., Liston, A., et al., 2023. Recombination variation shapes phylogeny and introgression in wild diploid strawberries. Mol. Biol. Evol., 40: msad049. DOI:10.1093/molbev/msad049 |
Feng, S., Ru, D., Sun, Y., et al., 2019. Trans-lineage polymorphism and nonbifurcating diversification of the genus Picea. New Phytol., 222: 576-587. DOI:10.1111/nph.15590 |
Garg, K.M., Chattopadhyay, B., 2021. Gene flow in Volant Vertebrates: species biology, ecology and climate change. J. Indian Inst. Sci., 101: 165-176. DOI:10.1007/s41745-021-00239-z |
Gernandt, D.S., Aguirre Dugua, X., Vázquez-Lobo, A., et al., 2018. Multi-locus phylogenetics, lineage sorting, and reticulation in Pinus subsection Australes. Am. J. Bot., 105: 711-725. DOI:10.1002/ajb2.1052 |
Hibbins, M.S., Hahn, M.W., 2022. Phylogenomic approaches to detecting and characterizing introgression. Genetics, 220: iyab173. DOI:10.1093/genetics/iyab173 |
Li, L., Abbott, R.J., Liu, B., et al., 2013. Pliocene intraspecific divergence and Plio-Pleistocene range expansions within Picea likiangensis (Lijiang spruce), a dominant forest tree of the Qinghai-Tibet Plateau. Mol. Ecol., 22: 5237-5255. DOI:10.1111/mec.12466 |
One Thousand Plant Transcriptomes Initiative, 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature, 574: 679-685. DOI:10.1038/s41586-019-1693-2 |
Pardo-Diaz, C., Salazar, C., Baxter, S.W., et al., 2012. Adaptive introgression across species boundaries in Heliconius butterflies. PLoS Genetics., 8: e1002752. DOI:10.1371/journal.pgen.1002752 |
Pease, J.B., Haak, D.C., Hahn, M.W., et al., 2016. Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biology., 14: e1002379. DOI:10.1371/journal.pbio.1002379 |
Rivas-González, I., Rousselle, M., Li, F., et al., 2023. Pervasive incomplete lineage sorting illuminates speciation and selection in primates. Science, 380: eabn4409. DOI:10.1126/science.abn4409 |
Ru, D., Sun, Y., Wang, D., et al., 2018. Population genomic analysis reveals that homoploid hybrid speciation can be a lengthy process. Mol. Ecol., 27: 4875-4887. DOI:10.1111/mec.14909 |
Schumer, M., Xu, C., Powell, D.L., et al., 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science, 360: 656-660. DOI:10.1126/science.aar3684 |
Smith, S.A., Walker-Hale, N., Walker, J.F., 2020. Intragenic conflict in phylogenomic data sets. Mol. Biol. Evol., 37: 3380-3388. DOI:10.1093/molbev/msaa170 |
Sun, Y., Abbott, R.J., Lu, Z., et al., 2018. Reticulate evolution within a spruce (Picea) species complex revealed by population genomic analysis. Evolution, 72: 2669-2681. DOI:10.1111/evo.13624 |
Wang, D., Sun, Y., Lei, W., et al., 2023. Backcrossing to different parents produced two distinct hybrid species. Heredity, 131: 145-155. DOI:10.1038/s41437-023-00630-9 |
Wen, D., Yu, Y., Zhu, J., et al., 2018. Inferring phylogenetic networks using PhyloNet. Syst. Biol., 67: 735-740. DOI:10.1093/sysbio/syy015 |



