b. Department of Bioengineering, Zhuhai Campus of Zunyi Medical University, Zhuhai 519041, Guangdong, China;
c. China-ASEAN College of Marine Sciences, Xiamen University Malaysia, Sepang 43900, Selangor, Malaysia;
d. School of Agriculture, Sun Yat-sen University, Shenzhen 518107, Guangdong, China;
e. School of Ecology, Sun Yat-sen University, Shenzhen 518107, Guangdong, China
Polyploidization is widely considered a major factor in the evolution and diversification of plants (Madlung, 2013). Previous studies have reported that over 15% of all known angiosperms have polyploid origins, and most of the studied angiosperms have had some form of polyploidization in their evolutionary histories (Wood et al., 2009). Polyploids can be formed from the hybridization of two different lineages and subsequent doubling of chromosomes (allopolyploids) or by genome doubling of a single lineage (autopolyploids). In addition, cryptic allopolyploid speciation may occur within morphologically uniform diploid-tetraploid species, e.g., Allium przewalskianum Regel (Liang et al., 2015). However, most of our understanding of polyploidization has been derived from crop species, including autopolyploids [e.g., Medicago sativa L. (Chen et al., 2020a), Solanum tuberosum L. (Bao et al., 2022), Saccharum spontaneum L. (Zhang et al., 2018)] and allopolyploids [e.g., Gossypium spp. (Chen et al., 2020b; Huang et al., 2020), Brassica spp. (Yang et al., 2016), Arachis spp. (Yin et al., 2020)]. Polyploidization has also been investigated in wild plants distributed in regional hot spots, e.g., in the Tibetan Plateau (Wu et al., 2022; Li et al., 2021). Knowledge of the origin and evolution of polyploidy is rare for tropical plant species and even rarer for plant species of intertidal zones, which provide valuable ecosystem services and are vulnerable to the changing environment.
Mangroves are a group of pantropical plant species that inhabit the intertidal zone (Tomlinson, 2016). They are highly adapted to saline and hypoxic environments and provide critical biological and ecological services in coastal ecosystems (Barbier et al., 2011; Duke and Schmitt, 2015). Natural hybridization is common among mangroves as their geographical distributions, habitats, flowering time, and pollinators often overlap (Ragavan et al., 2017). Previous studies have reported that hybridization in mangroves often occurs locally, and mangrove hybrids are simple F1s with low pollen viability and seed germination (Ragavan et al., 2017) and restricted dispersal beyond the hybrid sites (Lo, 2010). However, mangroves have experienced and survived several ancient geomorphological and environmental changes. Apart from contemporary demographic (e.g., overlapping distribution) and physiological (e.g., shared pollinators) traits, these ancient events can also induce secondary contact between closely related species, leading to interspecific hybridization and potentially the formation of new allopolyploid species (Abbott et al., 2013). One of the biodiversity hotspots of mangrove distribution (Duke, 2017), the Indo–West Pacific region (IWP, hereafter), has experienced several past climate-change events (Lohman et al., 2011), which has shaped the current distribution of many mangrove species (Wee et al., 2017). However, allopolyploid speciation in the mangroves instigated by these climate-change events in the IWP has been rarely reported.
Polyploids, particularly allopolyploids, can adapt more easily to changing environments by having two or more different genomes (Madlung, 2013). However, most nascent polyploid lineages face challenges like small population size and competition with their diploid relatives (Barker et al., 2016). Previous studies reported that one of the major factors for successful polyploid establishment is ecological niche differentiation (Baniaga et al., 2020), i.e., the polyploid and the progenitors have different resource needs or utilization strategies. This hypothesis is supported and contradicted by multiple lines of evidence. Some allopolyploids have been found to occupy divergent ecological niches from at least one of their progenitors (e.g., Han et al., 2022), whereas the ecological niches of some allopolyploids are intermediate or even non-divergent from that of its progenitors (e.g., Casazza et al., 2017). The roles of environmental heterogeneity and ecological niche differentiation during allopolyploid speciation in mangroves have yet to be investigated.
In this study, we focus on Acanthus, a genus native to the tropics and subtropics of the Old World (Europe, Asia, and East Africa), with approximately 30 known species. Three species, namely, Acanthus ilicifolius L., A. ebracteatus Vahl. and A. volubilis Wall. are true mangroves distributed in the IWP (Duke and Schmitt, 2015; Ragavan et al., 2015; Yang et al., 2015). Previous studies have indicated the three Acanthus mangroves share a common origin, as they form one clade distinct from the terrestrial Acanthus species (Acanthus leucostachyus Wall. ex Nees, A. mollis L., and A. montanus (Nees) T. Anderson) (Yang et al., 2015). A. volubilis can be easily distinguished from A. ilicifolius and A. ebracteatus by its unarmed and twining and delicate sprawling stems, un-serrated elliptical leaves, white corolla, and absence of bracteoles. A. ebracteatus and A. ilicifolius have similar habits and leaf morphology; nevertheless, the former species differs from the latter in the absence of bracteoles, white corolla, and smaller flowers and fruits (Ragavan et al., 2015). A. ilicifolius, as currently circumscribed, is the most abundant Acanthus true mangrove species in the IWP region (Ma et al., 2023), showing great variation in corolla morphology among populations. Multiple Acanthus species reported from the IWP region are currently treated as synonyms of A. ilicifolius. For example, A. xiamenensis R.T. Zhang was reported to be endemic to China (Zhang, 1985), but Wang and Wang (2007) treated this species as synonymous with A. ilicifolius. Debnath et al. (2013) reported a new mangrove species named A. albus Debnath, B.K. Singh & P. Giri from the Indian Sundarbans, but Ragavan et al. (2015) suggested further studies on this species to avoid complications in the identity of A. ilicifolius. Therefore, the species delimitation of A. ilicifolius remains unclear.
Our previous study of Acanthus ilicifolius based on chloroplast DNA markers revealed that populations from the eastern (i.e., the Pacific Ocean region) and western (i.e., the Indian Ocean region) coasts of the Thai–Malay Peninsula (TMP, hereafter) represent two well-differentiated lineages (Guo et al., 2020). We also observed some morphological differences between these two lineages in our fieldwork, which has been supported by exploring digital images from the Global Biodiversity Information Facility (GBIF: https://www.gbif.org/; accessed on January 2013–May 2023). Specifically, the western lineage of A. ilicifolius has a purple or deep purple corolla with a dark median band, whereas the corolla of the eastern lineage is violet to blue with a white to yellow median band. Similar morphological differences of A. ilicifolius populations across the TMP have been observed in other studies [e.g., western lineage: samples from the Andaman and Nicobar Islands of India (Ragavan et al., 2015), Indian Sundarbans (Chatterjee et al., 2023); eastern lineage: China (William Jackson Hooker's Chinese Plants collection (1869), accessed from https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:44873-1 on 20 May 2023); Java (Backer and Bakhuizen, 1965), New Guinea (Barker, 1986), Australia (Duke, 2006), and New Caledonia (Duke and Schmitt, 2015)]. A previous karyotype analysis reported the chromosome numbers of A. ilicifolius (N = 70) nearly two times larger than that of A. volubilis (N = 34) and A. ebracteatus (N = 38), indicating the tetraploid nature of A. ilicifolius samples (Suan, 1996).
Taken together, the molecular, morphological, and cytological differences as well as the geographic separation of the two lineages of Acanthus ilicifolius warrant further investigation into their evolutionary biology and biogeography. We hypothesized that 1) the Acanthus species distributed along the eastern coast of the TMP, identified so far as A. ilicifolius, is a new species that originated from polyploid speciation in the ancient past, and 2) the polyploid established successfully in this region by having a different ecological niche than its progenitors. We adopted an integrative approach to identify the allopolyploid that combined morphological, cytological, and molecular investigations of three Acanthus taxa collected from the IWP region. In addition to quantifying five plant traits and karyotype analysis, we reconstructed the phylogenetic relationships, evolutionary history, and biogeography of these three taxa using genetic information of eight low-copy nuclear genes and two chloroplast fragments. Based on current occurrence records and relevant climatic factors governing their distributions, we further mapped the climatic niches of Acanthus species and reconstructed their past and present potential distributions in this region. The main objectives of this study were to 1) determine the taxonomic status of Acanthus spp. in the IWP region and 2) identify the underlying factors shaping the interspecific relationships.
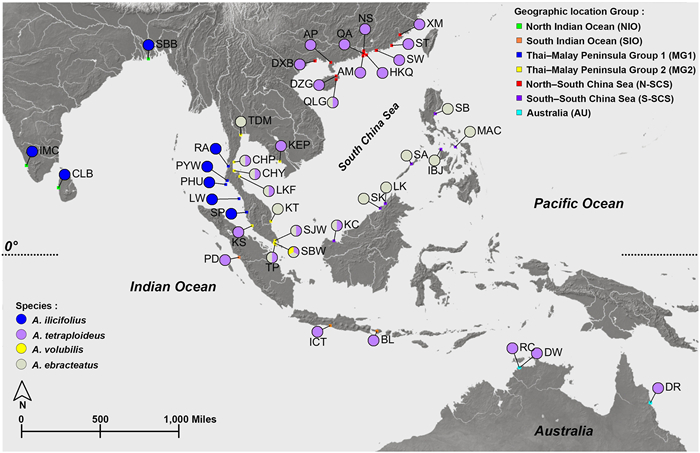
2. Material and methods 2.1. Sampling of plant materialsWe collected 493 plant samples of Acanthus species from 42 geographic sites across the IWP region between 2015 and 2022 (Table 1). A total of 306 samples of A. ilicifolius and 172 of A. ebracteatus were collected. Due to the restricted distribution of A. volubilis in the IWP region, we found the species only in Singapore, where 15 samples were collected. All samples were identified using published identification keys (Ragavan et al., 2015). The 42 sites were grouped into seven regions: North Indian Ocean (NIO), South Indian Ocean (SIO), West of the Thai–Malay Peninsula (MG1), East of the Thai–Malay Peninsula (MG2), North–South China Sea (N-SCS), South–South China Sea (S-SCS), and Australia (AU) (Fig. 1). Based on the findings of our previous study on A. ilicifolius (Guo et al., 2020), these seven regions were further classified into two lineages: the western lineage (NIO and MG1) and the eastern lineage (SIO, MG2, N-SCS, S-SCS, AU). The collected samples of A. ilicifolius (N = 306) were thus categorized into—101 samples from eight sites in NIO and MG1 regions representing the western lineage and 205 samples from 26 sites distributed in the other five regions representing the eastern lineage. The sample sizes varied from 6 to 15 individuals that were separated from each other by at least 10 m.
| Group | Site ID | Latitude (°) | Longitude (°) | Location | A. ebracteatus | A. tetraploideus† | A. ilicifolius‡ | A. volubilis |
| N-SCS | XM | 24.51 | 117.65 | Xiamen, Fujian, China | – | 10 | – | – |
| N-SCS | ST | 23.32 | 116.72 | Shantou, Guangdong, China | – | 8 | – | – |
| N-SCS | SW | 22.79 | 115.03 | Shanwei, Guangdong, China | – | 6 | – | – |
| N-SCS | NS | 22.75 | 113.62 | Nansha, Guangdong, China | – | 8 | – | – |
| N-SCS | QA | 22.43 | 113.64 | Qi'ao Island, Guangdong, China | – | 8 | – | – |
| N-SCS | HKQ | 22.28 | 113.93 | Hongkong, China | – | 12 | – | – |
| N-SCS | AM | 22.16 | 113.55 | Macau, China | – | 8 | – | – |
| N-SCS | AP | 21.45 | 109.97 | Anpu, Guangdong, China | – | 8 | – | – |
| N-SCS | DXB | 21.62 | 108.23 | Fangchenggang, Guangxi, China | – | 6 | – | – |
| N-SCS | DZG | 19.95 | 110.58 | Dongzhaigang, Hainan, China | – | 10 | – | – |
| N-SCS | QLG | 19.55 | 110.58 | Qingliangang, Hainan, China | 12 | 8 | – | – |
| S-SCS | SB | 15.76 | 121.56 | Sabang Beach, Philippines | 11 | – | – | – |
| S-SCS | MAC | 12.08 | 123.78 | Mactan, Philippines | 9 | – | – | – |
| S-SCS | IBJ | 11.82 | 122.17 | Ibajay, Aklan, Philippines | 15 | – | – | – |
| S-SCS | SA | 10.2 | 118.9 | Palawan, Philippines | 8 | – | – | – |
| S-SCS | LK | 5.82 | 116.02 | Kinarut, Sabah, Malaysia | 16 | – | – | – |
| S-SCS | SK | 5.28 | 115.46 | Kuala Penyu, Sabah, Malaysia | 11 | – | – | – |
| S-SCS | KC | 1.67 | 110.33 | Kuching, Sarawak, Malaysia | 10 | 4 | – | – |
| MG2 | KS | 3.34 | 101.26 | Kuala Selangor, Malaysia | – | 12 | – | – |
| MG2 | TP | 1.27 | 103.79 | Tanjung Piai, Johor, Malaysia | 7 | 4 | – | – |
| MG2 | SBW | 1.45 | 103.72 | Sungei Buloh Wetland Reserve, Singapore | 11 | 5 | – | 15 |
| MG2 | SJW | 1.71 | 103.79 | Batu Pahat, Johor, Malaysia | 15 | 3 | – | – |
| MG2 | KT | 3.82 | 103.33 | Kuantan, Malaysia | 15 | – | – | – |
| MG2 | LKF | 8.81 | 99.89 | Tha Sala, Nakhon Si Thammarat, Thailand | 8 | 8 | – | – |
| MG2 | CHY | 9.45 | 99.26 | Chaiya, Thailand | 6 | 3 | – | – |
| MG2 | CHP | 10.41 | 99.25 | Chumphon, Thailand | 8 | 4 | – | – |
| MG2 | TDM | 13.41 | 100 | Samut Songkhram, Thailand | 10 | – | – | – |
| MG2 | KEP | 10.49 | 104.32 | Kep, Cambodia | – | 9 | – | – |
| SIO | PD | 0.15 | 99.8 | Padang, Indonesia | – | 10 | – | – |
| SIO | ICT | −7.73 | 109.91 | Cilacap, Indonesia | – | 11 | – | – |
| SIO | BL | −8.34 | 115.09 | Bali, Indonesia | – | 8 | – | – |
| AU | RC | −12.38 | 130.86 | Rapid Creek, Australia | – | 8 | – | – |
| AU | DW | −12.37 | 130.92 | Darwin, Australia | – | 11 | – | – |
| AU | DR | −16.27 | 145.4 | Daintree River, Australia | – | 13 | – | – |
| MG1 | RA | 9.95 | 98.61 | Ranong, Thailand | – | 16 | – | |
| MG1 | PYW | 8.35 | 98.5 | Phangnga, Thailand | – | 13 | – | |
| MG1 | PHU | 7.92 | 98.34 | Phuket, Thailand | – | 8 | – | |
| MG1 | LW | 6.35 | 99.8 | Langkawi, Malaysia | – | 15 | – | |
| MG1 | SP | 4.84 | 100.63 | Kuala Sepetang, Malaysia | – | 12 | – | |
| NIO | IMC | 10.03 | 76.25 | Kerala, India | – | 9 | – | |
| NIO | CLB | 7.56 | 79.8 | Chilaw, Sri Lanka | – | 13 | – | |
| NIO | SBB | 21.85 | 89.77 | Katka, Sundarban, India | – | 15 | – | |
| In research region | 172 | 205 | 101 | 15 | ||||
| Note: †: Previously defined as eastern lineage of Acanthus ilicifolius, ‡: Previously defined as western lineage of A. ilicifolius. | ||||||||
|
| Fig. 1 Map of the sampling sites of four Acanthus species. A. ilicifolius, A. tetraploideus (previously identified as the eastern lineage of A. ilicifolius), A. ebracteatus, and A. volubilis in the Indo–West Pacific (IWP) region. The sampled populations are colored based on four species names, and the sampling locations are colored based on seven major biogeographic regions in the IWP. |
Morphological data of Acanthus species were obtained through fieldwork, online databases, and a literature survey (Duke, 2006; Duke and Schmitt, 2015; Ragavan et al., 2015; Herawati et al., 2020; Chatterjee et al., 2023). During the field survey, habit, plant height, the length and width of leaves, and the length, weight, and color of the corolla were measured, recorded, and photographed. Digital images of herbarium specimens were examined through three online databases: China Virtual Herbarium (CVH, https://www.cvh.ac.cn/), the Global Biodiversity Information Facility (GBIF, https://www.gbif.org/), and JSTOR (https://plants.jstor.org/). We also examined photographs of living plants from the following databases: Plants of the World Online (https://powo.science.kew.org/), Plant Photo Bank of China (PPBC, http://ppbc.iplant.cn/), eFlora of India (https://sites.google.com/site/efloraofindia/), The Plant Observatory (http://www.natureloveyou.sg/), Plant Resources of Southeast Asia (PROSEA, https://www.prota4u.org/prosea/), Australian Plant Image Index (https://www.anbg.gov.au/), and iNaturalist (https://www.inaturalist.org/).
2.3. Genome size estimation and karyotype analysisFor genome size analysis, we collected young, fresh leaves of Acanthus ebracteatus from four sites (QLG, TP, SK and SBW), of the western lineage of A. ilicifolius from four sites (SBB, RA, PHU and LW), and of the eastern lineage of A. ilicifolius from seven sites (XM, QA, QLG, KS, SBW, BL and DR) (Table 1 and Fig. 1). The number of plants analyzed per population varied from two to four individuals. Genome size was estimated by flow cytometry (FCM), using Oryza sativa L. (haploid genome size = 422 Mb) as an internal standard. We used LB01 (Dpooležel et al., 1989) as the nuclei isolation buffer, followed by filtration through a 40 μm nylon mesh and staining for 10 min under dark refrigeration with 25 μg/ml propidium iodide (PI). The mean red PI fluorescence of stained nuclei was quantified using a BD FACSVerse Flow Cytometer (USA) with a solid-state laser emitting at 488 nm. At least 5000 nuclei were collected for each sample. The genome size was calculated by using the following formula: Sample genome size = (sample G1 peak mean)/(standard G1 peak mean) × (Standard genome size) (D'hondt et al., 2011). Considering the difficulty of harvesting root tips from field samples for karyotype analysis, whole plant samples were collected from the RA site (A. ilicifolius western lineage sample) and QLG site (A. ilicifolius eastern lineage and A. ebracteatus samples). Plant materials were then transferred to a hydroponic solution for root development. Root tips were collected, and chromosome numbers were counted following Govindarajan and Subramanian (1983).
2.4. DNA extraction, amplification, and sequencingHealthy leaves, collected from 493 sampled individuals from 42 geographic sites (Table 1 and Fig. 1), were stored with silica gel until DNA isolation. Total DNA was extracted using a CTAB method (Doyle and Doyle, 1987). Eight low-copy nuclear genes [seven primer-pairs designed from the A. ilicifolius transcriptome data (SRR1793312) and one locus from a previous study (Yang et al., 2015)] and two plastid markers (trnS-trnG, 5′rps12-rpl20) were used for PCR amplification (Table S1). Low-copy nuclear genes were used because they contain more variable sites and have low levels of allelic dosages in genomes (Zhang et al., 2012). PCR products were sequenced directly using an ABI 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA). The sequences were assembled and manually edited using SEQMAN (DNASTAR, Madison, WI, USA) to identify the single nucleotide polymorphisms (SNPs).
For the eight nuclear genes, heterozygous sites were retained as degenerate bases with heterozygous peak threshold = 80% in SEQMAN. Sequences containing more than two degenerate bases were cloned into the pEASY-T1 Cloning Vector System (TransGen, Beijing, China), and 6–8 such clones were Sanger sequenced for phasing of heterozygous sites. Two alleles were obtained for each sample sequence, and the haplotypes were inferred using PHASE 2.1.1 embedded in DNASP v.5.10 (Librado and Rozas, 2009).
2.5. Phylogenetic analysis 2.5.1. Based on chloroplast genomesTo infer the origin of the Acanthus mangrove species in Acanthaceae, we sequenced the chloroplast genomes of four Acanthus mangrove species (i.e., the western lineage of A. ilicifolius, the eastern lineage of A. ilicifolius, A. ebracteatus, and A. volubilis) and the terrestrial A. montanus. The sequencing was done using the Illumina NovaSeq 6000 platform (Table S2), and the complete chloroplast genomes were assembled using the GetOrganelle software (Jin et al., 2020). The chloroplast genome sequences of the other two Acanthus species (A. mollis and A. leucostachyus), 21 species of the family Acanthaceae, and two outgroup species from the families Pedaliaceae (Sesamum indicum L.) and Bignoniaceae (Jacaranda mimosifolia D. Don) were downloaded from the NCBI database (Table S3). We performed HomBlocks (Bi et al., 2018) for multiple sequence alignment, and the maximum likelihood (ML) phylogenetic tree was constructed using IQ-tree (Minh et al., 2020) under the GTR + F + I + G4 model. To determine phylogenetic relationships between the Acanthus mangroves, we constructed independent haplotype networks based on chloroplast fragment sequences of all sampled individuals using the median-joining model (Bandelt et al., 1999) implemented in POPart (Leigh and Bryant, 2015). Continuous insertion-deletion (indel) events were treated as a single mutational event.
2.5.2. Based on nuclear gene sequencesFollowing the workflow of Rothfels (2021) and Han et al. (2022), we also constructed a nuclear phylogenetic tree to infer the speciation mode for the tetraploid species [i.e., the eastern lineage of Acanthus ilicifolius, as revealed in this study (see Section 3.1, and also by Suan, 1996)]. We first constructed haplotype networks of the nuclear datasets by using POPart with the median-joining model. Based on the nuclear gene haplotype networks, alleles of each tetraploid A. ilicifolius sample (N = 205) were then classified as sub-genome 1 (g1) and sub-genome 2 (g2) for each locus (according to the backbone built for the diploid Acanthus spp.). Finally, concatenated sequences were generated for the two sub-genomes. The concatenated sequence for each diploid Acanthus spp. (N = 288) was then generated using a degenerate base containing sequences. A total of 698 concatenated sequences were generated, from which we removed redundant sequences. The sequences shared between at least four individuals were kept for generating the phylogenetic tree. Finally, a total of 40 concatenated sequences, representing 90.7% of all sequences (633 of 698), were used to generate a maximum likelihood (ML) tree with 80% cutoff. The tree was built using IQ-tree under the TPM2+F + I model with 1000 bootstraps.
2.6. Analysis of population structure and genetic diversityTo infer the population structure of Acanthus ilicifolius, we used the sub-genome sequences of g1 and g2 for the tetraploid individuals and the degenerate base containing sequences of the diploid individuals to generate a single variant calling file (VCF) in PGDSpider v.2.1.1.5 (Lischer and Excoffier, 2012). Principal component analysis (PCA) was performed to assess the relatedness and clustering of the sub-genomes and diploid individuals using VCFTOOLS v.0.1.16 (Danecek et al., 2011) and PLINK v.1.90 (Purcell et al., 2007). We further used STRUCTURE v.2.3.4 to infer the number of ancestral genetic clusters (K) and their levels of admixture. The optimal K (set to vary from 1 to 10) was determined with the Delta K (Evanno et al., 2005). The results of the replicates at the ΔK were further processed using CLUMPP 1.1.2 (Jakobsson and Rosenberg, 2007) to assess the membership proportions for clusters identified in STRUCTURE. Furthermore, we calculated gene diversity (π) and population fixation statistics (FST) for each population using VCFTOOLS v.0.1.16.
2.7. Divergence time estimationTo estimate the divergence parameters between the Acanthus spp. (τs and θs), we used a multispecies coalescent (MSC) model (Xu and Yang, 2016) with a fixed species phylogeny. The MSC model was built using the Bayesian Phylogenetics and Phylogeography v.3.2 (BPP) program (Flouri et al., 2018) with the A00 parameters (species delimitation = 0, species tree = 0). Based on the divergence time estimates between the western and eastern lineages of A. ilicifolius in a previous study (Guo et al., 2020), we assigned a gamma prior G (6, 1000) for all θ parameters and τ0 ~ G (7, 200) for the age of the root, while the other divergence time parameters were assigned the uniform Dirichlet prior. The total number of MCMC iterations was run for 50, 000 generations, and we sampled every 10th generation with a burn-in period of 5000 generations. The analysis was run twice to confirm consistency between runs. A mutation rate μ of ~2 × 10−9 per site per year and a generation time of 5 years (He et al., 2022) were used to calculate the absolute divergence time and absolute population sizes following Tiley et al. (2020).
2.8. Environmental niche modelingTo check if the tetraploid lineage occupies an environmental niche that diverges from the diploid lineages, we first characterized their environmental niches and reconstructed their present and past distributions in the IWP region. Briefly, we downloaded occurrence records of Acanthus ilicifolius and A. ebracteatus from the Global Biodiversity Information Facility (accessed on January 2013–May 2023, https://doi.org/10.15468/dl.n5d3qs). Only occurrence records associated with images were retained for further identification based on morphological characters. Occurrence records were grouped as the tetraploid lineage of A. ilicifolius (east of the Thai–Malay Peninsula; Section 3.1), the diploid lineage of A. ilicifolius, and the diploid A. ebracteatus. After combining occurrence records and our sampling records, all records were cleaned for duplicates, points falling on the sea, and then spatially rarefied. A total of 389 points were retained for further analysis (tetraploid A. ilicifolius = 280, diploid A. ilicifolius = 88, diploid A. ebracteatus = 21). Nineteen bioclimatic variables were downloaded at 2.5 arc-minutes resolution from the PaleoClim database (http://www.paleoclim.org/; Brown et al., 2018) for current (averaged over 1979–2013) and two simulated paleoclimatic conditions: the Last Interglacial Maxima (LIG; 130 Kya) and the Last Glacial Maxima (LGM; 21 Kya). The collinearity among variables climate variables was determined based on the variance inflation factor threshold (VIF = 7), and the variables having VIF below this threshold were included in further analysis.
Environmental niches of the three lineages were characterized pairwise based on the centroid shift, overlap, unfilling, and expansion (COUE) framework (Guisan et al., 2014). Principal Component Analysis (PCA) was first conducted on the chosen bioclimatic variables to generate the gridded environmental space. By applying a kernel density function, we converted the occurrence points into smoothed densities of occurrences and plotted them in the environmental space. Observed niche overlap between the lineages was estimated using Schoener's index of niche breadth (D) and statistically evaluated using niche equivalency and similarity tests based on a 95% confidence interval. The niche overlap values were grouped following the classification provided by Rödder and Engler (2011). We further overlapped the environmental spaces of the three lineages and measured the unfilled niche of one lineage (U), the overlapping niche of both lineages (O), and the expanded niche of the other lineage (E). These analyses were performed using the ecospat package v.3.0 (Di Cola et al., 2017) in R.
We developed environmental niche models (ENM) using the spatially rarefied occurrence records and the selected bioclimatic variables, with Köppen–Geiger climate classes as the background. An ensemble model framework of eight algorithms was used. The models were calibrated with 70% of randomly selected data. The remaining 30% of data was used to evaluate individual model performance by estimating the true skill statistic (TSS) and the receiver operating characteristics curve (ROC) area. Models with high predictive accuracy (TSS > 0.7 and ROC > 0.8) were used in the ensemble model framework to identify areas in the IWP that were climatically suitable for the species during the five time periods. The continuous projections were converted to binary for comparative assessment across geological periods using the threshold value that maximized the TSS evaluation score. The ENMs were developed and evaluated using the biomod2 package (Thuiller et al., 2009) in R.
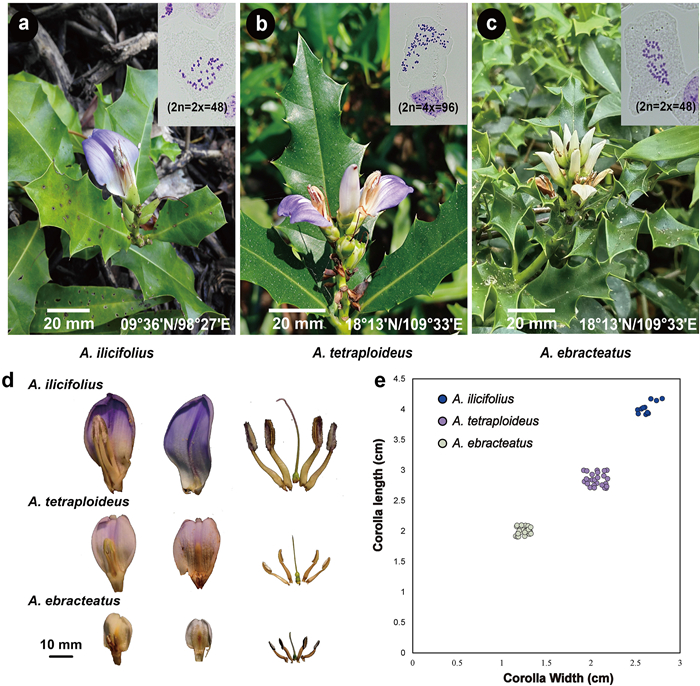
3. Results 3.1. Morphology and cytologyBased on observations of living plants and herbarium specimens, the two lineages of Acanthus ilicifolius distributed along the eastern and western coasts of the TMP showed similar variations in plant height (0.5–2 m), leaf size (13–18 × 3–5 cm), and leaf shape (i.e., elliptic, serrated, or dentate leaves). Nevertheless, differences in color and size of the corolla were observed between the two lineages (Figs. 2 and S1). The plants collected from the western side of the TMP (i.e., India, Sri Lanka, and western Thailand) have larger (3.8–4.2 × 2.5–2.8 cm) and purple or deep purple corollas with a dark median band, whereas those collected from the eastern side of the TMP (i.e., China, east Thailand, east Malesia, Singapore, Indonesia, and Australia) have smaller (2.7–3.0 × 1.9–2.2 cm) and purple or blue corollas with a white to yellow medium band. Flow cytometry (Fig. S2) and karyotype analysis (Fig. 2) revealed different ploidy levels of plant samples collected from the eastern and western sides of the TMP. The western samples (N = 9) have a mean genome size of 917.2 ± 20.2 Mb (mean ± SD) and chromosome number 2n = 2x = 48, whereas the eastern samples (N = 21) are tetraploid with a mean genome size of 1965.3 ± 37.8 Mb and chromosome number 2n = 4x = 96. The morphological characters of A. volubilis (i.e., twining and delicate sprawling stems and un-serrated elliptical leaves) and A. ebracteatus (i.e., white flowers, with corolla size varying from 1.8 to 2 × 1–1.3 cm) were consistent with previous reports (Ragavan et al., 2015). The genome size and chromosome number of A. ebracteatus samples (N = 13) were found to be 899.8 ± 13.84 Mb and 2n = 2x = 48, respectively. The chromosomes number 2n = 48 of the diploid Acanthus species (A. ebracteatus and western A. ilicifolius) was consistent with previous studies (Das et al., 1996; Narayanan, 1951). Based on morphological and cytological evidence and phylogenetic relationships between the eastern and western lineages of A. ilicifolius (Guo et al., 2020), we conclude that the two lineages should be treated as two distinct species. As the holotype of A. ilicifolius was collected from India (Linnaeus, 1753), which belongs to the western lineage, we named the tetraploid eastern lineage Acanthus tetraploideus H. Feng & Y.L. Huang, sp. nov. A key to Acanthus mangrove species in IWP and a description of the new species are provided in section 5 (Taxonomic treatment).
|
| Fig. 2 Morphology and karyotypes of three Acanthus species. Morphology and karyotype analyses of (a) A. ilicifolius, (b) A. tetraploideus and (c) A. ebracteatus; (d) Flower morphological differences between three Acanthus spp.; (e) Corolla length and width of A. ilicifolius (Phuket, Thailand, N = 10), A. ebracteatus (Hainan Island, China, N = 10, Guangxi, China, N = 8, Bangkok, Thailand, N = 10) and A. tetraploideus (Hainan Island, China, N = 9, Guangxi, China, N = 7, Guangdong, China, N = 9). |
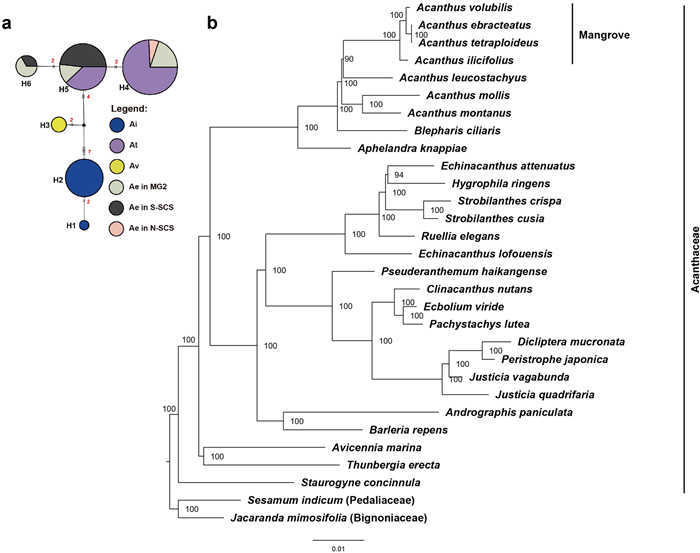
The seven Acanthus species (four true mangrove species and three non-mangrove species) and 21 Acanthaceae species constructed a well-supported phylogenetic tree based on their chloroplast genome information (Fig. 3b). The four mangrove species formed one clade with 100% bootstrap support. The relationship between tetraploid A. tetraploideus and diploid A. ebracteatus was strong within this mangrove clade with 100% bootstrap support. Compared to the European species A. mollis and A. montanus, the Asian species A. leucostachyus was more closely related to the mangrove clade. Among all Acanthaceae species, two species, namely Blepharis ciliaris (L.) B.L. Burtt and Aphelandra knappiae Wassh., were found to be the closest relatives of the Acanthus spp. The concatenated length of two cpDNA loci was 1260 bp, which contained 18 polymorphic sites (Table S1). Among the six cpDNA haplotypes (H1–H6, Fig. 3a), H1 and H2 were exclusive to A. ilicifolius, H3 was exclusive to A. volubilis, and H6 was exclusive to A. ebracteatus. The tetraploid A. tetraploideus and diploid A. ebracteatus shared the H4 and H5 haplotypes.
|
| Fig. 3 Phylogenetic analysis based on chloroplast DNA data. (a) Haplotype networks of chloroplast loci of four Acanthus species—A. ilicifolius (Ai, N = 172), A. ebracteatus (Ae, N = 101), A. tetraploideus (At, N = 205) and A. volubilis (Av, N = 15). Mutation steps (> 1) are shown on the line, indels with more than one nucleotide were considered one mutation, and node size is proportional to the number of haplotypes. The small black circle indicates the unobserved putative haplotype; (b) Phylogenetic tree of Acanthaceae species based on complete chloroplast genome data. Bootstrap values are labeled next to the node. |
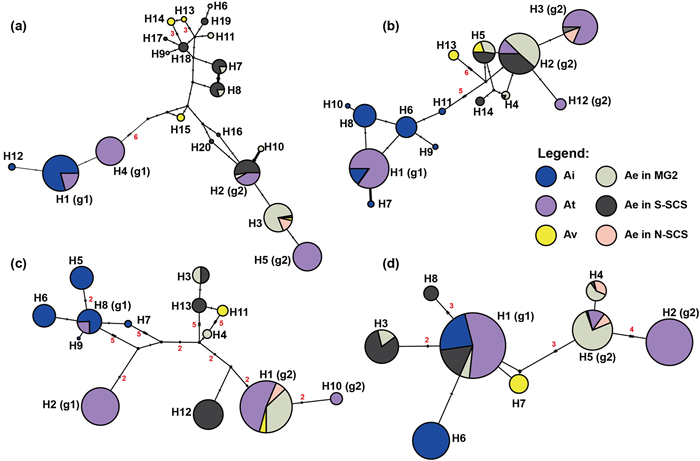
The alignment length of eight nDNA markers varied from 159 to 502 bp (with a concatenated length of 2421 bp), and polymorphic sites from 6 to 26 bp (with a concatenated length of 130 bp; Table S1). The maximum number of nDNA haplotypes was found in locus C48198 (N = 20) (Figs. 4 and S3). The haplotype networks revealed different patterns between the Acanthus spp. For most loci, dominant haplotypes were different between A. ebracteatus and A. ilicifolius, especially in loci C48198, C51040, and C72860, with at least 11, 6, and 10 mutation steps (Fig. 4a–c), respectively. Only the H1 haplotype of the C70897 locus was observed in both A. ebracteatus (mainly in the S-SCS region) and A. ilicifolius (Fig. 4d). For A. volubilis, the haplotypes of seven out of eight loci (except 70897) were mixed. For example, the H13 haplotype was mixed with H5 in C51040 and H15 with H14 or H13 in C48198. For locus C70897, the A. volubilis population had a unique haplotype H7.
|
| Fig. 4 Phylogenetic analysis based on nuclear DNA data. Haplotype networks of four nuclear loci of four Acanthus species—A. ilicifolius (Ai, N = 172), A. ebracteatus (Ae, N = 101), A. tetraploideus (At, N = 205) and A. volubilis (Av, N = 15): (a) C48198, (b) C51040, (c) C72860 and (d) C70897. Mutation steps (> 1) are shown on the line, indels with more than one nucleotide were considered one mutation, and node size is proportional to the number of haplotypes. The small black circle indicates the unobserved putative haplotype. |
In Acanthus tetraploideus, the haplotypes were mixed for all eight loci, with the haplotypes being close and/or shared with either A. ebracteatus or A. ilicifolius. For example, in locus C48198, the H1 and H4 haplotypes were close to A. ilicifolius, whereas H2 and H5 were close to A. ebracteatus. The unique haplotype in locus C72860 was seven and five mutations apart from A. ilicifolius and A. ebracteatus, respectively. The tetraploid A. tetraploideus showed a hybrid speciation history based on the relationship among different haplotypes. One ancestral progenitor (sub-genome1, g1) was inferred as closely related to A. ilicifolius, whereas the other progenitor (sub-genome2, g2) was closely related to A. ebracteatus (Figs. 4 and S3).
The ML tree based on 40 concatenated sequences of the two tetraploid sub-genomes and diploid Acanthus ilicifolius genomes showed two distinct clusters, with each cluster further divided into two (Ai and At-g1) and four (At-g2, Ae1, Ae2, and Av) clades respectively, where Ai = diploid A. ilicifolius, Ae = diploid A. ebracteatus, and At = tetraploid A. tetraploideus (Fig. 5a). In the first cluster, the Ai clade included A. ilicifolius accessions from the NIO and MG1 regions, while the At-g1 clade included A. tetraploideus sub-genome g1. In the second cluster, the sub-genome g2 of A. tetraploideus formed a monophyletic clade with 98% support and was sister to two A. ebracteatus accessions from the MG2 region. The A. ebracteatus accessions formed two clades—the Ae1 clade contained accessions from MG2, N-SCS (QLG) and two S-SCS (KC) accessions, and clustered with At-g2 with 90% bootstrap support; the other accessions of A. ebracteatus from MG2 (TP, SJW, SBW) and S-SCS regions formed the Ae2 clade. Two genotypes of A. volubilis were identified, which formed the Av clade but with low support.
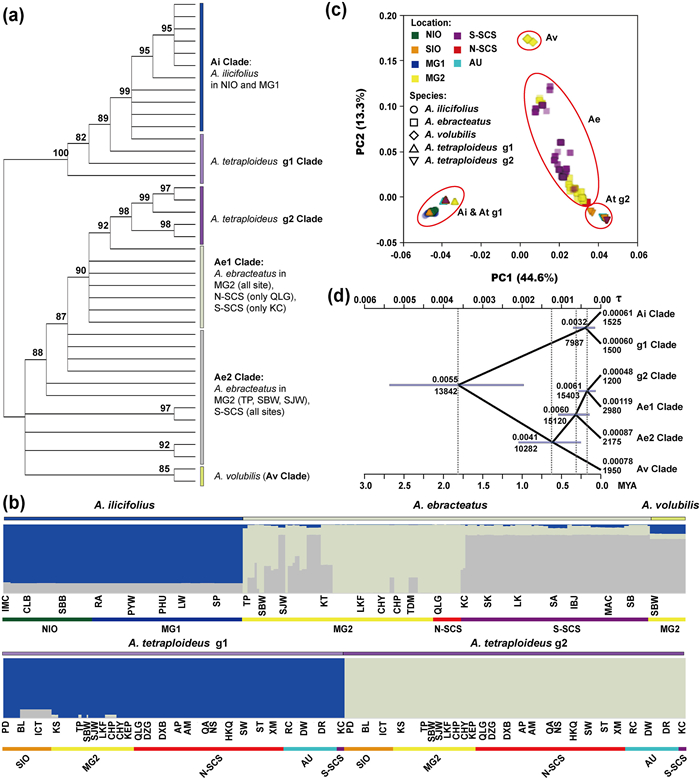
|
| Fig. 5 Genetic diversity and population structure analysis based on eight nuclear loci data. (a) Maximum Likelihood (ML) tree, (b) STRUCTURE analysis, and (c) principal component analysis based on eight nuclear loci data; (d) estimated divergence time with 95% confidence intervals. The upper and lower scale axes represent the MSC branch and absolute time, respectively. The upper and lower node values represent the MSC ancestral population size and absolute size. Four Acanthus species are represented: A. ilicifolius (Ai), A. ebracteatus (Ae), A. tetraploideus (At), and A. volubilis (Av). |
Population structure analysis revealed the optimal cluster number as three (Δ K = 3), and the three ancestry components (ac1–ac3) (Fig. 5b). For A. tetraploideus, the major ancestry components were ac1 and ac2 in g1 and g2, respectively, while the ac3 ancestry component was also present in g1 (BL and ICT accessions). For Acanthus ilicifolius, ac1, admixed with ac3, forming the major ancestry component in all accessions. For A. ebracteatus, three population-structure patterns emerged: (1) N-SCS and most of the MG2 accession showed a similar pattern to A. tetraploideus g2, with ac2 as the major ancestry component; (2) the S-SCS and part of the SJW accession had ac3 as the major ancestry component, with the occasional presence of the ac1 and ac2 ancestry components; and (3) accessions in TP, SBW, SJW, KT, TDM showed admixture of the ac2 and ac3 components. For A. volubilis, all accessions showed a similar pattern to the S-SCS accessions of A. ebracteatus but with more ac1 ancestry components.
PCA showed a population grouping pattern (Fig. 5c) consistent with STRUCTURE analysis. The first two components cumulatively explained 57.9% of the variation and identified five clusters. The Ai & As-g1 cluster comprised all A. ilicifolius and A. tetraploideus g1 accessions. For A. ebracteatus, individuals from MG2 and N-SCS accessions were more closely related to A. tetraploideus g2 than the S-SCS individuals, similar to population structure analysis. However, individuals from the S-SCS accession were separated into two groups. Most individuals were close to the MG2 group, but several individuals from the IBJ, SA, and MAC were clustered with the SJW and SBW individuals to form a separate group. Notably, the A. volubilis accessions were clustered as a separate group. Since the 16 populations of A. ebracteatus showed a mixed population structure between S-SCS and MG2 regions, we further classified these populations into three groups: AeG1 (TP, SJW, SBW), AeG2 (KT, LKF, CHY, CHP, TDM, and QLG) and AeG3 (All S-SCS populations).
Of the four Acanthus mangrove species, A. volubilis had the highest genetic diversity (π = 8.17 × 10−3), followed by A. ebracteatus (π = 6.07 × 10−3) and A. ilicifolius (π = 1.72 × 10−3). Sub-genome 2 (π = 1.53 × 10−3) of A. tetraploideus had higher genetic diversity than sub-genome 1 (π = 1.01 × 10−3) (Table 2). The highest genetic differentiation was found between A. ilicifolius and A. tetraploideus sub-genome2 (FST = 0.83), whereas the lowest FST value was observed between A. ebracteatus vs. A. tetraploideus sub-genome2 (FST = 0.26). Among the three groups of A. ebracteatus populations (AeG1, AeG2, and AeG3), the highest genetic diversity was observed in AeG1 (π = 6.12 × 10−3), followed by AeG3 (π = 5.94 × 10−3) and AeG2 (π = 1.10 × 10−3) The highest genetic differentiation was observed between AeG2 and AeG3 (FST = 0.30). Sub-genome 2 of A. tetraploideus showed the lowest genetic differentiation from AeG2 (FST = 0.36). Populations of A. volubilis showed the highest genetic divergence from AeG2 (FST = 0.71), followed by AeG1 (FST = 0.40) and AeG3 (FST = 0.38).
| Populations | FST between genetic groups | Pi (10−3) | |||||||
| Ae | AeG1 | AeG2 | AeG3 | At_g2 | At_g1 | Ai | Av | ||
| Ae | 0.00 | – | – | – | 0.26 | 0.50 | 0.52 | 0.40 | 6.07 |
| AeG1 | – | 0.00 | 0.25 | 0.15 | 0.50 | 0.71 | 0.69 | 0.40 | 6.12 |
| AeG2 | – | 0.25 | 0.00 | 0.30 | 0.36 | 0.77 | 0.79 | 0.71 | 1.10 |
| AeG3 | – | 0.15 | 0.30 | 0.00 | 0.45 | 0.56 | 0.56 | 0.38 | 5.95 |
| At_g2 | 0.26 | 0.50 | 0.36 | 0.45 | 0.00 | 0.79 | 0.83 | 0.80 | 1.53 |
| At_g1 | 0.50 | 0.71 | 0.77 | 0.56 | 0.79 | 0.00 | 0.48 | 0.76 | 1.01 |
| Ai | 0.52 | 0.69 | 0.79 | 0.56 | 0.83 | 0.48 | 0.00 | 0.74 | 1.72 |
| Av | 0.40 | 0.40 | 0.71 | 0.38 | 0.80 | 0.76 | 0.74 | 0.00 | 8.17 |
| Acanthus | – | – | – | – | – | – | – | – | 13.08 |
| Abbreviations: Ae: A. ebracteatus, At: A. tetraploideus, Ai: A. ilicifolius, Av: A. volubilis. The populations are identified in the text. FST: population fixation statistics; Pi: Gene diversity. | |||||||||
In the MSC model, we used the clade structure of the ML tree (((Ae2, (Ae1, g2)), Av), (Ai, g1)) (Fig. 5a) as a guide. Bottlenecks were observed for each clade, from ancestry node 7987–15, 403 to ca. 1200–2980 (Fig. 5d). A. ilicifolius diverged from other diploid species c. 1.8 MYA (95% confidence interval, CI: 0.98–2.69 MYA). A. volubilis diverged from A. ebracteatus and A. tetraploideus g2 clade at 0.61 MYA (0.25–1.05 MYA). A. tetraploideus g1 diverged from A. ilicifolius at 0.18 MYA (0.07–0.29 MYA), whereas A. tetraploideus g2 diverged from A. ebracteatus at 0.20 MYA (0.07–0.35 MYA).
3.4. Environmental nichesIn the environmental space, a low amount of niche overlap was found between the tetraploid and the two diploid species (D = 0.318 for Acanthus ilicifolius and D = 0.285 for A. ebracteatus), whereas a moderate degree of niche overlap was observed between the two diploid species (D = 0.438). The niche equivalency test showed that the climatic niches of the three Acanthus mangrove species were not equivalent (p > 0.05) (Fig. S4). Significant divergence of the climatic niches of the two diploid Acanthus species was observed (p = 0.057); however, the climatic niche of the tetraploid A. tetraploideus was significantly conserved with both Acanthus parental species (p = 0.001 for A. ilicifolius and p = 0.005 for A. ebracteatus). The COUE estimates also revealed high niche stability values between the tetraploid and the two diploid species (Table S5), indicating that the tetraploid A. tetraploideus has occupied most of the climate niches of A. ilicifolius and A. ebracteatus. However, a considerable amount of niche unfilling between the tetraploid and diploid species (U = 0.57 for A. ilicifolius and U = 0.49 for A. ebracteatus) also indicated that A. tetraploideus had occupied some unique climate niche outside the climatic envelopes of the two diploid Acanthus species.
In the geographic space, the ensemble projections of the three Acanthus mangrove species (A. tetraploideus, A. ilicifolius, and A. ebracteatus) also revealed that the tetraploid species could occupy 3.28% of the IWP region, higher than the diploid species (1.68% for A. ilicifolius and 0.84% for A. ebracteatus) (Fig. 6a). Overlay analysis identified that 2.64% of the IWP area was climatically suitable only for the tetraploid A. tetraploideus. Most of these suitable areas are distributed along the coast of southern China, Vietnam, Laos, eastern Thailand, parts of Indonesia and Malaysia, northern Philippines, southern Papua New Guinea, and northern Australia (Fig. 6b).
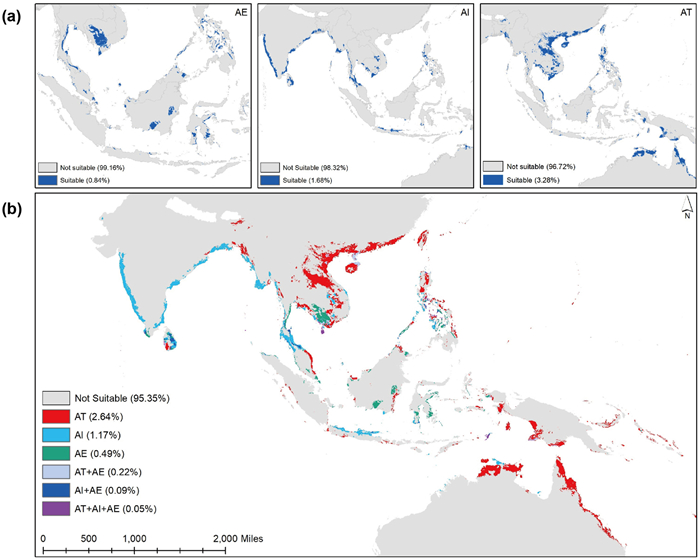
|
| Fig. 6 Potential distribution of three Acanthus species (AI: Acanthus ilicifolius, AE: A. ebracteatus, and AT: A. tetraploideus) in the Indo–West Pacific region: (a) three species individually, and (b) overlay map of the potential distributions of the three species. |
Projections across geological time revealed that climatically suitable habitat decreased from the LIG (0.58%) to the LGM (0.19%) (Fig. 7). We observed no overlap during the LIG between the climatically suitable areas of the three species, with A. ilicifolius having the maximum suitable areas (0.56%) and A. tetraploideus having the minimum (0.002%) (Fig. 7a). During the LGM, climatically suitable areas of both A. tetraploideus (0.08%) and A. ebracteatus (0.04%) increased, along with their overlapped distribution (0.07%) in the Sulawesi region (Fig. 7b).

|
| Fig. 7 Potential distribution of three Acanthus species (AI: A. ilicifolius, AE: A. ebracteatus, and AT: A. tetraploideus) in two paleoclimate conditions (a) during the Last Interglacial Maxima (LIG; 130 Kya) and (b) during the Last Glacial Maxima (LGM; 21 Kya). |
The species limits of Acanthus ilicifolius in the IWP region has long been ambiguous. The TMP has fragmented the geographic distribution of the species. Morphological, cytological, and molecular divergences have been reported between A. ilicifolius samples collected from the eastern and western sides of the TMP (Das et al., 1996; Suan, 1996; Duke and Schmitt, 2015; Ragavan et al., 2015). However, due to small morphological differences (i.e., corolla characters) and sparse sampling in these studies, the eastern and western lineages of A. ilicifolius have been treated as one species since 1753. The identity of the new Acanthus species in the IWP region, e.g., A. albus in India (Debnath et al., 2013) and A. xiamenensis (Zhang, 1985) in China, were also challenged due to their small morphological differences with A. ilicifolius (Ragavan et al., 2015). The present study adopted an integrated approach, including morphological, cytological, and molecular data to resolve the taxonomic ambiguity of the three true mangrove species of Acanthus, particularly focusing on A. ilicifolius. Our findings suggest that previously identified A. ilicifolius species distributed along the eastern side of the TMP should be regarded as a new species. The morphological (i.e., corolla color and size) and cytological (i.e., differences in chromosome number) differences observed between the eastern and western lineages of this species support previous observations (e.g., Backer and Bakhuizen, 1965; Barker, 1986; Suan, 1996; Ragavan et al., 2015). Moreover, molecular evidence based on two chloroplast and eight nuclear loci data indicate that all individuals (N = 205) from 26 sites of the eastern TMP were nearly identical (Figs. 3a and 5a–c), which might have originated from hybridization between A. ilicifolius distributed along the western coast of the TMP and A. ebracteatus. Based on these findings and the allotetraploid nature (2n = 4x = 96) of the lineage distributed on the eastern side of the TMP, we consider it a new species and have named it A. tetraploideus H. Feng & Y.L. Huang, sp. nov.
4.2. Origin of Acanthus tetraploideusMorphological analysis revealed that Acanthus tetraploideus has an intermediate or mixed phenotype between A. ilicifolius and A. ebracteatus. The flower size of A. tetraploideus is smaller than A. ilicifolius but larger than A. ebracteatus. The white to yellow colored median band in the corolla of A. tetraploideus is similar to A. ebracteatus, while the blue, purple, or light purple colored corolla is similar to A. ilicifolius. Morphological evidence indicated that A. tetraploideus was formed by hybridization between A. ilicifolius and A. ebracteatus. This "intermediate" or "mixed" phenotype has been reported in hybrid-derived speciation processes for several non-mangroves [e.g., Arachis spp. (Hancock et al., 2019), Trape spp. (Lu et al., 2022), Petrocosmea spp. (Qiu et al., 2011)] and mangrove species [e.g., Rhizophora spp., Bruguiera spp., and Sonneratia spp.; Ragavan et al., 2017].
Karyotype analysis revealed that the chromosome number of Acanthus tetraploideus was two times larger than that of the diploid A. ilicifolius and A. ebracteatus. The combined morphological and cytological evidence suggest that A. tetraploideus probably originated from hybridization between A. ilicifolius and A. ebracteatus, followed by chromosome doubling. Therefore, A. tetraploideus can be identified as an allopolyploid species. This is the first evidence of allopolyploidy among all true mangrove species, which can act as an important genetic model for understanding polyploid evolution under marine and estuarine tidal conditions.
An additional candidate progenitor for Acanthus tetraploideus is A. volubilis. A. volubilis is distributed in the IWP region and has the same ploidy level as A. ilicifolius and A. ebracteatus (Das et al., 1996). However, we were unable to investigate the morphological characteristics of A. volubilis due to its very restricted distribution in the IWP. Nevertheless, molecular evidence obtained from analyzing eight nuclear genes and complete chloroplast genomes of the Acanthus species further strengthened the hybrid speciation history of A. tetraploideus. A weak phylogenetic relationship with A. tetraploideus and low bootstrap support of the clade in the phylogenetic tree indicated that A. volubilis might not be one of the progenitors of the tetraploid species.
nDNA haplotype networks and phylogenetic analysis indicated that tetraploid species had two sub-genomes of ancestral progenitors—one closely related to Acanthus ilicifolius and another to A. ebracteatus. Phylogenetic trees generated from the chloroplast DNA revealed that among the two progenitors A. tetraploideus is more closely related to A. ebracteatus. The findings from the nuclear (inherited from parental) and chloroplast (inherited from maternal) DNA analyses indicated that A. ilicifolius and A. ebracteatus might be the paternal and maternal parents of A. tetraploideus, respectively. Whole genome sequencing technology allows a genome-wide method to reconstruct the origin of a polyploid species (Session and Rokhsar, 2013). However, reconstructing the origin of a polyploid species is particularly challenging when using scant loci, as each copy of the target locus (or set of loci) present in a polyploid sample need to be sequenced, and the distinct homologs need to be accurately reconstructed (Rothfels, 2021). Therefore, in this study, we used a phylogenetic-based method to reconstruct the origin of A. tetraploideus. Although used by other studies (Han et al., 2022; Lin et al., 2022), this method may lead to incorrect phasing. Therefore, caution should be exercised when interpreting our results, and we suggest future studies use a genome-wide method to verify the result.
Out of the 30 mangrove genera of the world, natural hybrids have been reported in the following genera: Rhizophora, Bruguiera, Ceriops, Lumnitzera, Sonneratia, Avicennia, and Acrostichum. Most of the 17 hybrids reported from these genera are limited to the F1 generation; therefore, the mangrove hybrids are often considered evolutionary dead-ends (Ragavan et al., 2017). A. tetraploideus is the only hybrid-derived species that reproduces by polyploidization. In addition, A. tetraploideus can reproduce through vegetative growth compared to other mangrove hybrids. Clonal reproduction may facilitate polyploid evolution by ameliorating the minority cytotype disadvantage and enhancing polyploid establishment ability through recruitment and spatial expansion of rare polyploids, especially in the absence of sexual reproduction (Baldwin et al., 2013). Therefore, it is possible that clonality in A. tetraploideus might help the populations establish at their early stage of formation.
4.3. Population structure and demographyPopulation structure analysis also supported the hybrid speciation history of Acanthus tetraploideus. STRUCTURE and PCA revealed that the two sub-genomes of the tetraploid species were clustered with two different progenitors with strong genetic differentiation. Moreover, we observed a strong population structure of A. ebracteatus populations in the IWP region, particularly in the S-SCS and MG2 regions. Among these population groups, the lowest genetic differentiation was observed between sub-genome 2 of the tetraploid species and the A. ebracteatus populations distributed along the eastern coast of the TMP (MG2) and one population from southern China (S-SCS). These findings indicate that the progenitors of the sub-genome 2 of A. tetraploideus might belong to these populations.
The geographic locations of the majority of these progenitor populations were found along the eastern (Acanthus ebracteatus) and western (A. ilicifolius) coasts of the TMP, indicating the possible role of this landmass in the hybrid speciation history of A. tetraploideus. The MSC model indicated that the two sub-genomes diverged from their respective progenitors during the Pleistocene epoch, and the confidence intervals of the divergence times covered the Last Interglacial Maxima (LIG). The epoch is characterized by glacial sea-level changes and repeated emergence and submergence of the landmasses, including the TMP, also known as the Sunda Shelf (Guo et al., 2018). These climatic and geological changes in the IWP region have shaped the current phylogeographic patterns of many mangroves (e.g., Sonneratia alba; Yang et al., 2017) and other marine species (Gaither et al., 2011). While previous studies have identified the Sunda Shelf as causing glacial vicariance of mangroves in the IWP [e.g., in Aegiceras corniculatum L.; Banerjee et al., 2022a], our study provided the first evidence of its possible role in facilitating hybridization and formation of an allopolyploid mangrove species.
The confidence intervals of the divergence times of the tetraploid sub-genomes from their respective progenitors covered the Last Interglacial Maxima (LIG) period, during which sea levels were 6 to 9 m higher than present (Hoffman et al., 2017). ENMs indicated that during the LIG and the subsequent LGM, climatically suitable habitats for the tetraploid and its diploid progenitors were restricted to certain glacial refugia distributed along the equators, similar to other coastal species, including mangroves (Banerjee et al., 2022b), from where postglacial recolonization occurred (Hodel et al., 2016). It is possible that further evolution of the tetraploid species occurred in these glacial refugia. Previous studies based on phylogenetic and spatially explicit approaches have identified the co-occurrence of several evolutionary processes and climate refugia (e.g., Smyčka et al., 2017; Garcia et al., 2022). Our ENM models further revealed that the tetraploid species could occupy larger areas than its diploid progenitors, and the pattern became more evident from the LIG to the LGM to the current climate conditions. These findings align with previous studies that suggested evolutionary processes confer distinct advantages to the polyploids that enable them to adapt more easily to changing environments and thrive in environments that pose challenges to their diploid progenitors (Madlung, 2013).
The ability of the tetraploid species to occupy larger and novel areas is also evident from its climatic niche expansion. Our analysis of the COUE framework along the two environmental gradients revealed that although the climatic niche of the tetraploid is not significantly divergent from that of its diploid progenitors, they are not equivalent, as the centroids of the climatic niche densities differ between them. The tetraploid species occupies nearly the entire niche space of the diploids and has expanded into novel environments. Our findings are consistent with previous studies that suggested climatic niche expansion, and not divergence, along with a shift in niche densities, can explain the co-occurrence of tetraploid and diploid species [e.g., in Arabidopsis arenosa L.; Molina-Henao et al., 2019)]. However, significant niche divergence was observed between the two diploid progenitors with moderate climatic niche overlap, thereby contradicting studies that reported that polyploids are more differentiated from their progenitors than the progenitors are from each other (Baniaga et al., 2020). This finding indicates that A. ilicifolius and A. ebracteatus can occupy non-equivalent and significantly divergent climatic spaces in the IWP region, and the tetraploid species originating from these progenitors can occupy even larger and novel areas.
However, the wider niche breadth and large distribution of a tetraploid species are often competitive with its parents (Baniaga et al., 2020). For Acanthus, A. tetraploideus is geographically isolated from A. ilicifolius but has overlapping distribution with A. ebracteatus. Compared to the wide distribution of A. tetraploideus, which ranges from the South China Sea (SCS) coast to the southwestern Pacific Ocean, A. ebracteatus is restricted to the eastern TMP and SCS regions. Although the IUCN Red data list identifies all Acanthus species as Least Concern (LC) (https://www.iucnredlist.org/), the populations of A. ebracteatus are declining in China, especially in areas where its distribution overlaps with that of A. tetraploideus. Indeed, the China Biodiversity Red List treated A. ebracteatus as Near Threatened (NT) (https://www.plantplus.cn/rep/protlist). We recommend priority conservation actions for A. ebracteatus populations, especially in the regions where the geographic distribution of A. ebracteatus overlaps with that of A. tetraploideus.
5. Taxonomic treatment 5.1. Key to Acanthus mangrove species in IWP1. Bracteoles present, corolla purple or deep purple, 2.5–4.5 cm ………………………………………………………………………….2
1. Bracteoles absent or minute, corolla white, 1.5–2.2 cm long…………………………………………….………………………3
2. Corolla with dark median band from top to bottom, 3.8–4.2 cm long……………………………………………………….. A. ilicifolius
2. Corolla with white to yellow median band, 2.7–3.0 cm long…………………………………………………... A. tetraploideus
3. Stem twining, stem axial spines absent, leaves elliptical, un-serrated……………………………………………………. A. volubilis
3. Stems erect or prostrate, stem axial spines present, leaf tip acute to obtuse with spiny edge, corolla with yellow median band.………… ………………………………….……. A. ebracteatus
5.2. Description of the new speciesAcanthus tetraploideus H. Feng & Y.L. Huang, sp. nov. (Fig. 8). 居中老鼠簕
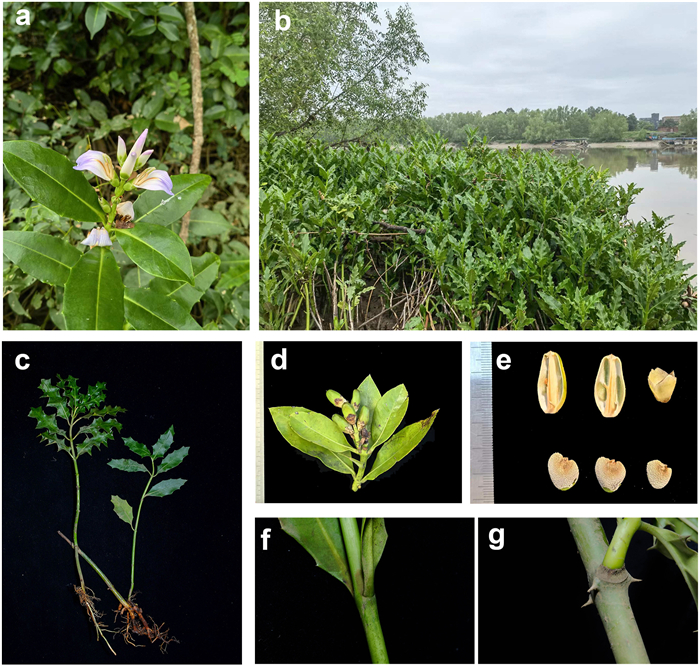
|
| Fig. 8 Morphology of Acanthus tetraploideus. (a) flower, (b) habitat, (c) variation in leaf shape, (d) fruit, (e) seed, stem spine absent (f) or present (g). |
urn: lsid: ipni.org: names: 77316490-1
Type. China. Hainan: Wenchang County, Tou-yuan village, 19.55°N, 110.58°E, landward edges of the mangroves, 10 Oct 2021, Hui Feng FH013 (holotype: SYS; isotype: PE, IBSC)
Diagnosis. Acanthus tetraploideus resembles A. ilicifolius in leaf shape and light to dark blue or violet flowers, but differ in having smaller (2.7–3.0 × 1.9–2.2 cm vs. 3.8–4.2 × 2.5–2.8 cm) corollas with white to yellow medium band (vs. dark purple band). A. tetraploideus is tetraploid (2n = 4x = 96), while A. ilicifolius is diploid (2n = 48). The geographic distribution of the two species is separated by the Thai–Malay Peninsula. A. tetraploideus is found in South China Sea, South Indian Ocean, and West Pacific Ocean, while A. ilicifolius is found around Andaman Sea, India, and Sri Lanka.
Description. Shrub: height up to 2 m. Stem: thick, green, light green, sparsely branched and stem axial spine either present or absent. Roots: occasionally above ground or prop roots on lower parts of reclining stem. Leaves: simple, opposite, lanceolate to broadly lanceolate, margin either entire or spiny and dentate, leaf base attenuate, leaf tip acute and narrowly pointed with or without spiny edge, Raceme: loose bearing to 16 pairs of flowers. Mature flower bud: ellipsoidal, 2.5–3 cm long; bract single, 6.5 × 6.5 mm; bracteoles 2, lateral, 4 × 2 mm; calyx four lobed, outer two lager in size, 1 × 0.8 cm, enclosing the flower bud, inner lateral lobes narrow, 1 by 0.5–0.8 cm, enclosed by upper and lower lobes. Corolla: blue, purple, or light purple. 2.5–3.5 cm × 1–2 cm, with white or yellow medium band; stamens 4, 1.5–2 cm long, sub equal with thick hairy connectives; anther green with maroon pigmentation, 6–7 × 1–1.5 mm, style 16 × 1 mm, Seeds 0.85–1.25 × 0.65–0.95 cm, 0.1–0.3 cm thick.
Phenology. Flowering and fruiting occur throughout the year
Distribution. Acanthus tetraploideus is wide-spread, occurring in China, Vietnam, Cambodia, east Thai–Malay Peninsula, Philippines, Indonesia, Papua New Guinea, Australia, and Vanuatu, commonly growing in landward edges of the mangroves just above the high tide mark, but also occurring in inner mangroves as the understory.
Additional specimens examined. CHINA: Fujian: Longhai, 6 May 2005, X.L. Hou & W.Q. Wang 9294 (AU); Yunxiao, May 1964, Z.X. Lin 0055 (AU), 25 June 2018, Y.M. Rao 1903 (AU), 2 May 2019, X.F. Zeng, ZXF47224 (AU), 12 July 2014, Tong et al. TONG-C-0030 (IGA); Dongshan, 22 March 1995, X.T. Zhang 3693 (AU). Guangdong: Dongguan, 14 July 2009, Q.R. Liu DJ20090714009 (BNU); Guangzhou: May 1951, H.D. Zhang 4751 (IBSC); Shenzhen, 21 April 2005, S.Z. Zhang & L.Q. Li 0460 (PE), 24 May 2006, Y.F. Deng 18765 (ISBC); Taishan, 8 May 1981, Z.X. Li 0528 (ISBC); Zhanjiang, 23 March 1952, X.Y. Hou 15058 (PE); Guangxi: Fangchen, 29 July 1937, X.R. Liang 69910(IBSC); Hepu, 26 May 1937, X.R. Liang 69324 (IBSC), 10 May 2011, Y.S. Huang & C.R. Lin yo252(IBK); Qinzhou, 2 April 2010, W.H. Wu W0037 (IBK). Hainan: Chengmai, 2 April 1984, Z.X. Li 880(IBSC); Haikou, 3 October 1983, Y.Z. Gao 492(ISBC); Qiongshan, 3 June 1981, P. Lin 8044 (AU); Qiongzhou, 19 April 1987, C.S. Wang 427 (PE); Wenchang, 26 September 1995, R.J. Wang 032(ISBC). Hong Kong, 16 April 1917, Y. Jiang 169 (ISBC), 17 August 1929, H.Y. Chen 259. VIETNAM: Hanoi: 21 January 1965, Sino-Vietnamese Expedition Team 2432 (PE). INDONESIA: Padang: 3 April 1994, D. Sureni 12 (ANDN), October 1999, Y. Mawardi 174 (ANDN); Lesser Sunda Islands: 2 March 1993, Kee. G. Sari Ros, I 41 (L); Seram Island: 11 October 1981, I. Muditha & Maskuri 221 (BO); Java: 27 September 1951, J. van Borssum 711 (BO). PAPUA NEW GUINEA: Lababia Island: 22 May 2008, B.J. Conn et al., 5252 (NSW); New Britain: 13 August 1954, A. Floyd 6542 (NL); Daru Island: 27 February 1936, L. Brass 6210 (NL). NEW CALEDONIA: 27 September 1925, A.U. Däniker 2192 (Z). AUSTRALIA: Queensland, 7 September 1981, I. Kukkonen 10824 (NL); East Arnhem: 3 August 1948, R.L. Specht 807 (US). VANUATU: Malekula, 12 October 1971, N. Hallé 6428 (NL).
AcknowledgmentsThis work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 32160051, 42076117, and 41776166), Guangdong Basic and Applied Basic Research Foundation (Grant Nos. 2022A1515012015, 2023A1515012772), and the Foreign Cultural and Educational Experts Project of the Ministry of Science and Technology (No. QNJ2021162001L). We acknowledge the assistance of the following people in fieldwork: Drs. Suhua Shi and Renchao Zhou (Sun Yat-sen University, China), Dr. Sining He (Southern University of Science and Technology, China), Dr. Haidan Wu (Northeast Normal University, China), Dr. Weixi Li (the University of Hong Kong, China), Mr. Xiaoyang Yang (South China Botanical Garden, China), Dr. Sonjai Havanond (Department of National Parks, Wildlife and Plant Conservation, Thailand), Mr. Chanop Jaengjai (Department of Marine and Coastal Resources, Thailand), Mr. Koh Kwan Siong (National Biodiversity Centre, Singapore), Ms. Fan Yang and Mr. Yuanhan Li (National University of Singapore, Singapore), and Dr. Abhishek Mukherjee (Indian Statistical Institute, India).
Data availability statement
The haplotype sequences of the eight nuclear and two chloroplast loci in this study have been deposited in GenBank under the accession numbers OR449523–OR449616. The complete chloroplast genome has been submitted to NCBI under accession numbers OR454362 for Acanthus tetraploideus, OR454360 for A. ilicifolius, OR454359 for A. ebracteatus, OR454363 for A. volubilis and OR454361 for A. montanus.
CRediT authorship contribution statement
Hui Feng: Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation. Achyut Kumar Banerjee: Writing – review & editing, Visualization, Methodology, Investigation, Formal analysis. Wuxia Guo: Writing – original draft, Validation, Software, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation. Yang Yuan: Validation, Software, Methodology, Investigation, Formal analysis, Data curation. Fuyuan Duan: Software, Methodology, Investigation, Formal analysis. Wei Lun Ng: Writing – review & editing, Resources, Investigation. Xuming Zhao: Resources, Methodology. Yuting Liu: Methodology, Formal analysis, Data curation. Chunmei Li: Visualization, Validation, Investigation, Data curation. Ying Liu: Writing – review & editing, Validation, Methodology, Investigation, Data curation. Linfeng Li: Writing – review & editing. Yelin Huang: Writing – review & editing, Validation, Supervision, Project administration, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors have no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.04.007.
Abbott, R., Albach, D., Ansell, S., et al., 2013. Hybridization and speciation. J. Evol. Biol., 26: 229-246. DOI:10.1111/j.1420-9101.2012.02599.x |
Backer, C., Bakhuizen, ven. den B.R., 1965. Acanthaceae. In: Flora of Java. The Netherlands: P. Noordhof, Groningen, pp. 44-593.
|
Baldwin, S.J., Husband, B.C., 2013. The association between polyploidy and clonal reproduction in diploid and tetraploid Chamerion angustifolium. Mol. Ecol., 22: 1806-1819. DOI:10.1111/mec.12217 |
Bandelt, H.J., Forster, P., Röhl, A., 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol., 16: 37-48. DOI:10.1093/oxfordjournals.molbev.a026036 |
Banerjee, A.K., Feng, H., Lin, Y., et al., 2022a. Phylogeographic pattern of a cryptoviviparous mangrove, Aegiceras corniculatum, in the Indo-West Pacific, provides insights for conservation actions. Planta, 255: 7. DOI:10.1007/978-3-030-93631-0_2 |
Banerjee, A.K., Feng, H., Lin, Y., et al., 2022b. Setting the priorities straight-Species distribution models assist to prioritize conservation targets for the mangroves. Sci. Total Environ., 806: 150937. DOI:10.1016/j.scitotenv.2021.150937 |
Baniaga, A.E., Marx, H.E., Arrigo, N., et al., 2020. Polyploid plants have faster rates of multivariate niche differentiation than their diploid relatives. Ecol. Lett., 23: 68-78. DOI:10.1111/ele.13402 |
Bao, Z., Li, C., Li, G., et al., 2022. Genome architecture and tetrasomic inheritance of autotetraploid potato. Mol. Plant, 15: 1211-1226. DOI:10.1016/j.molp.2022.06.009 |
Barbier, E.B., Hacker, S.D., Kennedy, C., et al., 2011. The value of estuarine and coastal ecosystem services. Ecol. Monogr., 81: 169-193. DOI:10.1890/10-1510.1 |
Barker, M.S., Arrigo, N., Baniaga, A.E., et al., 2016. On the relative abundance of autopolyploids and allopolyploids. New Phytol., 210: 391-398. DOI:10.1111/nph.13698 |
Barker, R.M., 1986. A taxonomic revision of Australian Acanthaceae. J. Adel. Bot. Gard., 9: 1-286. |
Bi, G., Mao, Y., Xing, Q., et al., 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics, 110: 18-22. DOI:10.1016/j.ygeno.2017.08.001 |
Brown, J.L., Hill, D.J., Dolan, A.M., et al., 2018. PaleoClim, high spatial resolution paleoclimate surfaces for global land areas. Sci. Data, 5: 1-9. DOI:10.4324/9780429429057-1 |
Casazza, G., Boucher, F.C., Minuto, L., et al., 2017. Do floral and niche shifts favour the establishment and persistence of newly arisen polyploids? A case study in an Alpine primrose. Ann. Bot., 119: 81-93. DOI:10.1093/aob/mcw221 |
Chatterjee, T., Chakrabarty, S., Roy, S., et al., 2023. Spatial and temporal variation of floral visitors and their visitation pattern on Acanthus ilicifolius L.: a case study from the mangrove ecosystem of Indian Sundarbans. J. Insect Conserv., 27: 493-503. DOI:10.1007/s10841-023-00472-0 |
Chen, H., Zeng, Y., Yang, Y., et al., 2020a. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun., 11: 2494. DOI:10.1038/s41467-020-16338-x |
Chen, Z.J., Sreedasyam, A., Ando, A., et al., 2020b. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet., 52: 525-533. DOI:10.1038/s41588-020-0614-5 |
D'hondt, L., Höfte, M., Van Bockstaele, E., et al., 2011. Applications of flow cytometry in plant pathology for genome size determination, detection and physiological status. Mol. Plant Pathol., 12: 815-828. DOI:10.1111/j.1364-3703.2011.00711.x |
Danecek, P., Auton, A., Abecasis, G., et al., 2011. The variant call format and VCFtools. Bioinformatics, 27: 2156-2158. DOI:10.1093/bioinformatics/btr330 |
Das, A., 1996. Karyotype analysis and 4C nuclear DNA estimation in three species of Acanthus, a mangrove associate from coastal Orissa. Cytobios, 87: 151-159. |
Debnath, H., Singh, B., Giri, P., 2013. A new mangrove species of Acanthus L. (Acanthaceae) from the Sunderban (India). Indian J. For., 36: 411-412. DOI:10.54207/bsmps1000-2013-99z0ec |
Di Cola, V., Broennimann, O., Petitpierre, B., et al., 2017. ecospat: an R package to support spatial analyses and modeling of species niches and distributions. Ecography, 40: 774-787. DOI:10.1111/ecog.02671 |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull, 19: 11-15. |
Dpooležel, J., Binarová, P., Lcretti, S., 1989. Analysis of nuclear DNA content in plant cells by flow cytometry. Biol. Plant. (Prague), 31: 113-120. DOI:10.1007/BF02907241 |
Duke, N.C., 2006. Australia's Mangroves: the Authoritative Guide to Australia's Mangrove Plants. Brisbane: University of Queensland, pp. 87-93.
|
Duke, N. C, Schmitt, K., 2015. Mangroves: unusual forests at the seas edge. Tropical Forestry Handbook. Berlin: Springer, pp. 1-24.
|
Duke, N.C., 2017. Mangrove floristics and biogeography revisited: further deductions from biodiversity hot spots, ancestral discontinuities, and common evolutionary processes. Mangrove Ecosystems: A Global Biogeographic Perspective. New York: Springer, pp. 17-53.
|
Evanno, G., Regnaut, S., Goudet, J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol., 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Flouri, T., Jiao, X., Rannala, B., et al., 2018. Species tree inference with BPP using genomic sequences and the multispecies coalescent. Mol. Biol. Evol., 35: 2585-2593. DOI:10.1093/molbev/msy147 |
Gaither, M.R., Bowen, B.W., Bordenave, T.R., et al., 2011. Phylogeography of the reef fish Cephalopholis argus (Epinephelidae) indicates Pleistocene isolation across the Indo-Pacific barrier with contemporary overlap in the coral triangle. BMC Evol. Biol., 11: 1-16. DOI:10.5402/2011/258431 |
García, M.B., Miranda, H., Pizarro, M., et al., 2022. Habitats hold an evolutionary signal of past climatic refugia. Biodivers. Conserv., 31: 1665-1688. DOI:10.1007/s10531-022-02419-4 |
Govindarajan, T., Subramanian, D., 1983. Karyomorphological studies in south Indian Acanthaceae. Cytologia, 48: 491-504. DOI:10.1508/cytologia.48.491 |
Guisan, A., Petitpierre, B., Broennimann, O., et al., 2014. Unifying niche shift studies: insights from biological invasions. Trends Ecol. Evol., 29: 260-269. DOI:10.1016/j.tree.2014.02.009 |
Guo, W., Banerjee, A.K., Ng, W.L., et al., 2020. Chloroplast DNA phylogeography of the holly mangrove Acanthus ilicifolius in the Indo-West Pacific. Hydrobiologia, 847: 3591-3608. DOI:10.1007/s10750-020-04372-1 |
Guo, Z., Guo, W., Wu, H., et al., 2018. Differing phylogeographic patterns within the Indo-West Pacific mangrove genus Xylocarpus (Meliaceae). J. Biogeogr., 45: 676-689. DOI:10.1111/jbi.13151 |
Han, T., Hu, Z., Du, Z., et al., 2022. Adaptive responses drive the success of polyploid yellow cresses (Rorippa, Brassicaceae) in the Hengduan Mountains, a temperate biodiversity hotspot. Plant Divers., 44: 455-467. DOI:10.1016/j.pld.2022.02.002 |
Hancock, W.G., Tallury, S.P., Isleib, T.G., et al., 2019. Introgression analysis and morphological characterization of an Arachis hypogaea × A. diogoi interspecific hybrid derived population. Crop Sci., 59: 640-649. DOI:10.2135/cropsci2018.07.0461 |
He, Z., Feng, X., Chen, Q., et al., 2022. Evolution of coastal forests based on a full set of mangrove genomes. Nat. Ecol. Evol., 6: 738-749. DOI:10.1038/s41559-022-01744-9 |
Herawati, W., Widodo, P., Palupi, D., 2020. Leaf morphological variation of Acanthus in some estuarine areas of Cilacap. IOP Conf. Ser. Earth Environ. Sci., 550: 012007. DOI:10.1088/1755-1315/550/1/012007 |
Hodel, R.G., de Souza Cortez, M.B., Soltis, P.S., et al., 2016. Comparative phylogeography of black mangroves (Avicennia germinans) and red mangroves (Rhizophora mangle) in Florida: Testing the maritime discontinuity in coastal plants. Am. J. Bot., 103: 730-739. DOI:10.3732/ajb.1500260 |
Hoffman, J.S., Clark, P.U., Parnell, A.C., et al., 2017. Regional and global sea-surface temperatures during the last interglaciation. Science, 355: 276-279. DOI:10.1126/science.aai8464 |
Huang, G., Wu, Z., Percy, R.G., et al., 2020. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet., 52: 516-524. DOI:10.1038/s41588-020-0607-4 |
Jakobsson, M., Rosenberg, N.A., 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23: 1801-1806. DOI:10.1093/bioinformatics/btm233 |
Jin, J., Yu, W., Yang, J., et al., 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol., 21: 1-31. DOI:10.1117/1.oe.59.1.016110 |
Leigh, J.W., Bryant, D., 2015. POPART: full-feature software for haplotype network construction. Methods Ecol. Evol., 6: 1110-1116. DOI:10.1111/2041-210X.12410 |
Li, M., Zheng, Z., Liu, J., et al., 2021. Evolutionary origin of a tetraploid Allium species on the Qinghai–Tibet plateau. Mol. Ecol., 30: 5780-5795. DOI:10.1111/mec.16168 |
Liang, Q., Hu, X., Wu, G., et al., 2015. Cryptic and repeated "allopolyploid" speciation within Allium przewalskianum Regel. (Alliaceae) from the Qinghai-Tibet plateau. Org. Divers. Evol., 15: 265-276. DOI:10.1007/s13127-014-0196-0 |
Librado, P., Rozas, J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451-1452. DOI:10.1093/bioinformatics/btp187 |
Lin, C., Xu, G., Jin, Z., et al., 2022. Molecular, chromosomal, and morphological evidence reveals a new allotetraploid fern species of Asplenium (Aspleniaceae) from southern Jiangxi, China. PhytoKeys, 199: 113. DOI:10.3897/phytokeys.199.81292 |
Linnaeus, C., 1753. Species Plantarum (Vol. vol. 2). Stockholm: Impensis Laurentii Salvii, pp. 939.
|
Lischer, H.E., Excoffier, L., 2012. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics, 28: 298-299. DOI:10.1093/bioinformatics/btr642 |
Lo, E., 2010. Testing hybridization hypotheses and evaluating the evolutionary potential of hybrids in mangrove plant species. J. Evol. Biol., 23: 2249-2261. DOI:10.1111/j.1420-9101.2010.02087.x |
Lohman, D.J., de Bruyn, M., Page, T., et al., 2011. Biogeography of the Indo-Australian archipelago. Annu. Rev. Ecol. Evol. Syst., 42: 205-226. DOI:10.1146/annurev-ecolsys-102710-145001 |
Lu, R.S., Chen, Y., Zhang, X.Y., et al., 2022. Genome sequencing and transcriptome analyses provide insights into the origin and domestication of water caltrop (Trapa spp., Lythraceae). Plant Biotechnol. J., 20: 761-776. DOI:10.1111/pbi.13758 |
Ma, D., Ding, Q., Zhao, Z., et al., 2023. Chloroplast genome analysis of three Acanthus species reveal the adaptation of mangrove to intertidal habitats. Gene, 873: 147479. DOI:10.1016/j.gene.2023.147479 |
Madlung, A., 2013. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity, 110: 99-104. DOI:10.1038/hdy.2012.79 |
Minh, B.Q., Schmidt, H.A., Chernomor, O., et al., 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol., 37: 1530-1534. DOI:10.1093/molbev/msaa015 |
Molina-Henao, Y.F., Hopkins, R., 2019. Autopolyploid lineage shows climatic niche expansion but not divergence in Arabidopsis arenosa. Am. J. Bot., 106: 61-70. DOI:10.1002/ajb2.1212 |
Narayanan, C., 1951. Nuclear behavior and chromosomal aberrations in mitosis of Acanthus ilicifolius and Asystasia coromandeliana. Indian J. Genet. Plant Breed., 11: 205-210. |
Purcell, S., Neale, B., Todd-Brown, K., et al., 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81: 559-575. DOI:10.1086/519795 |
Qiu, Z., Yuan, Z., Li, Z., et al., 2011. Confirmation of a natural hybrid species in Petrocosmea (Gesneriaceae) based on molecular and morphological evidence. J. Syst. Evol., 49: 449-463. DOI:10.1111/j.1759-6831.2011.00151.x |
Ragavan, P., Saxena, A., Mohan, P., et al., 2015. Taxonomy and distribution of species of the genus Acanthus (Acanthaceae) in mangroves of the Andaman and Nicobar Islands, India. Biodiversitas, 16: 225-237. |
Ragavan, P., Zhou, R., Ng, W.L., et al., 2017. Natural hybridization in mangroves–an overview. Bot. J. Linn. Soc., 185: 208-224. DOI:10.1093/botlinnean/box053 |
Rödder, D., Engler, J.O., 2011. Quantitative metrics of overlaps in Grinnellian niches: advances and possible drawbacks. Global Ecol. Biogeogr., 20: 915-927. DOI:10.1111/j.1466-8238.2011.00659.x |
Rothfels, C.J., 2021. Polyploid phylogenetics. New Phytol., 230: 66-72. DOI:10.1111/nph.17105 |
Session, A.M., Rokhsar, D.S., 2023. Transposon signatures of allopolyploid genome evolution. Nat. Commun., 14: 3180. DOI:10.1038/s41467-023-38560-z |
Smyčka, J., Roquet, C., Renaud, J., et al., 2017. Disentangling drivers of plant endemism and diversification in the European Alps–A phylogenetic and spatially explicit approach. Perspect. Plant Ecol. Evol. Systemat., 28: 19-27. DOI:10.1016/j.ppees.2017.06.004 |
Suan, J.C.P., 1996. Biological Studies of the Species of Acanthus in Singapore. National University of Singapore, Singapore.
|
Thuiller, W., Lafourcade, B., Engler, R., et al., 2009. BIOMOD–a platform for ensemble forecasting of species distributions. Ecography, 32: 369-373. DOI:10.1111/j.1600-0587.2008.05742.x |
Tiley, G.P., Poelstra, J.W., dos Reis, M., et al., 2020. Molecular clocks without rocks: new solutions for old problems. Trends Genet., 36: 845-856. DOI:10.1016/j.tig.2020.06.002 |
Tomlinson, P.B., 2016. The Botany of Mangroves, second ed. Cambridge: Cambridge University Press.
|
Wang, W., Wang, M., 2007. The Mangroves of China. Peking: Science Press.
|
Wee, A.K., Teo, J.X.H., Chua, J.L., et al., 2017. Vicariance and oceanic barriers drive contemporary genetic structure of widespread mangrove species Sonneratia alba J. Sm. in the Indo–West Pacific. Forests, 8: 483. DOI:10.3390/f8120483 |
Wood, T.E., Takebayashi, N., Barker, M.S., et al., 2009. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. U.S.A., 106: 13875-13879. DOI:10.1073/pnas.0811575106 |
Wu, S., Wang, Y., Wang, Z., et al., 2022. Species divergence with gene flow and hybrid speciation on the Qinghai–Tibet Plateau. New Phytol., 234: 392-404. DOI:10.1111/nph.17956 |
Xu, B., Yang, Z., 2016. Challenges in species tree estimation under the multispecies coalescent model. Genetics, 204: 1353-1368. DOI:10.1534/genetics.116.190173 |
Yang, J., Liu, D., Wang, X., et al., 2016. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet., 48: 1225-1232. DOI:10.1038/ng.3657 |
Yang, Y., Li, J., Yang, S., et al., 2017. Effects of Pleistocene sea-level fluctuations on mangrove population dynamics: a lesson from Sonneratia alba. BMC Evol. Biol., 17: 1-14. DOI:10.1186/s12862-016-0855-1 |
Yang, Y., Yang, S., Li, J., et al., 2015. Transcriptome analysis of the Holly mangrove Acanthus ilicifolius and its terrestrial relative, Acanthus leucostachyus, provides insights into adaptation to intertidal zones. BMC Genomics, 16: 605. DOI:10.1186/s12864-015-1813-9 |
Yin, D., Ji, C., Song, Q., et al., 2020. Comparison of Arachis monticola with diploid and cultivated tetraploid genomes reveals asymmetric subgenome evolution and improvement of peanut. Adv. Sci., 7: 1901672. DOI:10.1002/advs.201901672 |
Zhang, N., Zeng, L., Shan, H., et al., 2012. Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytol., 195: 923-937. DOI:10.1111/j.1469-8137.2012.04212.x |
Zhang, J., Zhang, X., Tang, H., et al., 2018. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet., 50: 1565-1573. DOI:10.1038/s41588-018-0237-2 |
Zhang, R., 1985. A new species of Acanthus Linn. Fujian (Family Acanthaceae). Wuyi Sci. J., 5: 237-239. |



