b. Ministry of Education Key Laboratory of Ecology and Resource Use of the Mongolian Plateau & Inner Mongolia Key Laboratory of Grassland Ecology, School of Ecology and Environment, Inner Mongolia University, Hohhot 010000, Inner Mongolia, China;
c. Department of Biology, University of Nebraska at Kearney, Kearney, NE 68849, USA;
d. CAS Key Laboratory of Mountain Ecological Restoration and Bioresource Utilization & Ecological Restoration and Biodiversity Conservation Key Laboratory of Sichuan Province, Chinese Academy of Sciences, Chengdu Institute of Biology, Chengdu 610042, Sichuan, China;
e. Center of Excellence in Phylogeny of Living Organisms, Department of Plant Science, College of Science, University of Tehran, P.O. Box 14155-6455, Tehran, Iran;
f. Institute of Botany, Academy of Sciences of the Republic of Uzbekistan, Tashkent 100047, Uzbekistan;
g. Faculty of Biology, Department of Botany and Plant Physiology, National University of Uzbekistan named after Mirzo Ulugbek, Tashkent 100174, Uzbekistan;
h. Faculty of Pharmacy, Kabul University, 1006 Kabul, Afghanistan
The Lamiaceae Martinov, commonly known as mints, have long served as a model system for evolutionary studies due to their morphological/biogeographical diversity and complexity (Bentham, 1832–1836; Cantino, 1992; Hedge, 1992; Harley et al., 2004; Mint Evolutionary Genomics Consortium, 2018; Rose et al., 2022). Lamiaceae are the sixth most species-rich family of angiosperms, with about 230 genera and 7000 species (Harley et al., 2004; Zhao et al., 2021a). The distribution of the family is virtually worldwide, but seven diversity centers are recognized: (1) The Mediterranean region and Southwest Asia, (2) Sub-Saharan Africa including Madagascar, (3) China, (4) Australia, (5) South America, (6) the southwest United States and Mexico, (7) and Southeast Asia (Hedge, 1992; Harley et al., 2004; Rose et al., 2022).
As one of the seven diversity centers of Lamiaceae, China contains at least 96 genera and 970 species (Xiang et al., 2017), of which seven genera (i.e., Hanceola Kudô, Heterolamium C.Y. Wu, Holocheila (Kudô) S. Chow, Loxocalyx Hemsl., Ombrocharis Hand.-Mazz., Rostrinucula Kudô, and Schnabelia Hand.-Mazz.) are endemic to China (Wu and Li, 1977; Li and Hedge, 1994). In addition, China is considered a diversity hotspot for several widespread genera such as Dracocephalum L., Elsholtzia Willd., Isodon (Schrad. ex Benth.) Spach, Nepeta L., Phlomoides Moench, Salvia L., and Scutellaria L. (Paton, 1990; Walker and Sytsma, 2007; Salmaki et al., 2012a; Hu et al., 2018, 2020; Zhao et al., 2017, 2021a, b, c, 2022; Chen et al., 2022a; Sun et al., 2022). During the past two decades, the circumscription of Lamiaceae and evolutionary relationships within the family have been gradually clarified based on a series of molecular phylogenetic studies (Scheen et al., 2010; Bendiksby et al., 2011; Drew and Sytsma, 2012; Li et al., 2016; Drew et al., 2017; Li and Olmstead, 2017; Zhao et al., 2021a). Concurrently, the phylogenetic position of several rare or endemic Chinese genera has been illuminated (Li et al., 2012; Xiang et al., 2013, 2018; Chen et al., 2014, 2016; Li et al., 2017a; Zhao et al., 2021d). To date, the species diversity and phylogenetic relationships of most large mint genera within China have been well documented (Yu et al., 2014; Yao et al., 2016; Li et al., 2017b; Hu et al., 2018, 2020; Zhao et al., 2017, 2020; Chen et al., 2021, 2022a, b), with only two large genera, Nepeta (42 spp. in China) (Wu and Li, 1977; Li and Hedge, 1994) and Phlomoides (58 spp. in China) (Li, 1985; Li and Hedge, 1994; Xiang et al., 2014; Zhao et al., 2021b, c, 2022, 2023a), lacking a comprehensive molecular phylogenetic treatment.
Phlomoides was established by Moench (1794), but it has generally been treated as a section of Phlomis L. Although Phlomoides was resurrected as a genus based on corolla shape and fruit structure (Adylov et al., 1986; Adylov, 1987; Kamelin and Machmedov, 1990a, b), the genus was not widely accepted (Ryding, 2008) until Scheen et al. (2010) confirmed it as a separate genus using molecular data. The separation of Phlomoides from Phlomis was later affirmed by studies with broader taxon sampling (Bendiksby et al., 2011; Salmaki et al., 2012a). Subsequent molecular phylogenetic studies demonstrated that at least seven genera should be transferred to Phlomoides (Scheen et al., 2010; Bendiksby et al., 2011; Mathiesen et al., 2011; Salmaki et al., 2012a; Zhao et al., 2023a, b), including Eremostachys Bunge, Lamiophlomis Kudô, Metastachydium Airy Shaw ex C.Y. Wu & H.W. Li, Notochaete Benth., Paraeremostachys Adylov et al., Pseuderemostachys Popov, and Pseudomarrubium Popov, making the redefined Phlomoides one of the largest and most heterogeneous genera of Lamiaceae, with ca. 150–170 spp. (Salmaki et al., 2012a, b; Zhao et al., 2021b, c, 2022, 2023a, b).
As currently defined, Phlomoides is mainly distributed in mountain steppe and (semi-) desert areas of three regions: Central Asia, the Iranian highlands, and China (Czerepanov, 1995; Salmaki et al., 2012b; Xiang et al., 2014; Zhao et al., 2022). In total, 58 species and 17 varieties can be found in China (Li, 1985; Li and Hedge, 1994; Xiang et al., 2014; Zhao et al., 2021b, c, 2022, 2023a), with 37 species and 9 varieties occurring in the Hengduan Mountains (HM) and Qinghai-Tibet Plateau region (QTP).
Phlomoides are quite variable in leaf, bract, calyx, and corolla morphology, as well as in habitat (Fig. 1). Phlomoides differs from its sister group, Phlomis, by the following set of characters: generally perennial herbs (vs. small shrubs or occasionally perennial herbs), cordate to triangular-ovate leaves, simple or laciniate to bipinnatisect (vs. lanceolate to oblong-lanceolate, not deeply lobed), upper corolla lips that are dome-shaped with hairy or fringed-incised apices (vs. laterally compressed, flattened, sickle-shaped, apices not fringed-incised), and a basic chromosome number of x = 11 (vs. x = 10) (Azizian and Cutler, 1982; Astanova, 1984; Ghaffari, 2006; Fang et al., 2007; Mathiesen et al., 2011; Salmaki et al., 2012a). The genus is widely regarded as taxonomically challenging due to some common characters shared among species, and because of considerable morphological variation in characters used to differentiate species. Currently, there is no definitive infrageneric classification and our knowledge of species delimitations within the genus is largely based on regional taxonomic work (e.g., Popov, 1926, 1940; Knorring, 1954; Hsuan, 1977; Rechinger, 1980; Li and Hedge, 1994; Sennikov and Lazkov, 2013, Lazkov and Sennikov, 2015; Ranjbar and Mahmoudi, 2015, 2017).

|
| Fig. 1 Morphological diversity and diagnostic characters of Phlomoides. A. P. ornata; B. P. speciosa; C. P. multifurcata; D. P. rotata; E. P. hamosa; F. P. sagittata; G. P. oreophila; H. P. jeholensis; I. Tuberous roots of P. tuberosa; J. P. karatavica; K. P. moluccelloides; L. Linear-tuberous roots of P. likiangensis; M. Flower of P. mongolica; N. Dissected corolla of P. moluccelloides, showing the posterior filaments with comblike-appendages; O. Dissected calyces of P. umbrosa; P. Bracts of P. ruptilis. A–C, E–P, photographed by Y. Zhao; D, photographed by Y.-P. Chen. |
In China, Flora Reipublicae Popularis Sinicae (Wu and Li, 1977) and Flora of China (Li and Hedge, 1994) are the two most important references for Lamiaceae taxonomy. However, at the time of those works, most Phlomoides as currently understood were treated as a section within Phlomis [i.e., Phlomis sect. Phlomoides (Moench) Briq.] while others were treated as separate genera (i.e., Lamiophlomis, Eremostachys, Metastachydium, and Notochaete). In addition, the most complete infrageneric classification system for Chinese Phlomoides was developed using only external morphology (Hsuan, 1977), and is not comprehensive in terms of taxon treatment. Based on leaf, trichome, calyx and stamen morphology, Hsuan (1977) divided Chinese Phlomoides into two subsections and 17 series (seven monotypic). This classification, however, tends to be ambiguous due to the substantial variation in morphological characters among the treated taxa. Furthermore, phylogenetic studies have substantially changed the concept of the genus (Scheen et al., 2010; Bendiksby et al., 2011; Salmaki et al., 2012a; Zhao et al., 2023a, b), with a much broader interpretation of Phlomoides now accepted. Thus, the current classification system is sorely outdated and a revised classification scheme for Phlomoides is necessary.
DNA-based phylogenetic analyses provide an effective complement to morphology in approaching taxonomic challenges. Taxonomic problems in several genera of Lamiaceae have been resolved with the support of molecular analyses using plastid DNA regions (e.g., atpB-rbcL, psbA-trnH, rpl16, rpl32-trnL, rps16, trnK, trnL-trnF, trnS-trnG, trnT-trnL, ycf1) and nuclear ribosomal nrITS and nrETS regions (Paton et al., 2004; Walker et al., 2004; Bräuchler et al., 2010; Drew and Sytsma, 2011, 2012; Pastore et al., 2011; Wilson et al., 2012; Xiang et al., 2013, 2018; Li et al., 2016; Yao et al., 2016; Hu et al., 2018; Chen et al., 2021). However, our current phylogenetic understanding of Phlomoides is limited, with only a few studies including representatives from this genus (Fang et al., 2007; Scheen et al., 2010; Bendiksby et al., 2011; Mathiesen et al., 2011; Salmaki et al., 2012a; Zhao et al., 2023a), and genetic data are only available for a few species of Phlomoides native to China. The lack of a detailed molecular phylogenetic framework of the genus leaves a gap in understanding species relationships as well as the evolutionary history of the genus. As one of the three diversity centers of Phlomoides, a molecular study focusing on taxa native to China is an important first step in elucidating the broader Phlomoides taxonomy.
Here, we use nine chloroplast markers (atpB-rbcL, psbA-trnH, rpl16, rpl32-trnL, rps16, trnK, trnL-trnF, trnS-trnG, trnT-trnL) and two nuclear DNA regions—nrETS and nrITS—to reconstruct the phylogeny of Chinese Phlomoides species. The infrageneric classification system proposed by Hsuan (1977) is compared with molecular results. Integrating molecular and morphological evidence, we aim to evaluate and revise the infrageneric classification of Chinese Phlomoides. The objectives of this study are to (1) present a comprehensive phylogenetic reconstruction of Phlomoides, with an emphasis on the Chinese taxa; (2) provide a phylogenetic background to test the infrageneric classification of Hsuan (1977) and revise the classification; (3) further contribute to a comprehensive phylogenetic framework for subfamily Lamioideae.
2. Materials and methods 2.1. Plant materialsThe ingroup included 111 samples representing 85 species and 6 varieties of Phlomoides and four individuals representing three Phlomis taxa (Phlomis herba-venti L. subsp. pungens (Willd.) Maire ex DeFilipps, P. fruticosa L., P. composita Pau), of which 51 species and 6 varieties (about 88% of total taxa) are distributed in China. Fresh leaves were collected and dried with silica gel, and voucher specimens were deposited in the Kunming Institute of Botany, Chinese Academy of Sciences. Materials of 34 individuals representing 31 species were sampled from herbarium specimens held at K, LE, and MW (Table S1). Two species each from tribes Lamieae (Lamium amplexicaule L. and L. album L.), Leucadeae (Leonotis leonurus (L.) R.Br. and Leucas mollissima Wall. ex Benth.), Leonureae (Leonurus cardiaca L. and L. japonicus Maxim.), Marrubieae (Marrubium vulgare L. and Pseudodictamnus mediterraneus Salmaki & Siadati) and three species of Paraphlomideae (Paraphlomis albida Hand. -Mazz., P. hispida C.Y. Wu and P. nana Y.P. Chen, C. Xiong & C.L. Xiang) were selected as an outgroup based on previous findings (Salmaki et al., 2012a; Zhao et al., 2021a).
Our taxon sampling was broad, encompassing the geographic range of the genus, including East Asia (most parts of China including the QTP and HM regions, Bhutan, Mongolia; 47 species), Central Asia (Kazakhstan, Kyrgyzstan, Tajikistan, Turkmenistan, Uzbekistan; 28 species), Southwest Asia (Afghanistan, Armenia, Iran; 9 species) and Europe (Ukraine, 1 species). This broad sampling offers the opportunity to reconstruct the backbone of the Phlomoides phylogeny and identify major lineages within the genus.
2.2. DNA extraction, amplification, and sequencingTotal genomic DNA was extracted either from silica-gel-dried leaf material using the modified CTAB method (Doyle and Doyle, 1987) or from herbarium specimens following Zeng et al. (2018). In this study, nine chloroplast DNA (cpDNA) markers (atpB-rbcL, psbA-trnH, rpl16, rpl32-trnL, rps16, trnK, trnL-trnF, trnS-trnG, trnT-L) and two nuclear DNA regions—nrITS and nrETS—were selected for phylogenetic reconstruction. Primers, mixtures and procedures for atpB-rbcL, psbA-trnH, and trnT-trnL followed Albaladejo et al. (2005), trnK and rpl32-trnL followed Salmaki et al. (2012a), trnS-trnG followed Scheen and Albert (2009), while rpl16, rps16, and trnL-trnF followed Chen et al. (2021). The primer pairs ETS-B (Beardsley and Olmstead, 2002) and 18S-IGS (Baldwin and Markos, 1998) were used for the amplification of nrETS, and Leu1 (Vargas et al., 1998) and ITS4 (White et al., 1990) for nrITS. PCR and sequencing protocols for two nrDNA markers followed Xiang et al. (2013).
Sequencing reactions were performed with the dideoxy chain termination method running on an ABI PRISM 3730 automated sequencer. All sequences used in this study together with their GenBank accession numbers are listed in Table S2.
2.3. Sequence alignment and phylogenetic analysesRaw sequences were assembled and edited using Geneious v.11.0.3 (Kearse et al., 2012) and aligned by the MAFFT v.7.308 (Katoh and Standley, 2013) with the G-INS-I algorithm. The final alignments were adjusted manually in PhyDE v.0.9971 (Müller et al., 2010). Three different datasets were analyzed: the cpDNA and nrDNA alignments separately, and an analysis with all data combined.
All datasets were analyzed using Bayesian inference (BI) and maximum likelihood (ML). The best-fit substitution model for each data set was selected by the jModelTest v.3.7 (Posada, 2008) under the Akaike information criterion (AIC) score. BI and ML analyses were conducted on the Cyberinfrastructure for Phylogenetic Research Science (CIPRES) Science Gateway v.3.3 (Miller et al., 2010), using MrBayes (Ronquist et al., 2012) and RAxML v.8.2.9 (Stamatakis, 2014), respectively. Details for parameter settings followed Zhao et al. (2023b). All phylogenetic trees with posterior probabilities (PP) and bootstrap values (BS) were exhibited and annotated in FigTree v.1.4.2 (Rambaut, 2014). The R package Phytools v.2.1.1 (Revell, 2012) was used to compare topological incongruence between the cpDNA and nrDNA trees.
2.4. Morphological dataTo test the taxonomic value of morphological characters (habit, basal leaves, calyces, bracts, corolla, and nutlets) traditionally used for the classification of Chinese species of Phlomoides, we carefully investigated those characters in the field and with herbarium specimens. We recorded the habit, the shapes of basal and stem leaves, and dissected corolla tubes. All images were taken by a digital single-lens-reflex camera, and a ruler was used during photography. Finally, we summarized these characters for each investigated species. Eight morphological characteristics were selected and mapped onto the tree to infer trait evolution within Phlomoides, including three vegetative characteristics [habit (herb/shrub), basal leaves (absent/present), and division of leaves (simple/laciniate)], four floral characteristics [calyx shape (tubular/campanulate or infundibular), bract shape (straight/hooked), upper corolla lip shape (galeate/erect) and upper corolla beard color (white/brown to black)] and one fruit trait [nutlet apex (dense simple long trichomes/glabrous or with sparse branched trichomes)].
3. Results 3.1. Sequences and alignment characterizationFor this study, 570 sequences were newly generated (Table S2). The cpDNA and nrDNA datasets used in this study were deposited in Figshare (http://10.6084/m9.figshare.23936628). The combined nrDNA dataset consisted of 124 individuals and 1253 aligned nucleotide position characters (695 nucleotides were variable), of which 703 positions were from the ITS1-5.8S-ITS2 region, while the nrETS region contributed 550 bp. Sequence lengths were 2317–2360 nucleotides (nt) in Phlomoides for the atpB-rbcL spacer, 327–372 nt for the psbA-trnH intergenic spacer, 1295–1305 nt for the rpl16 intron, 617–648 nt for the rpl32-trnL spacer, 912–942 nt for the rps16 intron, 947–954 nt for the trnK intron, 821–851 nt for the trnL-trnF spacer, 744–778 nt for the trnS-trnG spacer, and 696–724 nt for the trnT-trnL spacer. The concatenated and aligned plastid matrix consisted of 126 individuals and 9473 nucleotide position characters, of which 16.96% were variable in the dataset (Table 1). The resulting combined cpDNA and nrDNA dataset contained 10, 726 positions, of which 21.46% were variable sites (Table 1).
| Datasets | No. Taxa | Nucleotides (with ambiguous sites excluded) [bp] | GC content (%) | No. constant sites [bp] | No. variable sites [bp] | No. parsimony- informative sites [bp] |
| atpB-rbcL | 126 | 2424 | 39.3 | 2177 (89.81%) | 247 (10.19%) | 157 (6.48%) |
| psbA-trnH | 126 | 452 | 32.7 | 359 (79.42%) | 93 (20.58%) | 58 (12.83%) |
| rpl16 | 126 | 1387 | 36.3 | 1156 (83.35%) | 231 (16.65%) | 141 (10.17%) |
| rpl32-trnL | 126 | 908 | 31.3 | 664 (73.13%) | 244 (26.87%) | 148 (16.30%) |
| rps16 | 126 | 1013 | 35.7 | 864 (85.29%) | 149 (14.71%) | 89 (8.79%) |
| trnK | 126 | 956 | 34.2 | 745 (79.93%) | 211 (22.07%) | 130 (13.60%) |
| trnL-trnF | 126 | 894 | 36.1 | 757 (84.68%) | 137 (15.32%) | 82 (9.17%) |
| trnS-trnG | 126 | 592 | 33.7 | 474 (80.07%) | 118 (19.93%) | 65 (10.98%) |
| trnT-trnL | 126 | 847 | 29.5 | 670 (79.10%) | 177 (20.90%) | 104 (12.28%) |
| CP9 | 126 | 9473 | 35.6 | 7866 (83.03%) | 1607 (16.96%) | 974 (10.28%) |
| nrITS | 121 | 703 | 66.7 | 325 (46.23%) | 378 (53.77%) | 239 (34.00%) |
| nrETS | 119 | 550 | 63.3 | 233 (42.36%) | 317 (57.64%) | 223 (40.55%) |
| NR | 124 | 1253 | 65.0 | 558 (44.53%) | 695 (55.47%) | 462 (36.87%) |
| CP + NR | 126 | 10, 726 | 38.8 | 8424 (78.54%) | 2302 (21.46%) | 1436 (13.39%) |
The cpDNA, nrDNA, and combined cpDNA and nrDNA phylogenies all supported the monophyly of Phlomoides s.l. (Figs. 2, 3 and S1–S8). The relationships based on the cpDNA-derived tree (Figs. 2, S3 and S4) were better supported than the tree inferred using two nrDNA markers (Figs. S1, S5 and S6) as well as the combined cpDNA and nrDNA markers (Figs. S2, S7 and S8). Phylogenies derived from BI and ML analyses were generally concordant (Figs. S3–S8). The BI phylogeny from the combined cpDNA dataset is presented here, with nodal support values from both BI PP and ML BS (Fig. 2).

|
| Fig. 2 Phylogeny of Phlomoides inferred using Bayesian inference (BI), based on the combined plastid dataset. Support values displayed above branches follow the order BI-PP/ML-BS ("*" indicates PP = 1.00 or BS = 100%, solid circles indicate species occurring in China, hollow circles indicate species not in China). Sectional classification of Phlomoides is based on Kamelin and Machmedov (1990a), Sennikov and Lazkov (2013), Lazkov and Sennikov (2015), and Ranjbar and Mahmoudi (2015). Series classification of Phlomoides is based on Popov (1926, 1940), Knorring (1954) and Hsuan (1977). Multiple accessions of the same species are numbered according to Table S1. |

|
| Fig. 3 Select morphological characters mapped onto the Bayesian consensus tree. Symbols are filled squares (F) or squares (O). Characters are A. Habit: herb = O, shrub = F; B. Basal leaves: absent = O, present = F; C. Division of leaves: simple = O, laciniate to bipinnatisect = F; D. Calyx shape: tubular = O, campanulate or infundibular = F; E. Bracts shape: straight = O, hooked = F; F. Upper corolla lip shape: galeate = O; erect = F; G. Upper corolla beard color: white = O, brown to black = F; H. Nutlet apex: glabrous or with sparsely branched trichomes = O, dense, simple, long trichomes = F. |
The monophyly of Phlomoides s.l. (including traditionally circumscribed Eremostachys, Lamiophlomis, Metastachydium, Notochaete, Paraeremostachys, Pseuderemostachys, and Pseudomarrubium) was strongly supported (1.00 and 99% for the BI-PP and ML-BS, respectively; the values are listed in the same order below; Fig. 2), and the genus can be divided into six major clades. All clades had high support values, with Clades I and II collectively sister to a group that includes Clades III, IV, V, and VI. Monophyly for most traditionally defined sections (Filipendula (Popov) Adylov et al., Moluccelloides (Bunge) Sennikov, Paraeremostachys (Adylov et al.) Sennikov, Phlomoides (Popov) Adylov et al., and Thyrsiflorae (Rech.) Ranjbar & Mahmoudi) was not supported. In addition, the Chinese species of Phlomoides as well as most series recognized by Hsuan (1977) were not monophyletic.
Clade I ("Notochaete clade" sensu Salmaki et al., 2012a) consists of the former genus Notochaete (Fig. 2). Clade II (1.00/98%) contains 34 taxa (out of the 44 accessions) and is the core group of Chinese Phlomoides. Species in Clade II included members of Phlomoides sect. Phlomoides plus two accessions of Phlomoides rotata (Benth. ex Hook.f.) Mathiesen, which previously was defined as the monotypic genus Lamiophlomis. At the series level, 29 species from 11 series sensu Hsuan (1977) were included in this clade. Of the 11 series, five are monotypic but each contains only one individual in the present study (i.e., Atropurpureae, Jeholenses, Paohsingenses, Pedunculatae, Umbrosae), while the monophyly of the remaining six series (i.e., Dentosae, Franchetianae, Megalanthae, Melananthae, Tatsienenses, Tibeticae) was not supported. In addition, five unplaced species [Phlomis brevidentata H.W. Li (treated as a synonym of Phlomoides breviflora (Benth.) Kamelin & Makhm. in this study, see Taxonomic treatment section), Phlomoides breviflora, P. macrophylla (Benth.) Kamelin & Makhm., P. nyalamensis (H.W. Li) Y. Zhao & C.L. Xiang, and the recently described new species P. liangwangshanensis Y. Zhao, H.L. Zheng & C.L. Xiang] were recovered within this clade.
Clades III, IV, V, and VI collectively formed a clade. Clade III (Fig. 2; 1.00/100%) is comprised of seven taxa from three series of the section Phlomoides. Phlomoides tuvinica (A. Schroet.) Kamelin, Adylov & Makhm., which has not been placed in any series previously, was also recovered within Clade III. Clade IV (Fig. 2; 1.00/100%) is composed of species from two series. Two individuals of P. oreophila (Kar. & Kir.) Adylov, Kamelin & Makhm. were not sister to each other in the cpDNA phylogenetic tree (Fig. 2), but were resolved as monophyletic in the nrDNA phylogenetic tree (Fig. S1). Series Alpinae was nested within ser. Canescentes. Clade V contains eight taxa, the traditionally defined monotypic genus Metastachydium was sister to another subclade consisting of five species from series Tuberosae, plus P. adylovii Lazkov, which has not been assigned to a series. Most species within Clade VI were members of the traditionally defined genus Eremostachys, except for P. brachystegia (Bunge) Adylov, Kamelin & Makhm., which was placed within Phlomis s.l. Species within Clade VI are from six sections (Filipendula, Moluccelloides, Paraeremostachys, Pseuderemostachys (Popov) Lazkov, Thyrsiflorae, and Phlomoides) and 13 series, of which seven series are monotypic (Brachystegiae, Campanulatae, Fulgentes, Integrifoliae, Molucceliformes, Pauciflorae, Tuberosae). The monophyly of the remaining six series (Cordatae, Gymnocalyces, Laciniatae, Ovalifoliae, Rhodanthae, Speciosae) was not supported. In addition, P. sewerzovii (Herder) Mathiesen (formerly of the monotypic genus Pseuderemostachys) was sister to P. eremostachydioides (Popov) Y. Zhao & C.L. Xiang (formerly from the monotypic genus Pseudomarrubium), together embedded within Clade VI.
In the nrDNA phylogenetic tree (Figs. S1, S4 and S5), only Clades I, III, and VI were recovered as monophyletic. A comparison of plastid and nuclear trees was performed to identify discrepancies between them (Fig. S9). The cpDNA data supported Clade I as a sister to Clade II, but nrDNA data placed Clade I as a sister with all other five Clades. Another major difference is that Clade III is sister to a large clade composed of Clade IV, V, and VI in the cpDNA tree, but formed a sister group with Clade VI in the nrDNA tree. In addition, Clade II and Clade IV were grouped together in the nrDNA tree.
In the combined cpDNA and nrDNA phylogenetic tree (Figs. S2, S7 and S8), the monophyly of six major clades were supported as the same in the cpDNA trees (Figs. 2, S3 and S4), and the topology of most clades was also congruent with the cpDNA phylogenetic tree, but with the lower overall support values.
4. Discussion 4.1. Circumscription and phylogenetic relationships of PhlomoidesThis study represents the most comprehensive molecular phylogenetic survey of Phlomoides, in terms of both taxon sampling and number of markers, to date (Salmaki et al., 2012a; Zhao et al., 2023a, b). This affords the opportunity to examine the phylogeny, circumscription and infrageneric relationships of this taxonomically challenging group. Although several recent studies have clarified the boundaries of Phlomoides, no study has combined intensive sampling of both Phlomoides and related genera.
We confirmed the inclusion of seven traditionally defined genera (i.e., Eremostachys, Lamiophlomis, Metastachydium, Notochaete, Paraeremostachys, Pseuderemostachys and Pseudomarrubium) within Phlomoides. Monophyly of the broadly defined Phlomoides was well supported in all analyses, and six primary clades were inferred based on cpDNA data (Figs. 2, S3 and S4), yielding trees with higher resolution and better-supported relationships than the nrDNA phylogenetic tree (Figs. S1, S5 and S6) and combined cpDNA and nrDNA data (Figs. S2, S7 and S8). The recovered major clades were also supported by several morphological differences, such as the presence of basal leaves, trichomes on the apices of nutlets, and the color of trichomes within the upper corolla lip. Therefore, the BI topology from the combined cpDNA dataset will be the primary tree for discussing phylogenetic relationships.
Clade I comprises two species previously treated as the genus Notochaete (Fig. 2). Using three cpDNA sequences (trnL intron, trnL-trnF spacer, and rps16 intron), Scheen et al. (2010) found Notochaete hamosa Benth. (the type species of Notochaete) nested within Phlomoides, and later Mathiesen et al. (2011) formally transferred Notochaete into Phlomoides. This relationship was recovered by later studies based using additional DNA markers (Bendiksby et al., 2011; Salmaki et al., 2012a; Zhao et al., 2023a, b). The genus forms a separate lineage and has been defined as the "Notochaete" clade (Salmaki et al., 2012a) or "Notochaete" group (Zhao et al., 2023a), but Notochaete has not been placed within any section because of insufficient taxon sampling within Phlomoides. Our findings are congruent with previous studies but yield new information about the position of Notochaete, resolving it as sister to Clade II (Fig. 2), which in the cpDNA tree is composed mostly of Chinese species distributed in the Himalayan and Hengduan Mountains (Clade II; Fig. 2), or sister to all other species of Phlomoides in the nrDNA tree (Fig. S1).
Several features support the close relationship between Clades I and II in the cpDNA phylogeny. For example, most species in the two clades grow in forested habitats, and most species have nutlets that are glabrous and lack basal leaves. However, Clade I is sister to all other species of Phlomoides in the nrDNA phylogeny (Fig. S1). In this study, despite the position of the "Notochaete" group (Clade I) being discordant between the chloroplast and nuclear phylogenies, the monophyly of the two species is strongly supported. Morphologically, several distinct characters support the Notochaete group, including hooked floral bracts and calyx spines (vs. needlelike or lanceolate bracts and calyx teeth with needlelike spines at apex). In addition, these two species also have the smallest flowers within Phlomoides (less than 1 cm vs. 1.5–5 cm long). In light of these synapomorphies we will refer to this clade as P. sect. Notochaete (see Taxonomic treatment section) to accommodate its systematic position within the genus.
Clade II is composed of 29 species and 5 varieties that are mostly distributed in forests or alpine steppe regions of the QTP, Hengduan Mountains, and northern China. There are three potential synapomorphies for this clade: species generally have linear-tuberous or woody fibrous roots; lack basal leaves, and have glabrous nutlets. In addition, most species in this clade are tall herbs usually higher than 1 m, sometimes up to 2 m, while species in the other clades are much shorter. As shown in Fig. 2, at least five subclades can be recognized within Clade II based on the cpDNA phylogeny, but one species, P. pedunculata (Y.Z. Sun) Kamelin & Makhm., is sister to Clade III (0.67/35%) in the nrDNA phylogeny, indicating topological incongruence probably attributed to hybridization and chloroplast capture, as reported in Phlomis (the sister group of Phlomoides) (Albaladejo et al., 2005) and other genera in Lamiaceae (Drew and Sytsma, 2013; Drew et al., 2014; Deng et al., 2015; Walker et al., 2015; Hu et al., 2018; Celep et al., 2020). Geographically, many species within this clade have a sympatric distribution, and species in the clade are quite variable morphologically. Thorough morphological investigation and a detailed molecular population-level study are needed to resolve species relationships within this group.
Clades III, IV, V, and VI form a large clade sister to Clades I and II. All species in Clade III–VI are characterized by having basal leaves. Clade III contains six species that are distributed in steppe or alpine steppe regions of the QTP, Hengduan Mountains, and the Mongolia Plateau.
Clade IV consists of six species distributed in steppe or alpine steppe regions of Central Asia and the Mongolia plateau, and all species have cordate basal leaves. The corolla morphology of plants in this clade is distinct, i.e., the upper lip is bent at a right angle relative to the tube (Fig. 1G) (vs. upper lip usually straight or slightly curved downward, but not bent downward in other clades; Fig. 1M). In comparison with taxa from Clades III and V, the corolla of taxa in Clade IV have longer tubes and shorter limbs (tube three times vs. two times longer than limb). Within Clade IV, Phlomoides pratensis (Kar. & Kir.) Adylov, Kamelin & Makhm. and P. canescens have ovate-oblong floral leaves, while the other species in Clade IV have upper floral leaves narrowly linear, and lower floral leaves linear-lanceolate (Fig. 1G). Phlomoides pratensis has double-toothed calyx teeth (Fig. 1A), while the calyx teeth are rounded in the other five species. The nutlet morphology of species in this clade is also variable, with some taxa having sparse trichomes at the apex (i.e., P. alpina (Pall.) Adylov, Kamelin & Makhm., P. canescens, P. chinghoensis (C.Y. Wu) Kamelin & Makhm. and P. oreophila (Kar. & Kir.) Adylov, Kamelin & Makhm.) or glabrous nutlet apices (i.e., P. pratensis, P. koraiensis (Nakai) Kamelin & Makhm.). It is notable that P. koraiensis has six downward appendages at the filament base, which is a unique character within the genus; all other species within Clade IV have two appendages at the filament base or lack appendages (Y. Zhao pers. obs.).
Clade V is composed of eight species mainly distributed in steppe or alpine steppe regions of Central Asia and the Mongolian Plateau. Possible synapomorphies for this clade include tuberous roots that are globose to fusiform (Fig. 1I), +/−sagittate basal leaves, triangular floral leaves, and nutlets either glabrous or with branched trichomes. In this study, one undetermined individual (Phlomoides sp.), collected from Hebei Province in China, is morphologically similar to Phlomoides dentosa var. glabrescens (Danguy) C.L. Xiang & H. Peng. However, an individual of Phlomoides dentosa var. glabrescens, collected from Gansu Province, grouped with the type variety (P. dentosa var. dentosa) within Clade III, while Phlomoides sp. grouped with P. mongolica (Fig. 2; 1.00/100%; Fig. S1; 0.93/100%) within Clade V. After comparing the external morphology of the two specimens, we found that Phlomoides sp. has floral leaves similar to P. mongolica (floral leaves with no obvious petiole and blades often broad, length–width ratio less than 2), while P. dentosa var. glabrescens and P. dentosa var. dentosa have similar floral leaves (floral leaves with petiole ca. 5 mm long, blade often longer, length-width ratio about 2.5–4). Geographically, Phlomoides sp. and another morphologically similar species (P. mongolica) within Clade V are distributed in Hebei, Beijing, and eastern Inner Mongolia in China, while P. dentosa var. glabrescens and P. dentosa var. dentosa are distributed in western Inner Mongolia Gansu, Qinghai and Ningxia provinces in China. The collection of Phlomoides sp. likely represents a new species, but further morphological studies are needed before official recognition.
Clade VI is a well-supported clade (Fig. 2; 1.00/98%), with most species belonging to the previously defined genus Eremostachys. In our study, this clade consists of 35 species that are distributed in desert, desert steppe, or mountain steppe regions from Central Asia to western Asia. Morphologically, species in this clade usually have basal leaves (Fig. 1B and C), nutlets generally have long simple trichomes, and filaments often have comblike appendages at the base (Fig. 1N, arrow). Within clade VI, Phlomoides zenaidae (Popov) Adylov, Kamelin & Makhm. diverges first and is sister to the remaining taxa. Morphologically, this species can be distinguished from other species within the clade by having glabrous nutlets. In addition, the upper corolla lip morphology of P. zenaidae is unique. In other species within Clade VI, the posterior corolla lips have irregularly denticulate margins and bearded insides, but P. zenaidae has entire posterior corolla lips with sparse trichomes on the margins, similar to Phlomis. The next diverging lineage is P. sewerzovii. Morphologically, the upper lip of corolla of P. sewerzovii is erect and 2-lobed, with short stamens that are included within the corolla tube. These characters are rare in the genus with only three species, P. boraldaica A.L. Ebel, P. sagittata (Regel) C.L. Xiang & Y. Zhao (Fig. 1F), and P. sewerzovii, displaying these features (Fig. 3, character F). Phlomoides sewerzovii and P. boraldaica were members of P. sect. Pseuderemostachys, while P. sagittata was nested within Clade V. Clade VI is the largest major clade recovered, with six species distributed in China, and the remaining species ranging from central to western Asia. In comparison with species distributed in southwest China, the Hengduan Mountains, and the Himalayas, the species in this clade mostly grow in arid habitats and possess arid-adapted characters such as densely lanate stems, calyces, and nutlets (Fig. 1B), as well as napiform roots (Fig. 1J and K). Although several well-supported subclades can be recognized within this clade, relationships among species await more detailed taxon sampling.
4.2. Implications for infrageneric classification of PhlomoidesPrior to 1990, Phlomoides was placed within Phlomis s.l. as a section (Bentham, 1832–1836; Briquet, 1897; Hsuan, 1977), and Eremostachys was considered as a separate genus closely allied to Phlomis. Therefore, most names of the infrageneric categories (sections and series) of Phlomoides originated from Popov's (1940) classification system of Eremostachys and Popov's (1926) and Knorring's (1954) classification system of Phlomis sect. Phlomoides. In resurrecting the genus Phlomoides, Adylov et al. (1986) divided the genus into two sections, P. sect. Phlomoides and P. sect. Filipendula. Now, since the circumscription of Phlomoides has drastically changed, existing infrageneric classifications are no longer sufficient.
Molecular phylogenetic reconstructions of Phlomoides have gradually delimited the boundaries of the genus and identified allied genera (Pan et al., 2009; Scheen et al., 2010; Bendiksby et al., 2011; Mathiesen et al., 2011; Salmaki et al., 2012a; Zhao et al., 2023a, b). These insights were used as a basis for a revised classification of Phlomoides and several new sections were proposed. For example, based on Salmaki et al.'s (2012a) molecular phylogenetic analyses, Sennikov and Lazkov (2013) established three new sections, P. sect. Eremostachys, P. sect. Moluccelloides, and P. sect. Paraeremostachys. Later, Lazkov and Sennikov (2015) established the monotypic section P. sect. Pseuderemostachys based on its unique characters by having short stamens barely exserted form the corolla. Ranjbar and Mahmoudi (2015) proposed a new section, P. sect. Thyrsiflorae (Rech.) Ranjbar & Mahmoudi, which is consistent with Eremostachys sect. Thyrsiflorae Rech. f. To date, seven sections have been proposed within Phlomoides. Here, we did not adopt the concept of P. sect. Eremostachys (Bunge) Sennikov because this section was equal to the "Eremostachys laciniata core group" (Salmaki et al., 2012a), and members of this group were not clearly defined.
The classification scheme proposed by Hsuan (1977) divided Chinese Phlomoides (= Phlomis sect. Phlomoides) into two subsections and 17 series. This is the only infrageneric classification system for Phlomoides in China, but was largely based upon Popov's (1926) and Knorring's (1954) classifications. Hsuan (1977) recognized two subsections for Chinese Phlomoides, subsect. Anisostyleae M. Pop. (style unequally 2-cleft at apex) and subsect. Isostyleae M. Pop. (style equally 2-cleft at apex). In this study, we sampled 90 taxa of Phlomoides representing 28 series, but the monophyly of most series was not supported (Figs. 2 and S1). Another major difference between the proposed system of Hsuan (1977) and currently recognized Phlomoides is that several former genera are now treated within Phlomoides (i.e., Eremostachys, Lamiophlomis, Metastachydium, Notochaete). In the updated treatment of Lamiaceae in Flora of China (Li and Hedge, 1994), these genera were also segregated from Phlomoides.
In this study, we have taken a first step towards clarifying the circumscription of Phlomoides. This includes extensive sampling of Chinese taxa (51 species and 6 varieties, accounting for 88% of Chinese species) as well as 33 species from central Asia, western Asia, Europe, and Mongolia, representing all six aforementioned sections. In the resulting tree, six major clades of Phlomoides are recognized (Fig. 2). However, the species composition in each clade is not consistent with, nor do any of the six major clades match, previously established sections.
Establishing a tenable infrageneric classification for Phlomoides, as with any large and complex genus, is a necessary first step towards a stable taxonomy. Unfortunately, previous circumscriptions of Phlomoides have not resulted in a usable system, since neither the traditional sectional nor series classifications were supported here. Because this study is focused on taxa native to China, we only established one new section to accommodate the systematic placement of the "Notochaete" clade (Clade I) based on molecular data and unique morphological characters (see Taxonomic treatment section). Future studies involving increased taxon sampling, high-throughput sequencing, as well as morphological investigation are needed to provide more evidence for establishing a stable classification system for Phlomoides s.l.
4.3. The search for useful morphological charactersPhlomoides is morphologically very diverse, and it is difficult to identify clear synapomorphies for most clades from our phylogenetic analyses (except Clade I in cpDNA phylogenies; Fig. 2). The major reasons are that a detailed morphological study for the genus is lacking, and morphological characters supporting the clades found here are mostly unknown or limited. Although trends of some selected morphological characters corroborate the relationships based on the cpDNA phylogenetic tree, morphological synapomorphies are difficult to identify for most clades. The search for synapomorphies, especially in Clades II and VI, remains challenging given the ample morphological variation exhibited by species within each clade.
The traditional taxonomic framework of Chinese Phlomoides (Hsuan, 1977) is based on external morphological characters, such as the presence/absence of basal leaves, the indumentum on nutlets and leaves, flower size, calyx teeth shape, whether the 2-cleft style apices are equal/unequal, and whether stamens have basal appendages. The selection of these characters was based on observation of herbarium specimens, and some characters are highly variable in the field. During the past five years, we have investigated most species at the population-level in the field, dissected flowers and calyces, and observed and compared trichome morphology using light microscopy and scanning electron microscopy methods (unpublished data).
Here we mapped eight morphological characters (Fig. 3) onto the cpDNA tree; phylogenetic inferences imply that at least some of these characters have significant taxonomic utility. For example, habit (Fig. 3, character A) is the most reliable character to distinguish Phlomoides (herb; Fig. 4A) from the sister group Phlomis (shrub; Fig. 4B). Within Phlomoides, calyces with hooked spines (Fig. 3, character E; Fig. 4J) are unique to Clade I and an absence of basal leaves (Fig. 3, character B) is a potential synapomorphy of both the combined Clades I and II. Although P. rotata, P. tibetica (C. Marquand & Airy Shaw) Kamelin & Makhm., P. milingensis (C.Y. Wu & H.W. Li) Kamelin & Makhm. and P. atropurpurea (Dunn) Kamelin & Makhm. within Clade II usually have rosetted leaves, this character likely evolved as an adaptation to high alpine scree ecosystems. Clade VI is characterized by taxa having long simple trichomes at the apex of nutlets (Fig. 4O), while in Clades I–V, nutlet apices are usually glabrous or rarely have sparsely branched trichomes (Fig. 4P). This character is also correlated with geographical patterns; species in Clade VI are distributed in central to western Asia, while species in Clade I–V are mainly distributed in southwest China to the eastern Himalayas and eastern Central Asia.
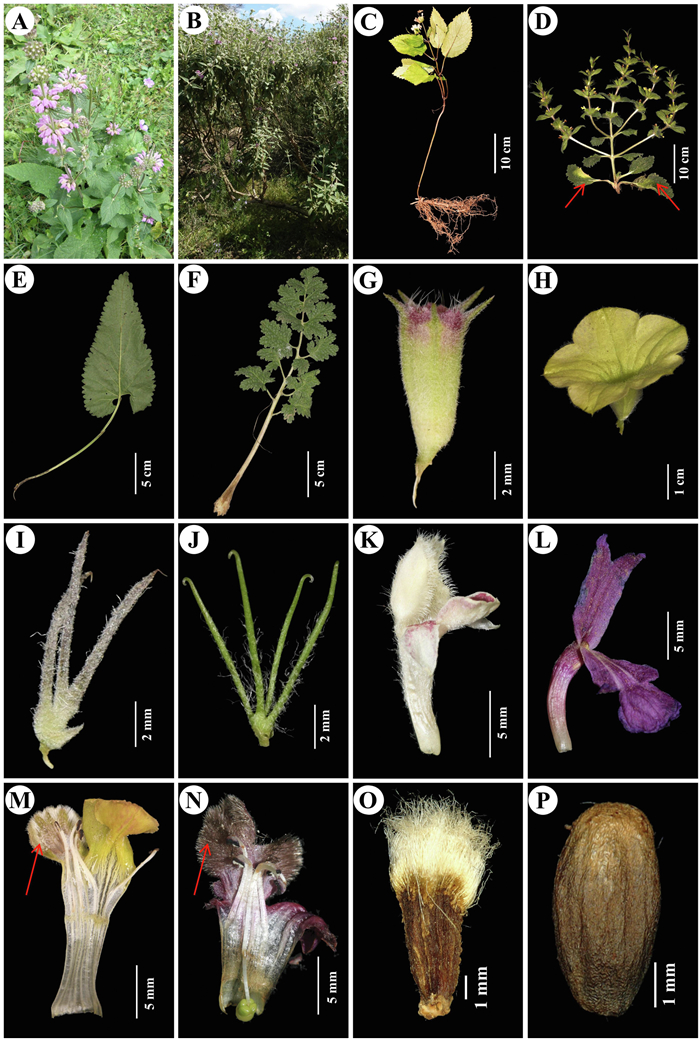
|
| Fig. 4 Selected morphological characters from Fig. 3. A. Phlomoides pratensis, representing an herb, B. Phlomis composita, representing a shrub; C. Phlomoides pedunculata, lacking basal leaves; D. P. multifurcata, arrow showing basal leaves; E. Simple basal leaf of P. mongolica; F. Bipinnatisect basal leaf of P. speciosa; G. Tubular calyx of P. umbrosa; H. Infundibular calyx of P. moluccelloides; I. Straight bracts of P. dentosa; J. Hooked bracts of P. hamosa; K. Galeate upper lip of P. ruptilis; L. Erect and 2-lobed upper lip of P. sagittata; M. Dissected corolla of P. multifurcata, arrow showing the white trichomes; N. Dissected corolla of P. nyalamensis, arrow showing the brown to black trichomes; O. Nutlet of P. moluccelloides, with dense simple long trichomes on the apex; P. Glabrous nutlet of P. franchetiana. A–B, photographed by C.-L. Xiang; C–P, photographed by Y. Zhao. |
Some characters are found to be confined to a few species. For example, campanulate or infundibular calyx tubes (Fig. 4H) only occur in P. isochila (Pazij & Vvied.) Salmaki and P. moluccelloides (Bunge) Salmaki, while calyx tubes in other species are tubular. Another example is that only four high-altitude endemic species in Clade II have brown to black hairs (Fig. 4N) inside the upper corolla lip. This is perhaps a morphological adaption to pollinators. To date, detailed morphological, anatomical, and palynological studies of the genus have not been conducted and available data is limited. The discovery of new and useful taxonomic characters is strongly needed for establishing a predictable infrageneric classification system.
5. Taxonomic treatmentBased on our molecular phylogenetic analyses, morphological investigation, specimen examination, as well as previous studies, we make the following nomenclatural updates, including a new section, three new combinations, and three new synonyms of Phlomoides in China.
Phlomoides Moench, Methodus: 403. 1794. – Type: P. tuberosa (L.) Moench.
= Eremostachys Bunge in Ledebour, Fl. Altaic. 2: 414. 1830. – Type: E. laciniate (L.) Bunge.
= Lamiophlomis Kudô in Mem. Fac. Sci. Taihoku Imp. Univ. 2: 210. 1929. – Type: L. rotata (Benth. ex Hook. f.) Kudô.
= Metastachydium Airy Shaw ex C.Y. Wu & H.W. Li in Acta Phytotax. Sin. 13(1): 73. 1975. – Type: M. sagittatum (Regel) C.Y. Wu & H.W. Li.
= Notochaete Benth. in Wallich, Pl. Asiat. Rar. 1: 63. 1830. – Type: N. hamosa Benth.
= Paraeremostachys Adylov, Kamelin & Makhm. in Novosti Sist. Vyssh. Rast. 23: 112. 1986. – Type: Pa. phlomoides (Bunge) Adylov, Kamelin & Makhm.
= Pseuderemostachys Popov in Novye Mem. Moskovsk. Obshch. Isp. Prir. 19: 148. 1941 ('1940'). – Type: Ps. sewerzovii (Herder) Popov.
= Pseudomarrubium Popov, Bot. Mater. Gerb. Bot. Inst. Komarova Akad. Nauk SSSR. 8: 75. 1940. – Type: Ps. eremostachydioides Popov.
Phlomoides sect. Notochaete Y. Zhao, Y. Salmaki & C.L. Xiang, comb. & stat. nov.钩萼草组(新拟) – Type: Phlomoides hamosa (Benth.) Mathiesen (≡ Notochaete hamosa Benth.).
Species in this section. – P. hamosa (Benth.) Mathiesen, P. longiaristata (C.Y. Wu & H.W. Li) Salmaki.
Notes. – Phlomoides hamosa and P. longiaristata were included in the former genus Notochaete (Li and Hedge, 1994) until Mathiesen et al. (2011) and Salmaki et al. (2012a) transferred them to Phlomoides. This section is characterized by having hooked calyx lobes (Fig. 4J), hooked bracts, and flowers less than 1 cm long (vs. calyx lobes and bracts abruptly or gradually narrowed to a short spinose apex (Figs. 1P and 4I), and flowers 1.5–5 cm long).
Phlomoides taronensis (C.Y. Wu) Y. Zhao & C.L. Xiang, comb. nov. & stat. nov.独龙草糙苏(新拟) ≡ Phlomis forrestii Diels. var. taronensis C.Y. Wu, Fl. Yunnan. 1: 612. 1977. – Type: CHINA. Yunnan Province: Gongshan, Taron-Taru divide, valley of Bucahwang, margin of bamboo thickets, elev. 2600 m, 3 September 1938. T.T. Yu 20094 (Holotype: KUN, 2008620!; Isotypes: A, 00001406!, PE, 00031197!).
Wu et al. (1977) considered this a variety of Phlomoides forrestii (= Phlomis forrestii) and differentiated from the typical variety (P. forrestii var. forrestii) by its ovate or elliptic floral leaves, subcordate, rounded or wedge-shaped base, the margin callous-crenate or toothed (vs. floral leaves ovate to elliptic, base cordate, margin crenate or serrate-crenate). At the same time, on one duplicate of the type specimen (PE, 00031197!), Wu et al. (1977) also commented that the variety taronensis is morphologically similar to P. melanantha (Diels) Kamelin & Makhm. (= Phlomis melanantha Diels), but can be distinguished by having stellate hairs on upper stems, and double-toothed calyx teeth. Phlomoides melanantha has glabrous stems and emarginated calyx apices. Originally, Wu also considered this collection a new species. On the holotype (KUN, 2008620!) duplicate (PE, 00031197!) sheets, he proposed a name "Phlomis taronensis C.Y. Wu sp. nov." on 22 April 1964, but subsequently, he published it as a variety of Phlomis forrestii (Wu et al., 1977). In the updated treatment of Lamiaceae in Flora of China (Li and Hedge, 1994), the variety taronensis was transferred to the typical variety without any explanation.
In this study, all three morphologically similar species (Phlomoides taronensis, P. megalantha, P. forrestii) were included for analyses. Phylogenetically, those species are grouped in Clade II (Fig. 2), but P. taronensis is sister to a subclade consisting of P. burmanica, P. melanantha, P. forrestii and P. atropurpurea. Based on our field investigations as well as specimen examinations, P. taronensis has lanceolate bracts (vs. rigid, subulate bracts in P. megalantha and P. forrestii). We here propose that Phlomis forrestii var. taronensis be elevated to species and formally transfer it to Phlomoides.
Phlomoides nyalamensis (H.W. Li) Y. Zhao & C.L. Xiang, comb. nov.聂拉木草糙苏(新拟) ≡ Phlomis nyalamensis H.W. Li in Fl. Xizang. 4: 157. 1985. – Type (designated here by Y. Zhao & C.L. Xiang): CHINA. Xizang: Nyalam County, Zhangmu, on the way from Lixin to Xuebugang, open spaces in forests, elev. 2700–2800 m, 29 June 1975, Qinghai-Xizang Comp. Exped. 6622 (Holotype: KUN, 1218985!; Isotype: KUN, 1218984!).
Li (1985) published the name based on specimens collected from Nyalam, Tibet (Xizang), China, and placed the species within Phlomis. Based on molecular phylogenetic studies (Pan et al., 2009; Mathiesen et al., 2011; Salmaki et al., 2012a), Xiang et al. (2014) proposed 11 combinations for Chinese Phlomoides but omitted this species. Here we propose a new combination.
Phlomoides nana (C.Y. Wu) Y. Zhao & C.L. Xiang, comb. nov.侏儒草糙苏(新拟) ≡ Phlomis nana C.Y. Wu, Fl. Xizang. 4: 154. 1985. – Type (designated here by Y. Zhao & C.L. Xiang): CHINA. Xizang Province: Nyalam County, Zhangmu, open spaces in forests, elev. 2700–2800 m, 14 August 1972, Tibetan Medic. Herb. Exped. 1180 (Holotype: PE, 0950649!; Isotype: PE, 0950648!).
Morphologically, Phlomoides nana is similar to P. nyalamensis and P. macrophylla, sharing similar emarginate calyx teeth and an upper corolla with brown to black hairs. But P. nana is readily distinguished from the latter two species by having short stems (shorter than 50 cm vs. stems usually 1–2 m tall).
Phlomoides dentosa (Franch.) Kamelin & Makhm. Bot. Zhurn. (Kiev) 75: 245. 1990. ≡ Phlomis dentosa Franch. Nouv. Arch. Mus. Hist. Nat., sér. 2 6: 123. 1883. – Type: CHINA. Inner Mongolia Province: June 1886, A. David 2731 (K, 000928261!).
= Phlomoides similis (Tscherneva) Kamelin & Makhm. Bot. Zhurn. (Kiev) 75: 243. 1990. ≡ Phlomis similis Tscherneva Rast. Tsentr. Azii 75: 243. 1990. syn. nov. – Type: CHINA. Qinghai Province: in fluxu superiore fl. Hoangho, oasis Guj-duj secus canales irrigatorios, elev. 2100 m, 14 June 1880, N.M. Przewalski s.n. (Holotype: LE, 01043000!; Isotype: LE, 01043001!).
In the protologue, Tscherneva (1970) stated that Phlomoides similis was morphologically similar to P. younghusbandii (Mukerjee) Kamelin & Makhm. and P. admirabilis (Tscherneva) Kamelin & Makhm., but differs by having triangular radical leaves with a cordate base and broad stems. However, we cannot find any differences between the type specimen of P. similis and P. dentosa. Geographically, P. similis is only known from the type locality (Qinghai Province), but P. dentosa is widely distributed in northern China (Inner Mongolia, Gansu and Qinghai). Here we synonymize the former species under P. dentosa.
Phlomoides dentosa (Franch.) Kamelin & Makhm. var. glabrescens C.L. Xiang & H. Peng ≡ Phlomis dentosa var. glabrescens Danguy, Bull. Mus. Natl. Hist. Nat. 17(5): 345. 1911. – Type: CHINA. Gansu Province: Si-Ning-Fou, elev. 2400 m, 18 July 1908, Anonymous 979 (P, 03284131!).
= Phlomoides admirabilis (Tscherneva) Kamelin & Makhm. ≡ Phlomis admirabilis Tscherneva, Rast. Tsentr. Azii 5: 56. 1970. syn. nov. – Type: CHINA. Gansu Province: near Kuan-gou-tschen, elev. 2300 m, 17 September 1901 (LE, 01041296!).
When publishing the species, Tscherneva (1970) indicated that Phlomoides admirabilis is closely related to P. dentosa var. glabrescens. Type specimens of both P. dentosa var. glabrescens and P. admirabilis were collected in Gansu, China at a similar altitude. After careful comparison, we cannot find any morphological differences based on type specimen examinations and field investigation. Thus, we treat P. admirabilis as a synonym of P. dentosa var. glabrescens.
Phlomoides breviflora (Benth.) Kamelin & Makhm. ≡ Phlomis breviflora Benth., Pl. Asiat. Rar. 1: 62. 1830. – Type: NEPAL. Gossain than (Gosainkund), Benth in Wall. Cat. Herb. Ind. n. 2066 (K, 001115039!). 短花草糙苏(新拟).
= Phlomis brevidentata H.W. Li, Fl. Xizang. 4: 157. 1985. syn. nov. – Type: CHINA. Xizang Province: Yadong, on the way from Yadong to Dingga, elev. 2850 m, 11 June 1975, Qinghai-Tibet Exped. 750283 (Holotype: KUN, 1218974!; Isotype: PE, 00835569!, 00835570!).
When publishing the species, the author noted that Phlomis brevidentata was similar to Phlomoides breviflora, but differs by having denser trichomes on the leaves, bracts and calyces, and with no appendages at the base of the posterior 2 stamens (Li, 1985). During field work, we found that trichome density varies among individuals of the same species at different life stages. Generally, younger individuals often have denser trichomes but these gradually fall off from the older leaves and stems. A probable reason that Phlomis brevidentata was considered a different species is that the type specimen was collected in the early flowering stage but the type specimen of Phlomoides breviflora was collected in the fruiting stage. As to the appendages at the base of the posterior 2 stamens, in the protologue of P. breviflora, Bentham (1830) only mentioned that "staminibus e tubo vix exsertis, filamentis nudis, " and never indicated that this species has appendages at the base of posterior stamens.
6. ConclusionsThis study presents the first molecular phylogenetic analyses of Phlomoides in which taxa representative of the entire genus are included, and the monophyly of the redefined Phlomoides s.l. is confirmed. Six major clades were recognized, but the monophyly for most previously defined sections and series was not recovered. Eight morphological characters were found to be mostly consistent with the phylogeny and to have taxonomic significance. In addition, based on molecular and morphological data, a section, three combinations as well as three synonyms are newly proposed. Future studies should focus on the entire genus to propose a taxonomic revision and reclassification for Phlomoides.
AcknowledgementsThis study was funded by the National Natural Science Foundation of China (No. 32161143015), International Partnership Program of Chinese Academy of Sciences (070GJHZ202211FN), the Natural Science Foundation of Yunnan Province (202001AS070016), the "Ten Thousand Talents Program of Yunnan" (Top-notch Young Talents Project, No. YNWR-QNBJ-2018-279), the CAS Interdisciplinary Team of the "Light of West China" program, and Yunnan Revitalization Talent Support Program "Innovation Team" project to CLX, the Iran National Science Foundation to YS (4001651), and the open research project of the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences. We thank Mr. Ming-Le Li for help with collecting plant materials. The authors thank to anonymous referees for giving some valuable comments on this paper.
Data availability statement
The datasets analyzed during the current study are available in the Figshare repository, http://10.6084/m9.figshare.23936628. Plant material and seeds used in this study can be obtained upon request from the corresponding author.
CRediT authorship contribution statement
Yue Zhao: Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Ya-Ping Chen: Methodology, Investigation, Formal analysis, Data curation. Bryan T. Drew: Writing – review & editing, Writing – original draft. Fei Zhao: Formal analysis. Maryam Almasi: Resources. Orzimat T. Turginov: Resources. Jin-Fei Xiao: Resources. Abdul G. Karimi: Resources. Yasaman Salmaki: Writing – review & editing, Writing – original draft, Resources, Funding acquisition. Xiang-Qin Yu: Writing – review & editing, Writing – original draft, Investigation, Formal analysis, Data curation. Chun-Lei Xiang: Writing – review & editing, Writing – original draft, Supervision, Resources, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.04.011.
Adylov, T.A., 1987. Labiatae. In: Adylov, T.A. (Ed.), Conspectus Florae Asiae Mediae, vol. 9. Science Publishers, Tashkent pp. 6-368.
|
Adylov, T.A., Kamelin, R.V., Machmedov, A.M., 1986. Zametki o semeistve Lamiaceae 1. Nov. Sist. Vyssh. Rast., 23: 110-114. |
Albaladejo, R.G., Aguilar, J.F., Aparicio, A., et al., 2005. Contrasting nuclear-plastidial phylogenetic patterns in the recently diverged Iberian Phlomis crinita and P. lychnitis lineages (Lamiaceae). Taxon, 54: 987-998. DOI:10.2307/25065483 |
Astanova, S.B., 1984. Chromosome numbers in the species of the families Alliaceae Asteraceae Caryophyllaceae Ebenaceae Linaceae Oleaceae Lamiaceae from Tadjikistan. Bot. Zhurn., 69: 1563-1564. DOI:10.2307/1222717 |
Azizian, D., Cutler, D.F., 1982. Anatomical, cytological and phytochemical studies on Phlomis L. and Eremostachys Bunge (Labiatae). Bot. J. Linn. Soc., 85: 249-281. DOI:10.1111/j.1095-8339.1982.tb00373.x |
Baldwin, B.G., Markos, S., 1998. Phylogenetic utility of the external transcribed spacer (ETS) of 18S–26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae). Mol. Phylogenet. Evol., 10: 449-463. DOI:10.1006/mpev.1998.0545 |
Beardsley, P.M., Olmstead, R.G., 2002. Redefining Phrymaceae: the placement of Mimulus, tribe Mimuleae, and Phryma. Am. J. Bot., 89: 1093-1102. DOI:10.3732/ajb.89.7.1093 |
Bendiksby, M., Thorbek, L., Scheen, A.C., et al., 2011. An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon, 60: 471-484. DOI:10.1002/tax.602015 |
Bentham, G., 1830. Synopsis of the genera and species of Indian Labiatae enumerated in the catalogue of the collections in Dr. Wallich's charge. In: Wallich, N., (Ed.), Plantae Asiaticae Rariores, vol. vol. 1. Treuttel and Wurtz, London, pp. 62-63.
|
Bentham, G., 1832-1836. Labiatarum Genera et species. London: James Ridgway. https://bibdigital.rjb.csic.es/idurl/1/12582.
|
Bräuchler, C., Meimberg, H., Heubl, G., 2010. Molecular phylogeny of Menthinae (Lamiaceae, Nepetoideae, Mentheae) – taxonomy, biogeography and conflicts. Mol. Phylogenet. Evol., 55: 501-523. DOI:10.1016/j.ympev.2010.01.016 |
Briquet, J.V., 1897. Labiatae. In: Engler, A., Prantl, K., (Eds). Die Naturlichen Pflanzenfamilien. Engelmann, W, Berlin pp. 132-375. https://doi.org/10.5962/bhl.title.4635.
|
Cantino, P.D., 1992. Toward a phylogenetic classification of the Labiatae. In: Harley, R.M., Reynolds, T. (Eds.). Advances in Labiatae Science, Royal Botanic Gardens, Kew, Richmond pp. 27-37.
|
Celep, F., Radars, E., Drew, B.T., 2020. Two new hybrid species of Salvia (S. × karamanensis and S. × doganii) from Turkey: evidence from molecular and morphological studies. Turk. J. Bot., 44: 647-660. DOI:10.3906/bot-2007-28 |
Chen, Y.P., Li, B., Olmstead, R.G., et al., 2014. Phylogenetic placement of the enigmatic genus Holocheila (Lamiaceae) inferred from plastid DNA sequences. Taxon, 63: 355-366. DOI:10.12705/632.8 |
Chen, Y.P., Drew, B.T., Li, B., et al., 2016. Resolving the phylogenetic position of Ombrocharis (Lamiaceae), with reference to the molecular phylogeny of tribe Elsholtzieae. Taxon, 65: 123-136. DOI:10.12705/651.8 |
Chen, Y.P., Liu, A., Yu, X.L., et al., 2021. A preliminary phylogenetic study of Paraphlomis (Lamiaceae) based on molecular and morphological evidence. Plant Divers., 43: 206-215. DOI:10.1016/j.pld.2021.03.002 |
Chen, Y.P., Turdimatovich, T.O., Nuraliev, M.S., et al., 2022a. Phylogeny and biogeography of the northern temperate genus Dracocephalum s. l. (Lamiaceae). Cladistics, 28: 429-451. DOI:10.1111/cla.12502 |
Chen, Y.P., Zhao, F., Paton, A.J., et al., 2022b. Plastome sequences fail to resolve shallow level relationships within the rapidly radiated genus Isodon (Lamiaceae). Front. Plant Sci., 13: 985488. DOI:10.3389/fpls.2022.985488 |
Czerepanov, S.K., 1995. Labiatae. In: Czerepanov, S.K. (Ed.), Vascular Plants of Russia and Adjacent States (The Former USSR). Cambridge University Press, Cambridge pp. 295-305.
|
Deng, T., Nie, Z.L., Drew, B.T., et al., 2015. Does the Arcto-Tertiary biogeographic hypothesis explain the disjunct distribution of Northern Hemisphere herbaceous plants? The case of Meehania (Lamiaceae). PLoS One, 10: e0117171. DOI:10.1371/journal.pone.0117171 |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull., 19: 11-15. |
Drew, B.T., Sytsma, K.J., 2011. Testing the monophyly and placement of Lepechinia in the tribe Mentheae (Lamiaceae). Syst. Bot., 36: 1038-1049. DOI:10.1600/036364411X605047 |
Drew, B.T., Sytsma, K.J., 2012. Phylogenetics, biogeography, and staminal evolution in the tribe Mentheae (Lamiaceae). Am. J. Bot., 99: 933-953. DOI:10.3732/ajb.1100549 |
Drew, B.T., Sytsma, K.J., 2013. The South American radiation of Lepechinia (Lamiaceae): phylogenetics, divergence times and evolution of dioecy. Bot. J. Linn. Soc., 171: 171-190. DOI:10.1111/j.1095-8339.2012.01325.x |
Drew, B.T., Cacho, N.I., Sytsma, K.J., 2014. The transfer of two rare monotypic genera, Neoeplingia and Chaunostoma, to Lepechinia (Lamiaceae), and notes on their conservation. Taxon, 63: 831-842. DOI:10.12705/634.6 |
Drew, B.T., González-Gallegos, J.G., Xiang, C.L., et al., 2017. Salvia united: the greatest good for the greatest number. Taxon, 66: 133-145. DOI:10.12705/661.7 |
Fang, L.Q., Pan, Y.Z., Gong, X., 2007. A karyomorphological study in the monotypic genus Lamiophlomis and five species in Phlomis (Lamiaceae). Acta Phytotax. Sin., 45: 627-632. DOI:10.1360/aps050192 |
Ghaffari, S.M., 2006. New or rare chromosome counts of some angiosperm species from Iran. Iran. J. Bot., 11: 185-192. |
Harley, R.M., Atkins, S., Budantsev, A.L., et al., 2004. Labiatae. In: Kadereit, J.W. (Ed.), The Families and Genera of Vascular Plants, vol. vol. 7. Springer Verlag, Berlin pp. 167-275. https://doi.org/10.1007/978-3-642-18617-2_11.
|
Hedge, I.C., 1992. A global survey of the biogeography of the Labiatae. In: Harley, R.M., Reynolds, T. (Eds.), Advances in Labiatae Science. Royal Botanic Gardens, Kew, Richmond pp. 7-17.
|
Hsuan, S.J., 1977. Phlomis. In: Wu, C.Y., Li, H.W. (Eds.), Flora Reipublicae Popularis Sinicae, vol. vol. 65. Science Press, Beijing pp. 428-478.
|
Hu, G.X., Takano, A., Drew, B.T., et al., 2018. Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann. Bot., 122: 649-668. DOI:10.1093/aob/mcy104 |
Hu, G.X., Liu, E.D., Wu, Z.K., et al., 2020. Integrating DNA sequences with morphological analysis clarifies phylogenetic position of Salvia grandifolia (Lamiaceae): an enigmatic species endemic to southwestern China. Int. J. Plant Sci., 181: 787-799. DOI:10.1086/709134 |
Kamelin, R.V., Machmedov, A.M., 1990a. Sistemaroda Phlomoides (Lamiaceae). Bot. Zhurn., 75: 241-250. |
Kamelin, R.V., Machmedov, A.M., 1990b. A new system of the genus Phlomis (Lamiaceae). Bot. Zhurn., 75: 1163-1167. |
Kearse, M., Moir, R., Wilson, A., et al., 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Knorring, O.E., 1954. Eremostachys Bunge, Phlomis L. In: Schischkin, B.K. (Ed.), Flora USSR. vol. vol. 21. Izdatel'stvo Akademii Nauk SSSR, Moskow pp. 1-108.
|
Lazkov, G.A., Sennikov, A.N., 2015. Taxonomic corrections and new records in vascular plants of Kyrgyzstan, 4. Memoranda. Soc. Fauna. Flora. Fenn, 91: 67-83. |
Li, H.W., 1985. Labiatae. In: Wu, C.Y. (Ed.), Flora Xizangica, vol. vol. 4. Science Press, Beijing pp. 98-224.
|
Li, H.W., Hedge, I.C., 1994. Lamiaceae. In: Wu, C.Y., Raven, P.H. (Eds.) Flora of China, vol. vol. 17. Beijing, Science Press; Missouri Botanical Garden Press, St. Louis pp. 50-299.
|
Li, B., Olmstead, R.G., 2017. Two new subfamilies in Lamiaceae. Phytotaxa, 313: 222-226. DOI:10.11646/phytotaxa.313.2.9 |
Li, B., Xu, W.X., Tu, T.Y., et al., 2012. Phylogenetic position of Wenchengia (Lamiaceae): a taxonomically enigmatic and critically endangered genus. Taxon, 61: 392-401. DOI:10.1002/tax.612010 |
Li, B., Cantino, P.D., Olmstead, R.G., et al., 2016. A large-scale chloroplast phylogeny of the Lamiaceae sheds new light on its subfamilial classification. Sci. Rep., 6: 34343. DOI:10.1038/srep34343 |
Li, J.C., Zhang, J.W., Zhang, D.G., et al., 2017a. Phylogenetic position of the Chinese endemic genus Heterolamium: a close relative of subtribe Nepetinae (Lamiaceae). J. Jpn. Bot., 92: 12-19. |
Li, P., Qi, Z.C., Liu, L.X., et al., 2017b. Molecular phylogenetics and biogeography of the mint tribe Elsholtzieae (Nepetoideae, Lamiaceae), with an emphasis on its diversification in East Asia. Sci. Rep., 7: 2057. DOI:10.1038/s41598-017-02157-6 |
Mathiesen, C., Scheen, A.C., Lindqvist, C., 2011. Phylogeny and biogeography of the lamioid genus Phlomis (Lamiaceae). Kew Bull., 66: 83-99. DOI:10.1007/s12225-011-9257-0 |
Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). IEEE, Piscataway pp. 45-52. https://doi.org/10.1109/GCE.2010.5676129.
|
Mint Evolutionary Genomics Consortium, 2018. Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol. Plant, 11: 1084-1096. DOI:10.1016/j.molp.2018.06.002 |
Moench, C., 1794. Methodus plantas horti botanici et agri Marburgensis, a staminum situ describendi. Officina Nova Libraria Academiae, Marburg pp. 1-780. https://doi.org/10.5962/bhl.title.304.
|
Muller, K., Muller, J., Quandt, D., 2010. PhyDe: Phylogenetic data editor, version 0.9971. Available online at: http://www.phyde.de/index.html (Accessed 23 November 2010).
|
Pan, Y.Z., Fang, L.Q., Hao, G., et al., 2009. Systematic position of Lamiophlomis and Paraphlomis (Lamiaceae) based on nuclear and chloroplast sequences. J. Syst. Evol., 47: 535-542. DOI:10.1111/j.1759-6831.2009.00050.x |
Pastore, J.F.B., Harley, R.M., Forest, F., et al., 2011. Phylogeny of the subtribe Hyptidinae (Lamiaceae tribe Ocimeae) as inferred from nuclear and plastid DNA. Taxon, 62: 1217-1329. |
Paton, A.J., 1990. A global taxonomic investigation of Scutellaria (Labiatae). Kew Bull., 45: 399-450. DOI:10.2307/4110512 |
Paton, A.J., Springate, D., Suddee, S., et al., 2004. Phylogeny and evolution of basils and allies (Ocimeae, Labiatae) based on three plastid DNA regions. Mol. Phylogenet. Evol., 31: 277-299. DOI:10.1016/j.ympev.2003.08.002 |
Popov, M.G., 1926. Phlomis vavilovii sp. n. and its allies. Contributions to the knowledge of the subgenus Phlomidopsis in Middle Asia. Bull. Inst. Pédol. Géobot. Univ. Asie. Centr., 13: 129-152. |
Popov, M.G., 1940. Opyt monografii roda Eremostachys Bunge. (An attempt of a monograph of the genus Eremostachys Bunge). Nov. Mem. Moskovsk. Obshch. Isp. Prir., 19: 1-166. |
Posada, 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol., 25: 1253-1256. DOI:10.1093/molbev/msn083 |
Rambaut, A., 2014. FigTree, v. 1.4.4. http://tree.bio.ed.ac.uk/software/figtree/. (Accessed 16 December 2019).
|
Ranjbar, M., Mahmoudi, C., 2015. A revision of Phlomoides sect. Thyrsiflorae (Lamiaceae). Webbia, 70: 237-245. DOI:10.1080/00837792.2015.1041794 |
Ranjbar, M., Mahmoudi, C., 2017. A taxonomic note on the Phlomoides sect. Filipendula (Phlomideae, Lamioideae, Lamiaceae) from Iran. Fedde. Repert., 128: 36-41. DOI:10.1002/fedr.201600025 |
Rechinger, K.H., 1980. Species novae generis Eremostachys (Labiatae). Plant Syst. Evol.: 127-131. |
Revell, L.J., 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol., 3: 217-223. DOI:10.1111/j.2041-210X.2011.00169.x |
Ronquist, F., Teslenko, M., Van, D.M.P., et al., 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol., 61: 539-542. DOI:10.1093/sysbio/sys029 |
Rose, J.F., Xiang, C.L., Sytsma, K.J., et al., 2022. A timeframe for mint evolution: towards a better understanding of trait evolution and historical biogeography in Lamiaceae. Bot. J. Linn. Soc., 200: 15-38. DOI:10.1093/botlinnean/boab104 |
Ryding, O., 2008. Pericarp structure and phylogeny of the Phlomis group (Lamiaceae subfam. Lamioideae). Bot. Jahrb. Syst., 127: 299-316. DOI:10.1127/0006-8152/2008/0127-0002 |
Salmaki, Y., Zarre, S., Ryding, O., et al., 2012a. Phylogeny of the tribe Phlomideae (Lamioideae: Lamiaceae) with special focus on Eremostachys and Phlomoides: new insights from nuclear and chloroplast sequences. Taxon, 65: 161-179. DOI:10.1002/tax.611012 |
Salmaki, Y., Zarre, S., Heubl, G., 2012b. The genus Phlomoides Moench (Lamiaceae; Lamioideae; Phlomideae) in Iran: an updated synopsis. Iran. J. Bot., 18: 207-219. DOI:10.22092/IJB.2012.101424 |
Scheen, A.C., Albert, V.A., 2009. Molecular phylogenetics of the Leucas group (Lamioideae; Lamiaceae). Syst. Bot., 34: 173-181. DOI:10.1600/036364409787602366 |
Scheen, A.C., Bendiksby, M., Ryding, O., et al., 2010. Molecular phylogenetics, character evolution and suprageneric classification of Lamioideae (Lamiaceae). Ann. Mo. Bot. Gard., 97: 191-219. DOI:10.3417/2007174 |
Sennikov, A.N., Lazkov, G.A., 2013. Taxonomic corrections and new records in vascular plants of Kyrgyzstan, 2. Memoranda. Soc. Fauna. Flora. Fenn., 89: 125-138. DOI:10.3897/BDJ.2.e1018 |
Katoh, K., Standley, D.M., 2013. MAFFT multiple sequence alignment software version 7.0: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Sun, Z.P., Zhao, Y., Bongcheewin, B., et al., 2022. Systematic placement of Elsholtzia griffithii (Nepetoideae, Lamiaceae), a new record from China. Phytotaxa, 548: 39-50. DOI:10.11646/PHYTOTAXA.548.1.3 |
Tscherneva, O.V., 1970. Phlomis. In: Grubov, V.I., Ivanina, L.I., Tscherneva, O.V. (Eds.), Rastieniya Centralnoi Azii. Po materialam Botanicheskogo Instituta im. VL Komarova (Plantae Asiae Centralis, secus materies Instituti botanici nomine VL Komarovii), vol. vol. 5. Nauka, Leningrad pp. 55-60.
|
Vargas, P., Baldwin, B.G., Constance, L., 1998. Nuclear ribosomal DNA evidence for a western North American origin of Hawaiian and South American species of Sanicula (Apiaceae). Proc. Natl. Acad. Sci. U.S.A., 95: 235-240. DOI:10.1073/pnas.95.1.235 |
Walker, J.B., Sytsma, K.J., 2007. Staminal evolution in the genus Salvia (Lamiaceae): molecular phylogenetic evidence for multiple origins of the staminal lever. Ann. Bot., 100: 375-391. DOI:10.1093/aob/mcl176 |
Walker, J.B., Sytsma, K.J., Treutlein, J., et al., 2004. Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am. J. Bot., 91: 1115-1125. DOI:10.3732/ajb.91.7.1115 |
Walker, J.B., Drew, B.T., Sytsma, K.J., 2015. Unravelling species relationships and diversification within the iconic California Floristic Province Sages (Salvia subgenus Audibertia, Lamiaceae). Syst. Bot., 40: 826-844. DOI:10.1600/036364415x689285 |
White, T.J., Bruns, T., Lee, S., et al., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M., Gelfand, D., Sninsky, J., et al. (Eds.), PCR Protocols: a Guide to Methods and Applications. Academic Press, San Diego pp. 315-322.
|
Wilson, T.C., Conn, B.J., Henwood, M.J., 2012. Molecular phylogeny and systematics of Prostanthera (Lamiaceae). Aust. Syst. Bot., 25: 341-352. DOI:10.1071/SB12006 |
Wu, C.Y., Li, H.W., 1977. Labiatae. In: Wu, C.Y., Li, H.W. (Eds.), Flora Reipublicae Popularis Sinicae, vol. vol. 65. Science Press, Beijing pp. 1-649.
|
Wu, C.Y., Li, H.W., Chen, J., et al., 1977. Lamiaceae. In: Wu, C.Y. (Ed.), Flora Yunnanica, vol. vol. 1. Science Press, Beijing pp. 497-817.
|
Xiang, C.L., Zhang, Q., Scheen, A.C., et al., 2013. Molecular phylogenetics of Chelonopsis (Lamiaceae: Gomphostemmateae) as inferred from nuclear and plastid DNA and morphology. Taxon, 62: 375-386. DOI:10.12705/622.11 |
Xiang, C.L., Dong, H.J., Hu, G.X., et al., 2014. Taxonomic notes on the genus Phlomoides (Lamiaceae: Lamioideae) from China. Plant Divers. Resour., 36: 551-560. DOI:10.7677/ynzwyj201413228 |
Xiang, C.L., Liu, Q.X., Peng, H., 2017. Lamiaceae. In: Chen, Y.Y. (Ed.), Species Catalogue of China, vol. vol. 1. Science Press, Beijing pp. 1-85. (in Chinese).
|
Xiang, C.L., Zhao, F., Cantino, P.D., et al., 2018. Molecular systematics of Caryopteris (Lamiaceae) and its allies with reference to the molecular phylogeny of subfamily Ajugoideae. Taxon, 67: 376-394. DOI:10.12705/672.7 |
Yao, G., Drew, B.T., Yi, T.S., et al., 2016. Phylogenetic relationships, character evolution and biogeographic diversification of Pogostemon s.l. (Lamiaceae). Mol. Phylogenet. Evol., 98: 184-200. DOI:10.1016/j.ympev.2016.01.020 |
Yu, X.Q., Maki, M., Drew, B.T., et al., 2014. Phylogeny and historical biogeography of Isodon (Lamiaceae): rapid radiation in south-west China and Miocene overland dispersal into Africa. Mol. Phylogenet. Evol., 77: 183-194. DOI:10.1016/j.ympev.2014.04.017 |
Zeng, C.X., Hollingsworth, P.M., Yang, J., et al., 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods, 14: 43. DOI:10.1186/s13007-018-0300-0 |
Zhao, F., Liu, E.D., Peng, H., et al., 2017. A new species of Scutellaria (Scutellarioideae, Lamiaceae) from Sichuan Province in southwest China. PeerJ, 5: e3624. DOI:10.7717/peerj.3624 |
Zhao, F., Li, B., Drew, B.T., et al., 2020. Leveraging plastomes of comparative analysis and phylogenomic inference within Scutellarioideae (Lamiaceae). PLoS One, 15: e0232602. DOI:10.1371/journal.pone.0232602 |
Zhao, F., Chen, Y.P., Salmaki, Y., et al., 2021a. An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol., 19: 2. DOI:10.1186/s12915-020-00931-z |
Zhao, F., Wu, Y.W., Drew, B.T., et al., 2021d. Systematic placement of the enigmatic Southeast Asian genus Paralamium and an updated phylogeny of tribe Pogostemoneae (Lamiaceae subfamily Lamioideae). Front. Plant Sci., 12: 646133. DOI:10.3389/fpls.2021.646133 |
Zhao, Y., Chen, Y.P., Liu, D.T., et al., 2021b. The identity of Phlomoides pararotata (Lamiaceae, Lamioideae). Phytotaxa, 524: 107-113. DOI:10.11646/phytotaxa.524.2.5 |
Zhao, Y., Chen, Y.P., Xiang, C.L., 2021c. Phlomoides liangwangshanensis (Lamiaceae), a new species from Yunnan, Southwest China. Phytotaxa, 491: 72-78. DOI:10.11646/phytotaxa.491.1 |
Zhao, Y., Chi, J.C., Chen, Y.P., et al., 2022. Three new records of Lamiaceae from China and Uzbekistan. Phytotaxa, 531: 111-119. DOI:10.11646/PHYTOTAXA.531.2.3 |
Zhao, Y., Zhao, F., Salmaki, Y., et al., 2023a. Home at last: systematic position of the genus Metastachydium (Lamiaceae) inferred from molecular phylogenetic analyses. Taxon, 72: 590-606. DOI:10.1002/tax.12935 |
Zhao, Y., Chen, Y.P., Yuan, J.C., et al., 2023b. Museomics in Lamiaceae: resolving the taxonomic mystery of the Pseudomarrubium. Curr. Plant Biol., 35–36: 100300. DOI:10.1016/j.cpb.2023.100300 |



