b. University of Chinese Academy of Sciences, Huairou District, Beijing 101408, China;
c. Plant Germplasm and Genomics Center, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China;
d. State Key Laboratory of Plant Diversity and Specility Crops, Institute of Botany, Chinese Academy of Sciences, Haidian District, Beijing 100093, China;
e. School of Pharmaceutical Sciences, Yunnan Key Laboratory of Pharmacology for Natural Products, and Yunnan College of Modern Biomedical Industry, Kunming Medical University, Kunming, Yunnan 650500, China;
f. Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Yezin, Nay Pyi Taw 05282, Myanmar
Tropical Southeast Asia is home to the second largest tropical rainforest in the world and harbors four out of 36 recognized biodiversity hotspots (Myers et al., 2000). This region consists of the continental Indochinese Peninsula and the Malay Archipelago, which is made up of more than 20, 000 islands (Lohman et al., 2011), and has experienced some of the most complex geological history in the world (de Bruyn et al., 2014; Zhang et al., 2023c). This complex history has resulted in exceptionally high plant diversity and rich endemism, for example in the Dipterocarpaceae and Orchidaceae, two families characteristic of Asian tropical rainforests. During the late Oligocene, as Australia drifted northward, it began to collide with Sundaland. The resulting volcanic activity and tectonic movements contributed to the formation of island chains connecting the regions east and west of the Wallace Line (Hall, 1998, 2011, 2013). Biogeographic studies on the genera Hedychium J. Koenig (Ashokan et al., 2022) and Artocarpus J.R. Forst. & G. Forst. (Williams et al., 2017) suggested that a variety of climatic events, including monsoonal intensification and sea-level fluctuations, have played significant roles in driving biodiversity patterns across Southeast Asia. This series of complex geological events and climatic changes provided the possibility of migration and dispersal of organisms in the region, but sampling difficulties across archipelagos have meant that some previous studies did not fully cover the distribution range of the taxa under investigation. Partly as a result of this complex history, the factors influencing biodiversity and historical patterns of species migration and dispersal across these and adjacent regions continue to be controversial (Tan et al., 2020; Zhang et al., 2023c).
Cymbidium Sw. (Orchidaceae: Epidendroideae) comprises approximately 60 species primarily distributed from Southwest China to the Malay Archipelago, extending into Sulawesi, New Guinea, and northeastern Australia. Many Cymbidium species (colloquially known as "Chinese Orchids") are prized as elegant ornamentals with delicate, fragrant flowers, and many Chinese Cymbidium species (e.g., C. ensifolium (L.) Sw., C. faberi Rolfe, C. goeringii (Rchb.f.) Rchb.f., C. kanran Makino, C. sinense (Andrews) Willd., and C. tortisepalum Fukuy.) have been cultivated for at least 2000 years, since before the time of Confucius (Liu et al., 2006; Du Puy and Cribb, 2007). Cymbidium spp. are highly vulnerable to illegal poaching and habitat loss, and are included in the Convention on International Trade in Endangered Species (CITES) and the Wild Plants Under National Key Protection in China. Further research into the phylogenetic relationships and evolutionary history of Cymbidium will benefit conservation efforts.
Cymbidium has undergone several revisions to its systematics and classification. Originally identified by European botanists in the 1600s, the first recognized Cymbidium species was Cymbidium aloifolium (L.) Sw. This was originally placed in the genus Epidendrum by Linnaeus in 1753 and later separated into the distinct genus Cymbidium by Olof Swartz. The circumscription of Cymbidium has been highly controversial since Swartz's time, with many species being transferred to other genera or remaining unresolved. Moreover, some Cymbidium species have been proposed to represent separate genera (e.g., Cyperorchis Blume and Iridorchis Blume), although these classifications have not been widely adopted. Hunt (1970) was the first to offer a revised and delimited genus, and several infrageneric classification systems followed his circumscription of Cymbidium (Liu et al., 2006, 2009; Du Puy and Cribb, 2007). In a recent revision, Liu et al. (2006) proposed a new classification system consisting of three subgenera, 16 sections, and 68 species. Subsequently, Du Puy and Cribb (2007) incorporated available phylogenetic data into a classification system consisting of 11 sections and 52 recognized species. In addition, Du Puy and Cribb (2007) suggested that 20 species and three varieties recognized by Liu et al. (2006) were narrowly circumscribed taxa requiring further study.
Because most Cymbidium phylogenies utilize relatively few DNA markers, phylogenetic relationships within the genus have not been well resolved (van den Berg et al., 2002; Yukawa et al., 2002; Zhang et al., 2002; Du Puy and Cribb, 2007; Sharma et al., 2012). Moreover, topological incongruence has been observed between phylogenies generated by nuclear ribosomal internal transcribed spacer (nrITS) and those by plastid DNA (ptDNA) (Zhang et al., 2021). The most recent plastome phylogeny strongly supported the presence of three major clades/subgenera within Cymbidium, and weakly supported the isolated species Cymbidium devonianum Paxton as the sister to the subgenus Jensoa (Zhang et al., 2023b). Meanwhile, more than ten recently-derived species represented by multiple individuals were not fully delimited, resulting from incomplete lineage sorting, plastome capture, or artificial hybridization in some high value cultivars (Zhang et al., 2023b). Plastome datasets can be used to reconstruct a robust Cymbidium phylogeny when heterogeneous samples are excluded, and this can then be used to infer the evolutionary and biogeographical history of the genus.
Cymbidium exhibits particularly high diversity and species richness across Southwest China, Indo-Burma, and the Sunda Shelf, which belong to the Indo-Burma and Sundaland biodiversity hotspots, respectively. These two biodiversity hotspot regions serve as important connections to neighboring areas, including Wallacea, the Philippines, and the Himalayas (Tan et al., 2020; Zhang et al., 2023c). Although Cymbidium is a diverse, widely-distributed genus, its biogeographical history remains unclear because phylogenetic relationships of Cymbidium have not been well resolved using nrITS and ptDNA markers (Yukawa et al., 2002; Zhang et al., 2002; van den Berg et al., 2002; Zhang et al., 2021). Previous phylogenetic analyses of Orchidaceae suggest that Cymbidium likely originated between the late Oligocene to early Miocene (Givnish et al., 2015; Li et al., 2019; Kim et al., 2020; Zhang et al., 2023a). Moreover, Du Puy and Cribb (2007) suggested that Cymbidium was unlikely to have originated from either the Himalayas or Australia, largely based on the phylogeny by Yukawa et al. (2002) including 37 taxa of Cymbidium using nrITS and ptDNA matK datasets. Considering the lack of Cymbidium fossils, the origin and dispersal history of the genus is unlikely to be resolved without a well-sampled, dated, and robust phylogeny.
Around 90% of species in subfamily Epidendroideae (ca. 22, 000 species) are epiphytic (Chase et al., 2015; Zotz et al., 2021), and epiphytism has evolved independently multiple times in this subfamily (Chomicki et al., 2015; Zhang et al., 2023a). Research suggests that epiphytism likely evolved in Southeast Asia and represents an adaptation to regional conditions in Epidendroideae (Givnish et al., 2016). Generally, epiphytic/lithophytic species have hard, thick, leathery leaves lacking protruding veins. This phenotype results from having a large volume of spongy mesophyll and is highly associated with crassulacean acid metabolism (CAM) photosynthesis, an adaptation to water-deficient environments and a driver of species diversification (Du Puy and Cribb, 2007). However, Cymbidium contains several terrestrial species, despite its the most recent common ancestor likely being epiphytic (Zhang et al., 2023a). Currently, a satisfactory evolutionary framework for studying the factors driving the transition from epiphytism to terrestrialism in Cymbidium is lacking. Studies of the epiphytic to terrestrial transition in the orchids suggest that the transition may be influenced by climatic fluctuations, which may be advantageous for niche colonization in orchids, e.g., Hexalectris Raf. (Sosa et al., 2016) and Galeandra Lindl. (Martins et al., 2018). Thus, there is a pressing need to comprehensively study the epiphytic–terrestrial transition in Cymbidium in order to shed light on the potential factors influencing it.
Here, we sought to provide a robust phylogeny to serve as the basis for understanding the evolutionary history and inferring the underlying drivers of the rich biodiversity of Cymbidium. We included complete plastome sequences from 56 Cymbidium species, representing approximately 90% of all recognized species, to construct a comprehensive phylogeny. The main goals of this study were to: 1) clarify phylogenetic relationships within Cymbidium; 2) reconstruct the evolutionary history of Cymbidium through both time and space; and 3) estimate species diversification associated with paleoclimatic factors and life forms.
2. Materials and methods 2.1. Plant sampling, DNA isolation, and sequencingOur phylogenetic analyses are based on plastome sequences from 67 species, including 56 Cymbidium species covering all 11 recognized sections (Du Puy and Cribb, 2007). We newly sequenced 17 plastomes, representing 13 Cymbidium species and four outgroups. The remaining 43 plastome sequences of Cymbidium and three species of other genera of tribe Cymbidiinae are derived from Zhang et al. (2023b). A total of 11 outgroup species were used, including ten species from nine genera of tribe Cymbidiinae (Freudenstein and Chase, 2015), as well as Cremastra appendiculata (D. Don) Makino from tribe Epidendreae. We also investigated whether adding more outgroups improves the phylogenetic resolution of Cymbidium by using the plastome dataset from Zhang et al. (2023b), which consists of 237 individuals representing 50 Cymbidium species. All newly sequenced plastomes have been submitted to GenBank (Table S1).
Leaf tissue was collected from living herbarium specimens, plants cultivated in a greenhouse, or from the wild. Genomic DNA was extracted from dry leaves using a modified CTAB method (Doyle and Doyle, 1987). Purified DNA was fragmented to 350–500 bp for library construction, following standard protocols (NEBNext Ultra II DNA Library Prep Kit for Illumina), and as described by Zeng et al. (2018). The 150 bp pair-end reads were generated using an Illumina Hi-Seq 2500 Sequencing System.
2.2. Plastome assembly and annotationClean raw reads were used for de novo assembly of complete plastomes using the GetOrganelle v.1.7.7.0 toolkit (Jin et al., 2020). To confirm the accuracy of the de novo assemblies, plastomes were visualized using Bandage v.0.9.0 (Wick et al., 2015). Plastomes were automatically annotated using CPGAVAS2 (Shi et al., 2019) and PGA (Qu et al., 2019), and subsequently manually checked and adjusted with Geneious v.2021.1.1 (Biomatters, Auckland, New Zealand), with emphasis on the start and stop codons of protein coding genes, as well as pseudogenes. Plastome maps were visualized using the Organellar Genome DRAW v.1.3.1 (Greiner et al., 2019).
2.3. Phylogenetic analysesAfter excluding one IR region, we aligned plastome sequences in MAFFT v.7.505 (Katoh and Standley, 2013), using the default options, to create a whole genome alignment dataset (WGM). This dataset was trimmed by removing gaps with trimAl v.1.2 (Capella-Gutiérrez et al., 2009) to create two matrices: whole genome alignment matrix-T1 (WGM-T1) using the "-gt 0.9 -cons 60" option, and whole genome alignment matrix-T2 (WGM-T2) using the "-nogaps" option. An additional alignment matrix (WGM-O) was created from the Cymbidium plastome dataset from Zhang et al. (2023b), but with the outgroups from this study. We also created a protein coding sequence (CDS) supermatrix dataset. For this purpose, we extracted 80 CDS genes from annotated plastomes using the "get_annotated_regions_from_gb.py" script (https://github.com/Kinggerm/PersonalUtilities). Each CDS gene was aligned using MAFFT, then all 80 CDS alignments were concatenated.
Maximum Likelihood (ML) analyses were conducted for all datasets with RAxML v.8.2.10 (Stamatakis et al., 2008), using the GTR+Γ+I model to determine the best-scoring ML tree and 1000 replicates to obtain Maximum Likelihood bootstrap support (MLBS) values of branches/nodes. MLBS values ≥ 70 were considered well supported. Bayesian Inference (BI) analyses were conducted with MrBayes v.3.2.7 (Ronquist and Huelsenbeck, 2003). The best DNA substitution model was chosen for each DNA dataset by estimating the Bayesian Information Criterion (BIC) in jModeltest 2 (Darriba et al., 2012). Markov chain Monte Carlo (MCMC) runs were conducted with 10, 000, 000 generations by sampling every 1000 generations. The average standard deviation of split frequencies for each dataset was lower than 0.005 and Potential Scale Reduction Factor of Convergence Diagnostic for the datasets was 1.00, which showed that the number of generations for the dataset was sufficient. The first 25% of the trees was discarded as burn-in, and the remaining trees were used to generate a majority-rule consensus tree. BI Posterior Probability (BIPP) values ≥ 0.95 were considered well supported. The trees were visualized using the online tool tvBOT (https://www.chiplot.online/tvbot.html) (Xie et al., 2023).
2.4. Divergence time estimatesThe plastome dataset used for divergence time estimates consisted of 170 Orchidaceae species, covering all five recognized subfamilies and 14 tribes (including 56 Cymbidium species), with Agave americana L. as the outgroup. A total of 108 plastome sequences were downloaded from GenBank (Table S1). Whole plastome sequences were aligned with one IR region excluded. ML analysis was performed under GTR+Γ+I model in RAxML v.8.2.10 (Stamatakis et al., 2008). Because Cymbidium fossils have yet to be recorded, divergence time estimates are based on Orchidaceae species from more distant genera that have representative fossils. Time calibrations in Orchidaceae widely rely on three unambiguous Orchidaceae fossils: Dendrobium winikaphyllum Conran, Bannister & Lee; Earina fouldenensis Conran, Bannister & Lee (Conran et al., 2009); and Meliorchis caribea S.R. Ramírez & al. (Ramirez et al., 2007). To set more specific and reasonable time intervals, according to the divergence time at the genus level of Orchidaceae, most of which were lower than 50.0 Ma (Givnish et al., 2015; Zhang et al., 2023a), the lower bounds of the calibration constraint were set to 50.0 Ma, conservatively. In total, four calibration constraints were set: 1) the upper and lower bounds of the Orchidaceae crown age were set to 80.0 Ma and 100.0 Ma, respectively, which is based on the crown age of Orchidaceae in Zhang et al. (2023a) (101.52 Ma, 95% HPD: 97.08–102.56 Ma) and in Givnish et al. (2016) (90.0 Ma, 95% HPD: 79.7–99.5 Ma); 2) the upper and lower bounds of the Goodyera clade crown age were set to 15.0 Ma and 50.0 Ma, respectively, which is based on a Miocene amber-encased M. caribea pollinarium from the Dominican Republic; the upper and lower bounds of the both 3) Earina-sister and 4) the Australian Dendrobium crown age were set to 20.0 Ma and 50.0 Ma, which is based on fossil leaves of Miocene orchids (Dendrobium Sw. and Earina Lindl.) from New Zealand.
The divergence times were estimated with MCMCTree in PAML v.4.10.5 (Yang, 2007). In the control file, we used the independent rates model (clock = 2) and GTR + G model (model = 7). The RootAge was set to ' < 1.28'. Two independent MCMC runs were conducted, each consisting of 400, 000 iterations, with sampling performed every 10 iterations. The first 16, 000 iterations were discarded as burn-in. The acceptance proportions for parameters varied from 31.0% to 41.0% in the two independent analyses. The mcmcfile was used for calculating the effective sample sizes (ESS) values (> 200) in Tracer v.1.7.2 (Rambaut et al., 2018), to validate the convergences. Chronograms were visualized using the online tool tvBOT (https://www.chiplot.online/tvbot.html) (Xie et al., 2023). The chronogram that preserved the five sister taxa from subtribe Cymbidiinae as outgroups was used to perform subsequent biogeographical and diversification analyses.
2.5. Ancestral area reconstructionWe used distribution information, geographic characteristics, and floristic regions of Cymbidium and five sister outgroup species (Table S1) (Turner et al., 2001; van Welzen et al., 2003; Thomas et al., 2012; Sirichamorn et al., 2014; Liu et al., 2021) to define six geographical regions: A) East Asia, mainly the Himalayas, Mountains of Southwest China, and Indo-Burma; B) India and Sri Lanka; C) Sundaland; D) the Philippines, including Palawan; E) Wallacea; and F) New Guinea, northeastern Australia, and adjacent islands. Ancestral regions and biogeographical dispersal events were inferred using R package BioGeoBEARS (Matzke, 2018). The BioGeoBEARS package included the three most commonly used models in historical biogeography: the dispersal–extinction–cladogenesis (DEC) model (Ree and Smith, 2008), the dispersal–vicariance analysis (DIVA) model (Ronquist, 1997), and the Bayesian Inference of Historical Biogeography for Discrete Areas (BayArea) model (Landis et al., 2013). Additionally, the founder-event speciation can be counted within the three models in BioGeoBEARS, which was implemented with the + j parameter. All six models were fitted to our data and the best-fit model was evaluated using Akaike Information Criterion (AIC) values. Results of the best-fit model were visualized as pie charts at each node of the chronogram, with each pie chart indicating the relative proportion of support for each geographical region received for that node.
2.6. Time-dependent diversification analysesTo estimate the diversification rate shifts of Cymbidium, the diversification rates of each clade were estimated using the chronogram in BAMM v.2.5.0 (Rabosky et al., 2013, 2014a; Rabosky, 2014; Shi and Rabosky, 2015). Based on the classification system by Du Puy and Cribb (2007), the clade-specific sampling fractions of clades Ⅰ, Ⅱ, Ⅲ, and Ⅵ were set to 1, 0.92, 0.95, and 0.85, respectively. The BAMM program was run for 50, 000, 000 iterations, sampling every 1000 iterations. Convergence of the BAMM results was verified by calculating the ESS values (> 200) using R-package 'BAMMtools v.2.1.10' (Rabosky et al., 2014b), after the first 10% of posterior samples were discarded as burn-in.
2.7. Paleoenvironmental-dependent diversification analysesGlobal climate fluctuations and plate tectonics are well-known to have impacted the evolution of Southeast Asian floras (de Bruyn et al., 2014; Zhang et al., 2023c). Here, we hypothesized that global cooling and monsoonal intensification likely influenced the diversification of Cymbidium. To test this hypothesis, four paleoenvironment-dependent diversification rate models devised by Condamine et al. (2013) were implemented in R package RPANDA v.2.2 (Morlon et al., 2016). The annual mean temperatures of the paleoenvironments were downloaded from Zachos et al. (2008). The hematite/goethite ratios representing Asian monsoonal intensity were downloaded from Clift et al. (2014).
2.8. Life form-dependent diversification analysesThe life forms of Cymbidium species were coded as either epiphytic or terrestrial based on literature and our field observations (Table S1) (Liu et al., 2006; Du Puy and Cribb, 2007). To determine whether life form is associated with changes in diversification rate, eight Binary State Speciation and Extinction (BiSSE) models (Maddison et al., 2007) were implemented in R package 'diversitree' (FitzJohn, 2012). The best model was determined according to the lowest AIC value. Finally, the ancestral life form of Cymbidium was predicted by using Brownian evolution in the "ER" model (Schluter et al., 1997) with R package 'phytools' (Revell, 2012).
3. Results 3.1. Plastome characteristics of the plastid genomesThe plastomes of the 13 Cymbidium and four outgroup species sequenced here exhibit typical quadripartite architecture, with a large single copy (LSC: 84, 179 bp–86, 879 bp), a small single copy (SSC: 13, 285 bp–18, 397 bp), and two inverted repeat (IR: 25, 269 bp–27, 122 bp) regions (Table 1). The boundaries of the LSC/IR and SSC/IR regions are consistent across the majority of the 17 species. The plastome size varies from 149, 481 bp to 159, 502 bp, and the GC content varies from 36.40% to 37.00% (Table 1). The 17 plastomes contain between 104 and 114 unique genes, including 70–80 CDS, 30 tRNA, and four rRNA genes. Among these, six full-length CDS (ndhB, rpl2, rpl23, rps7, rps19, and ycf2), three partial CDS (rpl22, rps12, and ycf1), four rRNA, and seven tRNA genes are duplicated in the IR regions.
| Species | %GC | Sequence Length (bp) | LSC (bp) | IR (bp) | SSC (bp) | Gene Content (Protein-Coding/tRNA/rRNA) | Pseudogenes/Loss |
| C. aloifolium | 36.80% | 156, 140 | 85, 298 | 27, 001 | 16, 840 | 75/30/4 | ndhA, ndhD, ndhE, ndhF, ndhH; ycf15 |
| C. atropurpureum | 36.90% | 154, 337 | 85, 786 | 26, 715 | 15, 121 | 74/30/4 | ndhA, ndhD, ndhE, ndhG, ndhH, ndhI, ycf15 |
| C. cyperifolium | 37.00% | 152, 735 | 85, 341 | 25, 598 | 16, 198 | 75/30/4 | ndhA, ndhD, ndhF, ndhG; ndhH, ycf15 |
| C. elegans | 36.70% | 156, 504 | 85, 317 | 26, 683 | 17, 821 | 80/30/4 | ycf15 |
| C. floribundum | 36.80% | 153, 930 | 84, 396 | 25, 669 | 18, 196 | 77/30/4 | ndhD, ndhF, ndhH; ycf15 |
| C. hartinahianum | 37.00% | 150, 365 | 84, 179 | 25, 785 | 14, 616 | 74/30/4 | ndhA, ndhC, ndhD, ndhF, ndhG; ndhH, ycf15 |
| C. hookerianum | 36.80% | 154, 142 | 84, 543 | 26, 372 | 16, 855 | 77/30/4 | ndhD, ndhF, ndhG; ycf15 |
| C. mannii | 37.00% | 153, 024 | 84, 666 | 26, 951 | 14, 456 | 74/30/4 | ndhA, ndhB, ndhC, ndhD, ndhF, ndhH; ycf15 |
| C. pubescens | 36.70% | 153, 386 | 85, 981 | 25, 269 | 16, 867 | 70/30/4 | ndhA, ndhB, ndhC, ndhD, ndhF, ndhG, ndhH, ndhI, ndhJ; rpl20, ycf15 |
| C. roseum | 36.80% | 155, 838 | 84, 798 | 27, 122 | 16, 796 | 77/30/4 | ndhF, ndhG, ndhI; ycf15 |
| C. sanderae | 36.80% | 156, 030 | 84, 767 | 26, 682 | 17, 899 | 80/30/4 | ycf15 |
| C. whiteae | 36.80% | 156, 319 | 84, 949 | 26, 706 | 17, 958 | 80/30/4 | ycf15 |
| C. erythrostylum | 36.90% | 152, 575 | 84, 212 | 26, 949 | 14, 465 | 71/30/4 | ndhA, ndhB, ndhC, ndhD, ndhE, ndhF; ndhG, ndhI, ndhJ, ycf15 |
| Thecostele alata | 36.40% | 154, 092 | 85, 076 | 26, 069 | 16, 878 | 72/30/4 | ndhA, ndhB, ndhD, ndhF, ndhH, ndhI, ndhJ, ndhK; ycf15 |
| Grammatophyllum speciosum | 36.90% | 156, 648 | 85, 015 | 26, 688 | 18, 257 | 80/30/4 | ycf15 |
| Eulophia graminea | 37.00% | 149, 481 | 85, 078 | 25, 559 | 13, 285 | 70/30/4 | ndhA, ndhB, ndhC, ndhD, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK; ycf15 |
| Cremastra appendiculata | 37.00% | 159, 502 | 86, 879 | 27, 113 | 18, 397 | 80/30/4 | ycf15 |
The WGM dataset was 159, 655 bp in length, including 33, 821 variable sites and 16, 265 parsimony-informative sites; the WGM-T1 dataset was 117, 771 bp in length, including 24, 860 variable sites and 12, 833 parsimony-informative sites; the WGM-T2 dataset was 93, 367 bp in length, including 16, 086 variable sites and 8175 parsimony-informative sites; and the CDS supermatrix dataset was 73, 855 bp in length, including 11, 667 variable sites and 5421 parsimony-informative sites.
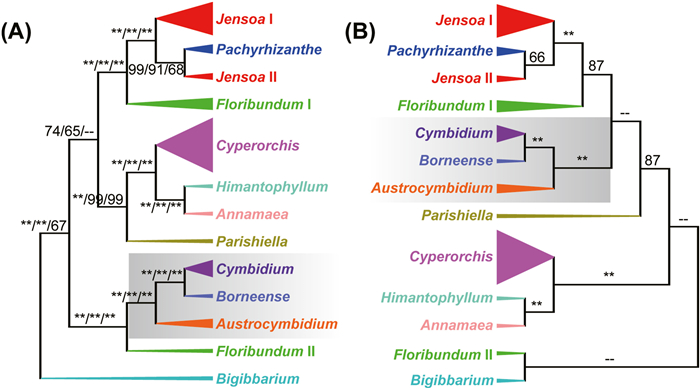
All analyses fully support monophyly of Cymbidium (MLBS = 100/BIPP = 1.00). Phylogenetic relationships between major clades within Cymbidium are well-resolved using the WGM, WGM-T1, and WGM-T2 datasets, with identical topologies (Fig. 1 and S1–S3). These datasets support four major clades, as well as nine monophyletic sections. Only two sections, Jensoa and Floribundum, are not supported as monophyletic groups. The first divergent clade (Clade I) is C. devonianum (MLBS = 100/BIPP = 1.00), which is sister to the remaining three clades. The following clade (Clade II) contains 12 species (MLBS = 100/BIPP = 1.00), corresponding to the monophyletic subclades Austrocymbidium (3 spp.) (MLBS = 100/BIPP = 1.00), Cymbidium (7 spp.) (MLBS = 100/BIPP = 1.00), Borneense (1 spp., Cymbidium borneense J.J. Wood), and Floribundum (1 spp., C. elongatum). The sister relationship between Clades III and IV is moderately supported by both the WGM (MLBS = 74/BIPP = 1.00) and WGM-T1 (MLBS = 65/BIPP = 0.99) datasets. Within Clade III, subclade Pachyrhizanthe is sister to subclade Jensoa II (MLBS = 99/BIPP = 1.00). In addition, the Pachyrhizanthe: Jensoa II cluster is sister to subclade Jensoa I (MLBS = 100/BIPP = 1.00). Subclade Floribundum I is the most basal and sister to the (Pachyrhizanthe: Jensoa II): Jensoa I cluster (MLBS = 100/BIPP = 1.00). Within Clade IV, subclade Cyperorchis is sister to a group consisting of subclade Himantophyllum and subclade Annnamaea (MLBS = 100/BIPP = 1.00). Subclade Parishiella is the most basal of Clade IV. The WGM-O dataset strongly supports four major clades (MLBS = 100), with C. devonianum being the first divergent clade (Fig. S4). With differences among the remaining three clades in the topology, Clade III is the second divergent clade (MLBS = 92), and Clade II is sister to Clade IV with weak support (MLBS = 69).

|
| Fig. 1 Maximum Likelihood phylogeny of Cymbidium based on the WGM dataset, including 56 species. MLBS and BIPP values are shown above and below branches, respectively. Scale bar represents the number of substitutions per site for the ML tree with branch length (top-left). |
However, the topology of the trees based on the CDS supermatrix dataset differs from those based on the WGM, WGM-T1, and WGM-T2 datasets (Figs. 2 and S5). Notably, the CDS supermatrix dataset does not support subclade Bigibbarium as the most basal or sister to the other three clades. Instead, the CDS supermatrix dataset supports subclade Bigibbarium as sister to subclade Floribundum II, albeit with weak support (MLBS = 56/BIPP = 0.92), and the Bigibbarium: Floribundum II cluster as sister to the other subclades (Fig. S5). In addition, the (Cymbidium: Borneense): Austrocymbidium cluster is sister to the ((Jensoa II: Pachyrhizanthe): Jensoa I): Floribundum I cluster according to the CDS supermatrix phylogeny (MLBS = 31/BIPP = 1.00) (Fig. S5), although it is sister to subclade Floribundum II according to the other three datasets (MLBS = 100/BIPP = 1.00). Finally, the CDS supermatrix dataset supports subclade Parishiella as basal to the ((Cymbidium: Borneense): Austrocymbidium): (((Jensoa II: Pachyrhizanthe): Jensoa I): Floribundum I) cluster (MLBS = 87/BIPP = 1), although it is basal to Clade IV according to the other three datasets (MLBS = 100/BIPP = 1.00).
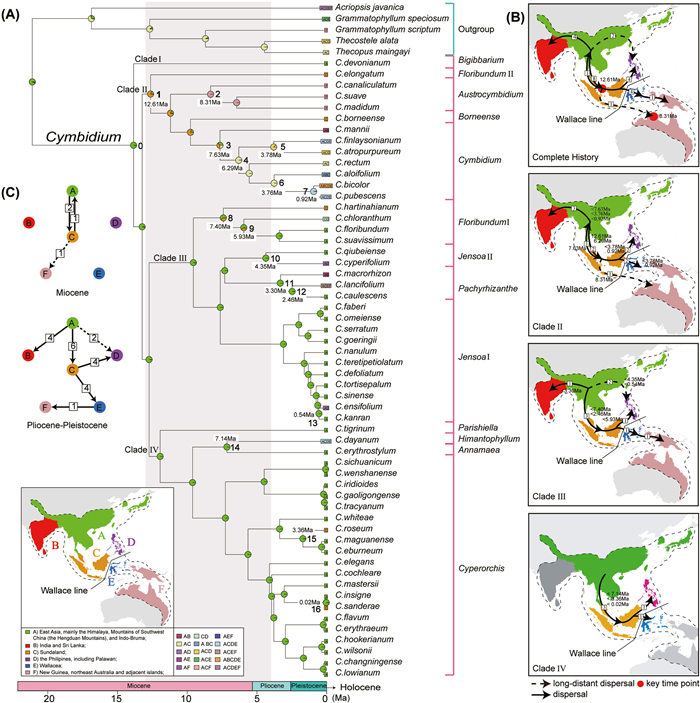
3.3. Estimation of divergence timesThe stem age of Cymbidium was estimated as 21.10 Ma (95% Highest Posterior Density (HPD): 12.20–29.80 Ma) during the early Miocene, and the crown age as 13.80 Ma (95% HPD: 8.12–19.20 Ma), which represents the split between Clade I and the other three clades (Fig. S6; Table 2). Clade II was estimated to have diverged at 13.20 Ma (95% HPD: 7.76–18.36 Ma) during the middle Miocene. Clades III and Clade IV were estimated to have diverged at 12.70 Ma (95% HPD: 7.45–17.70 Ma). Within Clade II, subclade Floribundum II diverged at 12.62 Ma (95% HPD: 7.38–17.56 Ma) and subclade Austrocymbidium diverged at 11.19 Ma (95% HPD: 6.52–15.71 Ma). The split between subclade Borneense and subclade Cymbidium was estimated to have occurred at 9.75 Ma (95% HPD: 5.62–13.91 Ma). Within Clade III, subclade Floribundum I diverged at 9.55 Ma (95% HPD: 5.38–13.94 Ma), the stem age of subclade Jensoa I was estimated to be 7.62 Ma (95% HPD: 4.15–11.57 Ma), and the split between subclade Jensoa II and Pachyrhizanthe was dated to 7.30 Ma (95% HPD: 3.93–11.17 Ma). Within Clade IV, subclade Parishiella diverged at 11.93 Ma (95% HPD: 6.94–16.72 Ma), the subclade Cyperorchis diverged at 9.60 Ma (95% HPD: 5.45–13.94 Ma), and subclades Himantophyllum and Annamaea diverged at 7.14 Ma (95% HPD: 3.54–11.27 Ma).
| Clade | Stem age | 95% HPD of stem age | Crown age | 95% HPD of crown age |
| Cymbidium | 21.10 | 12.23–29.84 | 13.83 | 8.12–19.23 |
| Clade I | 13.83 | 8.12–19.23 | 13.83 | 8.12–19.23 |
| Clade II | 13.22 | 7.76–18.36 | 12.62 | 7.38–17.56 |
| Subclade Floribundum II | 12.62 | 7.38–17.56 | 12.62 | 7.38–17.56 |
| Subclade Austrocymbidium | 11.19 | 6.52–15.71 | 8.31 | 4.53–12.45 |
| Subclade Borneense | 9.75 | 5.62–13.91 | 9.75 | 5.62–13.91 |
| Subclade Cymbidium | 9.75 | 5.62–13.91 | 7.64 | 4.32–11.16 |
| Clade III | 12.72 | 7.45–17.70 | 9.55 | 5.38–13.94 |
| Subclade Floribundum I | 9.55 | 5.38–13.94 | 7.40 | 4.00–11.29 |
| Subclade Jensoa II | 7.30 | 3.93–11.17 | 4.35 | 1.75–7.95 |
| Subclade Pachyrhizanthe | 7.30 | 3.93–11.17 | 3.30 | 1.55–5.83 |
| Subclade Jensoa I | 7.62 | 4.15–11.57 | 3.09 | 1.63–5.04 |
| Clade IV | 12.72 | 7.45–17.70 | 11.93 | 6.94–16.72 |
| Subclade Parishiella | 11.93 | 6.94–16.72 | 11.93 | 6.94–16.72 |
| Subclade Himantophyllum | 7.14 | 3.54–11.27 | 7.14 | 3.54–11.27 |
| Subclade Annamaea | 7.14 | 3.54–11.27 | 7.14 | 3.54–11.27 |
| Subclade Cyperorchis | 9.60 | 5.45–13.94 | 7.25 | 4.04–10.79 |
The crown age of the monophyletic subclade Austrocymbidium, which is only distributed in Australia, was estimated to be 8.31 Ma (95% HPD: 4.53–12.45 Ma). The crown age of the terrestrial cluster (Jensoa II: Pachyrhizanthe): Jensoa I was estimated to be 7.62 Ma (95% HPD: 4.15–11.57 Ma). Finally, the terrestrial species Cymdidium hartinahianum and C. borneense originated at 7.40 Ma (95% HPD: 4.00–11.30 Ma) and 9.75 Ma (95% HPD: 5.62–13.91 Ma), respectively.
3.4. Reconstruction of the ancestral areaAccording to the best fit model (BAYAREALIKE + J model, AIC = 249.4) (Table S2), Cymbidium was inferred to have originated in region A (East Asia, mainly the Himalayas, Mountains of Southwest China, and Indo-Burma) during the early Miocene (node 0, Fig. 3A). In total, 25 dispersal events were identified, including four events during the Miocene and 21 during the Pliocene-Pleistocene (Fig. 3B and C).
|
| Fig. 2 Phylogenetic topologies of Cymbidium using (A) WGM and (B) CDS datasets. MLBS values from WGM, WGM-T1, WGM-T2, and CDS are shown above branches. The pentastar indicates that the MLBS value was 100 and the hyphen indicates that the MLBS value was less than 60. |
|
| Fig. 3 Ancestral area reconstruction and dispersal history of Cymbidium. (A) The BioGeoBEARS results according to the BAYAREALIKE + J model, based on the extant distribution of Cymbidium. The node age represents the time of dispersal events. The gray shadows represent the time period of peak monsoon intensity. (B) Dispersal history of Cymbidium and Clades Ⅱ, Ⅲ and Ⅵ. Arrows indicate dispersal events and direction, dotted arrows indicate long-distance dispersal events, boxed numbers represent the number of dispersal events, and red circles represent the key time point of dispersal events. (C) Dispersal history during the Miocene and Pliocene-Pleistocene periods. |
We identified the most dispersal events between regions A and C (Sundaland). These included eight emigration events from region A to Sundaland, plus one immigration event from Sundaland to region A (node 3, Fig. 3A), occurring only in the ancestor of subclade Cymbidium. We also detected four dispersal events from region A to region B (India and Sri Lanka), but none from Sundaland to region B. The species in the Philippines had two sources, including four dispersal events from Sundaland and two from region A. To the east of the Wallace Line, we detected four dispersal events from Sundaland to region E (i.e., Wallacea) and one from Wallacea to region F (New Guinea, northeast Australia, and adjacent islands) (node 12, Fig. 3A), as well as one long-distance dispersal event from Sundaland to region F (node 2, Fig. 3A).
We observed primarily unidirectional dispersal events, with only one immigration event occurring in the opposite direction from Sundaland to region A (node 3, Fig. 3A). Emigration occurred primarily from region A, with a total of 14 dispersal events observed. Only one immigration event to region A was observed. The second largest number of emigration events occurred from Sundaland, which serves as a connector between region A and adjacent regions, such as the Philippines, Wallacea, and Australia. Notably, one long-distance dispersal event was observed from Sundaland to Australia, followed by in situ diversification within Australia and the formation of the monophyletic subclade Austrocymbidium (node 2, Fig. 3A).
3.5. Estimation of time-dependent diversification ratesThe BAMM analyses failed to identify any significant rate shifts in the best diversification shift configuration (Fig. 4A). Cymbidium had a high speciation rate during the early stage in the middle Miocene and this has steadily decreased to the present day (Fig. 4A). In contrast, the extinction rate has slowly increased from origination to the present (Fig. S7B). Consequently, the net diversification rate has steadily decreased through time (Fig. S7C).
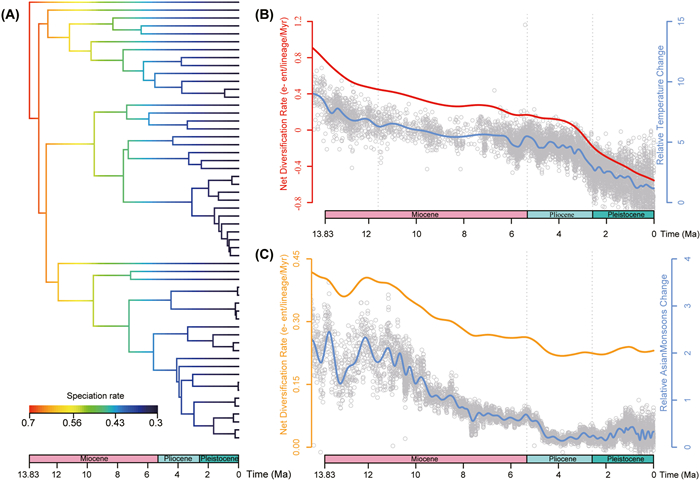
|
| Fig. 4 Diversification dynamics of Cymbidium. (A) BAMM analysis showed that Cymbidium originated in the middle Miocene with a high speciation rate followed by decreasing speciation rate to the present day. Palaeoenvironment-dependent diversification rate analysis showed that the net diversification rate of Cymbidium positively correlates with (B) paleotemperatures (data from Zachos et al. (2008)) and (C) Asian monsoon intensity (data from Clift et al. (2014)). |
To determine whether the diversification patterns differed between epiphytic and terrestrial species, we analyzed the speciation rate, extinction rate, and net diversification rate of the largest terrestrial and epiphytic clades in Cymbidium (i.e., the terrestrial cluster consisting of subclades Jensoa I, Jensoa II, and Pachyrhizanthe; the epiphytic cluster consisting of subclades Parishiella, Himantophyllum, Annamaea, and Cyperorchis). The speciation rate of the terrestrial clade was quite similar to that of the entire genus (Fig. S7A), while the extinction rate was slightly lower (Fig. S7B). The net diversification rate of the terrestrial clade has been slightly higher than the entire genus since approximately 3.8 Ma (Fig. S7C). The epiphytic clade has had a higher speciation rate and extinction rate since 10 Ma, but the net diversification rate has been lower than that of the entire genus since 1.8 Ma (Fig. S7). The BiSSE results showed that the difference in speciation rate between epiphytic and terrestrial species was not significant (Table S3, AIC = 280.51).
3.7. Estimation of paleoenvironment-dependent diversification ratesPaleoenvironment-dependent analyses indicated that both the Asian monsoon intensity and historical temperature were positively correlated with the net diversification rate of Cymbidium (Fig. 4B and C; Table S4). According to AICc values, the overall best model (Table S4, model 12, AICc = 280.719) suggested that speciation rate varied exponentially with temperature change and exhibited constant extinction rate. The second-best paleotemperature-dependent model suggested that speciation rate varied linearly with temperature change and exhibited constant extinction rate (Table S4, model 16, AICc = 280.819). The speciation rate of the second-best model was lower than that of the best model but was still positively correlated with temperature. In the monsoon-dependent model, the best (Table S4, model 3, AICc = 284.365) and second-best (Table S4, model 7, AICc = 284.376) models suggested that the positive relationship between speciation rate and monsoon intensity exhibits a similar trend with two parameters. These results demonstrate that the decreasing net diversification rates of Cymbidium over time may be related to climate fluctuation and the cooling trend since the middle Miocene.
3.8. Reconstruction of the ancestral state of life formOur analyses predict that the ancestral Cymbidium life form was epiphytic. A state shift from epiphytic to terrestrial occurred at least three times: the node at which subclade Borneense and subclade Cymbidium split (9.75 Ma), the node at which C. hartinahianum and the rest of the species in subclade Floribundum I split (7.40 Ma), and the node at which subclade Floribundum I and the (Jensoa II: Pachyrhizanthe): Jensoa I cluster split (9.55 Ma) (Fig. 5).
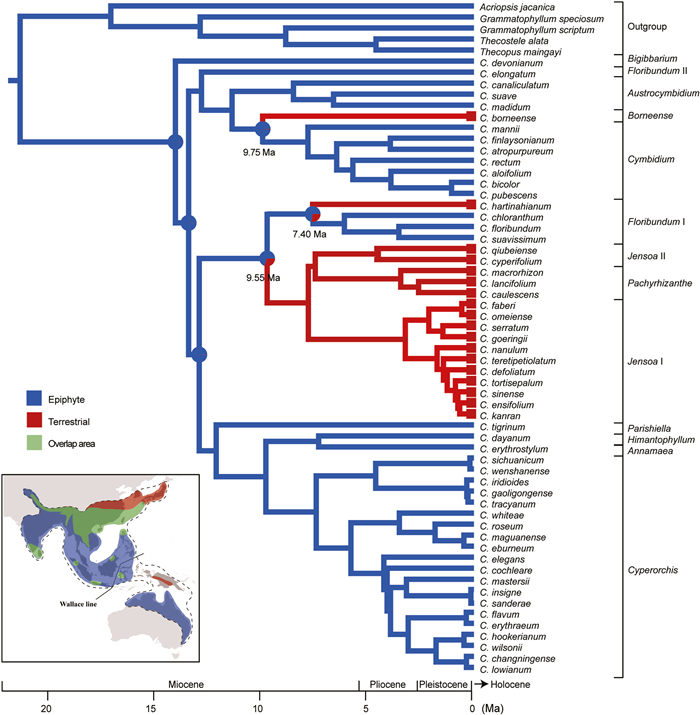
|
| Fig. 5 Reconstruction of life form evolution in Cymbidium. Characters were coded according to Liu et al. (2006), Du Puy and Cribb (2007) and field observations. Pie charts represent relative probabilities for each ancestral state shift. The distribution areas of the epiphytic species, the terrestrial species, and the overlapping areas of the two are marked on the map in blue, red, and green, respectively (bottom-left). |
Previous studies failed to fully resolve the phylogenetic relationships within Cymbidium using nrITS and ptDNA markers separately or combined (van den Berg et al., 2002; Yukawa et al., 2002; Zhang et al., 2002; Du Puy and Cribb, 2007; Sharma et al., 2012; Zhang et al., 2021). Earlier studies using nrITS and ptDNA matK data with a sample size of up to 75% of all Cymbidium species supported four to five major clades (van den Berg et al., 2002; Yukawa et al., 2002), which is inconsistent with traditional classification systems (Liu et al., 2006). In the most recent study, Zhang et al. (2021) reconstructed a phylogeny for 70 Cymbidium samples (representing about 50 recognized species) using nrITS and seven ptDNA markers. Their results showed that the phylogenetic backbone was not well resolved using either nrITS or concatenated ptDNA datasets, and that the topology was incongruent between nrITS and ptDNA phylogenies. The topological incongruence likely resulted from the relatively low number of informative sites in both the nrITS and ptDNA datasets. In the most recent Orchidaceae phylogenomic study using low-copy nuclear genes with 16 selected species of Cymbidium (Zhang et al., 2023a), however, topological incongruences were recovered between nuclear datasets (with six different number gene-sets) and plastome datasets (Fig. S8). The conflicting placement of some samples may have resulted from hybridization/introgression or incomplete lineage sorting (Yu et al., 2013; Zhang et al., 2021). Incomplete species samplings and missing data can also cause topological incongruence between analyses.
In this study, plastome phylogenies support four major Cymbidium clades, in agreement with the nrITS + matK dataset used by Yukawa et al. (2002) and the plastome datasets used by Zhang et al. (2023b). In addition, the relationships between and within the four major clades are well-resolved for the first time. Notably, we sampled 35 of the 36 taxa included in the Yukawa et al. (2002) study (we did not include Cymbidium aliciae), and found that they all cluster into the same clades as in this earlier study. In Yukawa et al. (2002), Clades I, III, and IV formed a clade, with moderate support, according to parsimony and neighbor-joining analyses. The plastome phylogeny of Zhang et al. (2023b) including 50 taxa showed that the genus consisted of three major clades and the isolated species C. devonianum. However, the phylogenetic relationships among those four clades were not well-resolved.
Increasing the sampling fraction of ingroups and sister outgroups can improve the accuracy of phylogenetic inferences (Hedtke et al., 2006; Heath et al., 2008). In this study, we included more sister outgroups (ten species from subtribe Cymbidiinae and one species from tribe Epidendreae). The phylogenies based on the WGM, WGM-T1, WGM-T2 and WGM-O datasets fully support that C. devonianum (Clade I) was the first to diverge. These results indicate that the inclusion of sufficient and close outgroups can improve phylogenetic resolution in the placement of C. devonianum. They also confirm the key role of outgroups in phylogenetic reconstruction, which is useful for further molecular phylogeny. Within Clades II and IV, C. elongatum and C. tigrinum are supported as sister to the remaining species, with support values ranging from 85 to 100 in two trimmed WGM datasets. This relationship is completely collapsed in the CDS supermatrix dataset, indicating that C. elongatum and C. tigrinum may be treated as separate clades. Therefore, it appears that Cymbidium experienced a rapid radiation during the early stages of diversification, and conserved DNA regions may fail to reveal clear evolutionary relationships among them. In the nuclear phylogenomic study (Zhang et al., 2023a), the species tree of the 299 gene-set obtained nearly congruent clades with the plastome tree (Fig. S8D), with the exception that C. devonianum fell into Clade II as sister to C. aloifolium. The placement of C. devonianum requires further investigation. Future nuclear phylogenomics research on Cymbidium should increase species sampling and be compared with plastome phylogenies to achieve a comprehensive understanding of the evolutionary history of Cymbidium.
Compared to the two systematic classifications of Cymbidium recently published (Liu et al., 2006; Du Puy and Cribb, 2007), our results are largely in agreement with the classification of Du Puy and Cribb (2007). However, in our study two sections in this classification are not supported as monophyletic: subclades Floribundum and Jensoa (Figs. 1 and 2). In addition, Du Puy and Cribb (2007) commented on ten taxonomically-confused or little-known species that were included in this study: Cymbidium caulescens, C. mannii, C. changningense, C. defoliatum, C. flavum, C. gaoligongense, C. maguannense, C. serratum, C. sichuanicum, and C. teretipetiolatum. In agreement with Du Puy and Cribb (2007), the current phylogenetic results indicate that C. defoliatum, C. mannii, and C. teretipetiolatum should be treated as separate species. Our analyses groups the remaining species into one clade, including C. caulescens as sister to C. lancifolium, C. changningense sister to C. lowianum, C. flavum sister to C. erythraeum, C. gaoligongense sister to C. tracyanum (as well as C. iridioides), C. maguannense sister to C. eburneum, C. serratum sister to C. goeringii, and C. sichuanicum sister to C. wenshanense. The phylogenetic relationships among these species may provide important evidence for taxonomic revision of Cymbidium in a future study.
4.2. Evolutionary and biogeographic history of CymbidiumWe dated the origin of Cymbidium to the early Miocene (21.10 Ma), which is older than some estimates (Givnish et al., 2015; Zhang et al., 2023a), but later than that of Kim et al. (2020). These discrepancies are likely because our robust and large-scale phylogeny is based on an increased sample size. Our ancestral area reconstruction indicated that the most likely ancestral region for Cymbidium is the region spanning northern Indo-Burma to the eastern Himalayas, as the earliest divergent species (C. devonianum) and more than 60% of extant species are widely distributed across this region. Subsequently, Cymbidium underwent dispersal from the continental Indochinese Peninsula to the Malay Archipelago, and vice versa. Previous studies, including those of van den Berg et al. (2002) and Du Puy and Cribb (2007), suggested that Cymbidium originated in Southeast Asia or from Indo-Burma to the Malay Archipelago. Our results, in contrast, strongly indicate that Cymbidium is more likely to have orignated from the eastern Himalayas to the eastern Indo-Burma region, whereas Cymbidium species in the Malay Archipelago are likely derived from Indo-Burmese ancestors.
Cymbidium exhibit uneven distribution patterns from Southeast Asia to northeast Australia. For instance, subclade Cymbidium ranges widely across Southeast Asia, whereas subclade Austrocymbidium is represented by only a few species restricted to northeast Australia. Geographically, Southeast Asia is closely connected with the Himalayas, Mountains of Southwest China (i.e, the Hengduan Mountains (Liu et al., 2022)), and Australia. The biogeographical history of Cymbidium indicates that plate tectonics and the climate dynamics of Southeast Asia created oppotunities for dispersal and shaped the biogeographic pattern of the genus. The ancestor of subclade Cymbidium dispersed to the Sunda Shelf during the middle Miocene, and experienced species contraction and expansion in mainland Asia and Borneo during the late Miocene. Beginning in the late Oligocene, the Australian Plate began to collide with Sundaland (Hall, 1998, 2009, 2011, 2013). This geological process facilitated the emergence of islands and the formation of land bridges, and the exposed Sunda Shelf provided channels for organisms to migrate and spread (Hall, 1998, 2009). In addition, plate tectonics are thought to have disrupted the Indonesian throughflow, initiating the East Asian monsoon and resulting in a moist monsoonal climate across Sundaland and East Asia, which was highly suitable for Cymbidium (Morley, 2012). These factors contributed to the divergence of subclades in Southeast Asia, such as subclades Cymbidium, Borneense, and Floribundum II. Meanwhile during this period, the ancestor of subclade Austrocymbidium underwent long-distance dispersal to Australia via island chains. This event may be attributed to historical contingency, as the dust-like seed may have swept across the island chains and arrived at Australia by chance (Givnish et al., 2016).
Biogeographical inference suggests that the dispersal of Cymbidium to the adjacent regions (Borneo, Philippines, and Sulawesi) primarily occurred during the Pliocene-Pleistocene period, including four dispersal events crossing the Wallace Line (Fig. 3C). Volcanic activity and repeated sea level fluctiations during this period may have faciliteted the dispersal of Cymbidium between mainland Asia and island Southeast Asia. Climate-driven fluctuations in sea level played a crucial role in the formation of island chains, facilitating species interchange and subsequent isolation (Cannon et al., 2009; Hanebuth et al., 2011). Additionally, volcanic activity produced island chains as stepping stones between Sundaland and adjacent regions (Hall, 2009). For example, subclade Cymbidium spread from Borneo to the Philippines and Sulawesi, forming its current biogeographic pattern in Southeast Asia within 5 Ma. In addition, C. cyperfolium and C. ensifolium dispersed from mainland Asia to the Philippines via island chains during periods of low sea level. Taken together, it is clear that a complex geographical history and fluctuating climates encouraged species migration and diversification, with sea-level fluctuations, tectonic movements, and volcanic activity also impacting species dispersal patterns and promoting isolation events (Thomas et al., 2012, 2017; Guo et al., 2015; Williams et al., 2017; Kartonegoro et al., 2022; Zhou et al., 2022).
4.3. Species diversification and evolution of life forms in CymbidiumThe eastern Himalayas and northern Indo-Bruma are not only the birthplace of Cymbidium, but also boast the highest Cymbidium species diversity, containing half of all Cymbidium species (Du Puy and Cribb, 2007). It is commonly accepted that climate changes have played an important role in species diversification (Spicer, 2017; Ye et al., 2022). Our results show that Cymbidium has exhibited a high net diversification rate, followed by a decelerating speciation rate and an accelerating extinction rate. The positive relationship between net diversification rate and global temperature and monsoon intensity indicates that Cymbidium should be well-suited to tropical regions, thriving in warm and humid environments. As such, the initial high net diversification rate may have benefited from favorable hydrothermal conditions brought on by monsoons. The eastern Himalayas and northern Indo-Burma are situated in the central region impacted by the uplift of the Himalayas (Ding et al., 2017) and the intensification of the Asian monsoon system, which is characterized by the interaction of three Asian monsoons (East Asia monsoon, South Asia monsoon, and Northwest Pacific monsoon) (Wang and LinHo, 2002; Jiang et al., 2017). A 'supermonsoon' period was established around 3.0–14.0 Ma and marked by a distinct increase in annual precipitation (Clift et al., 2014; Farnsworth et al., 2019). The impact of monsoons on other plant lineages has been extensively explored (Jiang et al., 2017; Kong et al., 2017; Ding et al., 2020; Ashokan et al., 2022; Ye et al., 2022). Several studies have reported the profound influence of Asian monsoons on the diversification patterns of various plant groups. For example, the paleotropical woody bamboo subtribe, Melocanninae, shares a similar pattern with the impact of South Asian Monsoon intensification since the early Miocene (Zhou et al., 2022). Investigations of Hedychium provide evidence of Himalayan orogeny and monsoonal intensification shaping the evolutionary trajectories of this genus (Ashokan et al., 2022). In the karst ecosystems of southern China, monsoons have been identified as a key driver of diversification in Primulina (Kong et al., 2017). All highlight the significant role of mountain uplift and monsoon intensification in shaping regional biodiversity dynamics. We speculate that monsoonal intensification during the Miocene likely played a crucial role in the diversification of Cymbidium. In this scenario, Cymbidium had a high initial speciation rate. However, after the middle Miocene, there was a continuous decline in temperature (Zachos et al., 2008; Holbourn et al., 2018; Westerhold et al., 2020). We hypothesize that this climatic change may have influenced the living environment of Cymbidium, resulting in a decrease in the speciation rate and an increase in the extinction rate, ultimately leading to a decrease in net diversification rate.
Additionally, we found that the extinction rate of the terrestrial clade is lower than that of the entire lineage (Fig. S7B), suggesting that the epiphytic to terrestrial transition may represent an adaptation to a cooling climate. Climate fluctuations during the Miocene resulted in more severe environmental conditions, including reduced precipitation (Farnsworth et al., 2019) and colder temperatures (Holbourn et al., 2018) from 6.3 to 9.4 Ma. This period coincided with the time point associated with habit changes (Fig. 5). In addition, compared to epiphytic Cymbidium species, terrestrial species exhibit adaptations to cooler conditions. For example, certain Cymbidium species are distributed across higher altitudes and latitudes, e.g., C. goeringii and C. kanran in Japan and Korea (Du Puy and Cribb, 2007) (Fig. 5). The transition from epiphytic to terrestrial habits likely conferred advantages, enabling these species to adapt to cooler environments and occupy a wider range of northern niches. Several studies on orchids and ferns have demonstrated similar evolutionary patterns. For instance, re-terrestrialization in microsoroid ferns (Polipodiaceae) allowed these plants to colonize new ecological niches (Chen et al., 2023) and the orchid genus Galeandra (Martins et al., 2018) and Hexalectris (Sosa et al., 2016) responded to the expansion of savannas in South America. These examples highlight the ecological flexibility and adaptive strategies employed by different plant lineages in response to changing environmental conditions.
Although the results from trait-dependent diversification rates analysis indicated that there were no significant differences in diversification rate between epiphytic and terrestrial species (Table S3), changing life form may have improved environmental adaptability and allowed Cymbidium species to occupy new inches. Trait-dependent diversification is detected less often when phylogenetic trees have fewer tips or younger root ages (Helmstetter et al., 2023), and methodological biases associated with our small datasets could have lead to ambiguous evolutionary histories. Within the orchid family, the adaptive evolution of traits does not necessarily accelerate the diversification rate. For instance, the change from C3 to CAM photosynthesis has been demonstrated to act as an evolutionary "gateway" trait allowing Malagasy Bulbophyllum species to widen their ecological niches, despite a lack of significant differences in the diversification rate between these two photosynthetic types (Gamisch et al., 2021).
5. ConclusionsIn this study, we generated a robust and relatively comprehensive plastome-based phylogeny of Cymbidium covering all recognized sections and including around 90% of known species. The relationships between and within the four major clades are well-resolved for the first time. Cymbidium most likely originated in the region of East Asia in the early Miocene (~21.10 Ma), mainly in the Himalayas, Mountains of Southwest China (the Hengduan Mountains), and Indo-Burma. Our findings indicate that dispersal of Cymbidium species to Southeast Asia was facilitated by various geoclimatic factors, such as the uplift of the Himalayas and subsequent tectonic movements, sea-level fluctuations, and volcanic activities. The diversification of Cymbidium coincided with monsoonal intensification in the early Miocene, while global cooling since the middle Miocene may have contributed to a steadily decreasing net diversification rate. The transition from epiphytic to terrestrial habits potentially facilitating the adaptation to cooling environments and the colonization of northern niches, yet without significant effect on diversification rate. The results of this study provide further insight into the evolutionary history of plants in Southeast Asia. However, further studies should be conducted on more taxa to better understand floristic exchanges between mainland Asia and Southeast Asia, as well as the adaptation strategies of plants to climate changes in the Southeast Asian region.
AcknowledgementsWe are grateful to all members of the Biodiversity Research Group, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences; to Jia-Lin Huang and Shi-Bao Zhang for providing plant materials; to Chun-Yan Lin, Jing Yang, Ji-Xiong Yang, Bin-Wen Yuan, and Chun-Xia Zeng for conducting the molecular experiments; to Yun-Hong Tan, Yao-Wu Xing, Zhi-Jie You, and Le Zhang for their technical assistance and discussion; and Richard T. Corlett for his valuable comments and corrections in language. This study was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31000000), The 14th Five-Year Plan of the Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (XTBG-1450101), the Science and Technology Basic Resources Investigation Program of China (2021FY100200), the Key Basic Research Program of Yunnan Province, China (202101BC070003), and the Yunnan Revitalization Talent Support Program "Young Talent" and "Innovation Team" Projects, and Ecological and Environmental Conservation Program from the Department of Ecology and Environment of Yunnan Province.
Author contributions
De-Zhu Li, Jun-Bo Yang and Wen-Bin Yu conceived of and designed the study. Xiao-Hua Jin, Ji-Dong Ya, Jun-Bo Yang, and Wen-Bin Yu contributed plant materials. Wen-Bin Yu performed the plastome assembly, annotation, and phylogenetic analyses. Xiao-Hua Jin identified habit characteristics. Hai-Yao Chen, Xin Yao, and Lu Lu performed the time calibration, ancestral area reconstruction, diversification analysis, and ancestral state reconstruction. Lin Wang generated the distribution map. Hai-Yao Chen and Wen-Bin Yu wrote the manuscript. All authors read and approved the final version of manuscript.
Data availability
Data will be made available on request.
Declaration of competing interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.03.001.
Ashokan, A., Xavier, A., Suksathan, P., et al., 2022. Himalayan orogeny and monsoon intensification explain species diversification in an endemic ginger (Hedychium: Zingiberaceae) from the Indo-Malayan Realm. Mol. Phylogenet. Evol., 170: 107440. DOI:10.1016/j.ympev.2022.107440 |
Cannon, C.H., Morley, R.J., Bush, A.B.G., 2009. The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc. Natl. Acad. Sci. U.S.A., 106: 11188-11193. DOI:10.1073/pnas.0809865106 |
Capella-Gutiérrez, S., Silla-Martínez, J.M., Gabaldón, T., 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 25: 1972-1973. DOI:10.1093/bioinformatics/btp348 |
Chase, M.W., Cameron, K.M., Freudenstein, J.V., et al., 2015. An updated classification of Orchidaceae. Bot. J. Linn. Soc., 177: 151-174. DOI:10.1111/boj.12234 |
Chen, C.C., Hyvönen, J., Schneider, H., 2023. Re-terrestrialization in the phylogeny of epiphytic plant lineages: microsoroid ferns as a case study. J. Syst. Evol., 61: 613-626. DOI:10.1111/jse.12899 |
Chomicki, G., Bidel, L.P.R., Ming, F., et al., 2015. The velamen protects photosynthetic orchid roots against UV-B damage, and a large dated phylogeny implies multiple gains and losses of this function during the Cenozoic. New Phytol., 205: 1330-1341. DOI:10.1111/nph.13106 |
Clift, P.D., Wan, S., Blusztajn, J., 2014. Reconstructing chemical weathering, physical erosion and monsoon intensity since 25 Ma in the northern South China Sea: a review of competing proxies. Earth Sci. Rev., 130: 86-102. DOI:10.1016/j.earscirev.2014.01.002 |
Condamine, F.L., Rolland, J., Morlon, H., 2013. Macroevolutionary perspectives to environmental change. Ecol. Lett., 16: 72-85. DOI:10.1111/ele.12062 |
Conran, J.G., Bannister, J.M., Lee, D.E., 2009. Earliest orchid macrofossils: early Miocene Dendrobium and Earina (Orchidaceae: Epidendroideae) from New Zealand. Am. J. Bot., 96: 466-474. DOI:10.3732/ajb.0800269 |
Darriba, D., Taboada, G.L., Doallo, R., et al., 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods, 9: 772. DOI:10.1038/nmeth.2109 |
de Bruyn, M., Stelbrink, B., Morley, R.J., et al., 2014. Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity. Syst. Biol., 63: 879-901. DOI:10.1093/sysbio/syu047 |
Ding, L., Spicer, R.A., Yang, J., et al., 2017. Quantifying the rise of the Himalaya orogen and implications for the South Asian monsoon. Geology, 45: 215-218. |
Ding, W.N., Ree, R.H., Spicer, R.A., et al., 2020. Ancient orogenic and monsoon-driven assembly of the world's richest temperate alpine flora. Science, 369: 578-581. DOI:10.1126/science.abb4484 |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull., 19: 11-15. |
Du Puy, D., Cribb, P., 2007. The genus Cymbidium. Royal Botanic Gardens, Kew.
|
Farnsworth, A., Lunt, D.J., Robinson, S.A., et al., 2019. Past East Asian monsoon evolution controlled by paleogeography, not CO2. Sci. Adv., 5: eaax1697. DOI:10.1126/sciadv.aax1697 |
FitzJohn, R.G., 2012. Diversitree: comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol., 3: 1084-1092. DOI:10.1111/j.2041-210X.2012.00234.x |
Freudenstein, J.V., Chase, M.W., 2015. Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann. Bot., 115: 665-681. DOI:10.1093/aob/mcu253 |
Gamisch, A., Winter, K., Fischer, G.A., et al., 2021. Evolution of crassulacean acid metabolism (CAM) as an escape from ecological niche conservatism in Malagasy Bulbophyllum (Orchidaceae). New Phytol., 231: 1236-1248. DOI:10.1111/nph.17437 |
Givnish, T.J., Spalink, D., Ames, M., et al., 2016. Orchid historical biogeography, diversification, Antarctica and the paradox of orchid dispersal. J. Biogeogr., 43: 1905-1916. DOI:10.1111/jbi.12854 |
Givnish, T.J., Spalink, D., Ames, M., et al., 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B-Biol. Sci., 282: 20151553. DOI:10.1098/rspb.2015.1553 |
Greiner, S., Lehwark, P., Bock, R., 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res., 47: W59-W64. DOI:10.1093/nar/gkz238 |
Guo, Y.Y., Luo, Y.B., Liu, Z.J., et al., 2015. Reticulate evolution and sea-level fluctuations together drove species diversification of slipper orchids (Paphiopedilum) in South-East Asia. Mol. Ecol., 24: 2838-2855. DOI:10.1111/mec.13189 |
Hall, R., 1998. The plate tectonics of Cenozoic SE Asia and the distribution of land and sea. In: Hall, R., Holloway, J.D. (Eds.), Biogeography and Geological Evolution of SE Asia. Backhuys Publishers, Leiden. pp. 99-131.
|
Hall, R., 2009. Southeast Asia's changing palaeogeography. Blumea, 54: 148-161. DOI:10.3767/000651909X475941 |
Hall, R., 2011. Australia-SE Asia collision: plate tectonics and crustal flow. In: Hall, R., Cottam, M.A., Wilson, M.E.J. (Eds.), The SE Asian Gateway: History and Tectonics of the Australia-Asia Collision. Geological Society, London. pp. 75-109.
|
Hall, R., 2013. The palaeogeography of Sundaland and Wallacea since the late Jurassic. J. Limnol., 72: 1-17. |
Hanebuth, T.J.J., Voris, H.K., Yokoyama, Y., et al., 2011. Formation and fate of sedimentary depocentres on Southeast Asia's Sunda Shelf over the past sea-level cycle and biogeographic implications. Earth Sci. Rev., 104: 92-110. DOI:10.1016/j.earscirev.2010.09.006 |
Heath, T.A., Hedtke, S.M., Hillis, D.M., 2008. Taxon sampling and the accuracy of phylogenetic analyses. J. Syst. Evol., 46: 239-257. |
Hedtke, S.M., Townsend, T.M., Hillis, D.M., 2006. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst. Biol., 55: 522-529. DOI:10.1080/10635150600697358 |
Helmstetter, A.J., Zenil-Ferguson, R., Sauquet, H., et al., 2023. Trait-dependent diversification in angiosperms: patterns, models and data. Ecol. Lett., 26: 640-657. DOI:10.1111/ele.14170 |
Holbourn, A.E., Kuhnt, W., Clemens, S.C., et al., 2018. Late Miocene climate cooling and intensification of southeast Asian winter monsoon. Nat. Commun., 9: 1584. DOI:10.1038/s41467-018-03950-1 |
Hunt, P.F., 1970. Notes on Asiatic orchis 5. Kew Bull., 24: 93-94. |
Jiang, C., Tan, K., Ren, M.X., 2017. Effects of monsoon on distribution patterns of tropical plants in Asia. Chin. J. Plant Ecol., 41: 1103-1112. DOI:10.17521/cjpe.2017.0070 |
Jin, J.J., Yu, W.B., Yang, J.B., et al., 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol., 21: 1-31. DOI:10.1117/1.oe.59.1.016110 |
Kartonegoro, A., Mota de Oliveira, S., van Welzen, P.C., 2022. Historical biogeography of the Southeast Asian and Malesian tribe Dissochaeteae (Melastomataceae). J. Syst. Evol., 60: 237-252. DOI:10.1111/jse.12752 |
Katoh, K., Standley, D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Kim, Y.K., Jo, S., Cheon, S.H., et al., 2020. Plastome evolution and phylogeny of Orchidaceae, with 24 new sequences. Front. Plant Sci., 11: 22. DOI:10.3389/fpls.2020.00022 |
Kong, H.H., Condamine, F.L., Harris, A.J., et al., 2017. Both temperature fluctuations and East Asian monsoons have driven plant diversification in the karst ecosystems from southern China. Mol. Ecol., 26: 6414-6429. DOI:10.1111/mec.14367 |
Landis, M.J., Matzke, N.J., Moore, B.R., et al., 2013. Bayesian analysis of biogeography when the number of areas is large. Syst. Biol., 62: 789-804. DOI:10.1093/sysbio/syt040 |
Li, Y.X., Li, Z.H., Schuiteman, A., et al., 2019. Phylogenomics of Orchidaceae based on plastid and mitochondrial genomes. Mol. Phylogenet. Evol., 139: 106540. DOI:10.1016/j.ympev.2019.106540 |
Liu, J., Lindstrom, A.J., Nagalingum, N.S., et al., 2021. Testing the causes of richness patterns in the paleotropics: time and diversification in cycads (Cycadaceae). Ecography, 44: 1606-1618. DOI:10.1111/ecog.05910 |
Liu, J., Milne, R.I., Zhu, G.F., et al., 2022. Name and scale matter: clarifying the geography of Tibetan Plateau and adjacent mountain regions. Global Planet. Change, 215: 103893. DOI:10.1016/j.gloplacha.2022.103893 |
Liu, Z.J., Chen, S.C., Cribb, P.J., 2009. Cymbidium Swartz, Nova Acta Regiae Soc. Sci. Upsal., ser. 2, 6: 70. 1799. In: Wu, Z.Y., Peter, H.R., Hong, D.Y. (Eds.), Flora of China 25. Science Press, Beijing & Missouri Botanical Garden Press, St. Louis pp. 260-280.
|
Liu, Z.J., Chen, S.C., Ru, Z.Z., 2006. The Genus Cymbidium in China. Science Press, Beijing.
|
Lohman, D.J., de Bruyn, M., Page, T., et al., 2011. Biogeography of the Indo-Australian archipelago. Annu. Rev. Ecol. Evol. Syst., 42: 205-226. DOI:10.1146/annurev-ecolsys-102710-145001 |
Maddison, W.P., Midford, P.E., Otto, S.P., 2007. Estimating a binary character's effect on speciation and extinction. Syst. Biol., 56: 701-710. DOI:10.1080/10635150701607033 |
Martins, A.C., Bochorny, T., Perez-Escobar, O.A., et al., 2018. From tree tops to the ground: reversals to terrestrial habit in Galeandra orchids (Epidendroideae: Catasetinae). Mol. Phylogenet. Evol., 127: 952-960. DOI:10.1016/j.ympev.2018.06.041 |
Matzke, N.J., 2018. nmatzke/BioGeoBEARS: BioGeoBEARS: BioGeography with Bayesian (and likelihood) Evolutionary Analysis with R Scripts (v1.1.1). Zenodo https://doi.org/10.5281/zenodo.1478250.
|
Morley, R.J., 2012. A review of the Cenozoic palaeoclimate history of Southeast Asia. In: Gower, D.J., Johnson, K.G., Richardson, J.E., et al., (Eds.), Biotic Evolution and Environmental Change in Southeast Asia. Cambridge University Press, Cambridge. pp. 79-114.
|
Morlon, H., Lewitus, E., Condamine, F.L., et al., 2016. RPANDA : an R package for macroevolutionary analyses on phylogenetic trees. Methods Ecol. Evol., 7: 589-597. DOI:10.1111/2041-210X.12526 |
Myers, N., Mittermeier, R.A., Mittermeier, C.G., et al., 2000. Biodiversity hotspots for conservation priorities. Nature, 403: 853-858. DOI:10.1038/35002501 |
Qu, X.J., Moore, M.J., Li, D.Z., et al., 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 15: 50. DOI:10.1186/s13007-019-0435-7 |
Rabosky, D.L., 2014. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One, 9: e89543. DOI:10.1371/journal.pone.0089543 |
Rabosky, D.L., Donnellan, S.C., Grundler, M., et al., 2014a. Analysis and visualization of complex macroevolutionary dynamics: an example from Australian scincid lizards. Syst. Biol., 63: 610-627. DOI:10.1093/sysbio/syu025 |
Rabosky, D.L., Grundler, M., Anderson, C., et al., 2014b. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol., 5: 701-707. DOI:10.1111/2041-210X.12199 |
Rabosky, D.L., Santini, F., Eastman, J., et al., 2013. Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nat. Commun., 4: 1958. DOI:10.1038/ncomms2958 |
Rambaut, A., Drummond, A.J., Xie, D., et al., 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol., 67: 901-904. DOI:10.1093/sysbio/syy032 |
Ramirez, S.R., Gravendeel, B., Singer, R.B., et al., 2007. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature, 448: 1042-1045. DOI:10.1038/nature06039 |
Ree, R.H., Smith, S.A., 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol., 57: 4-14. DOI:10.1080/10635150701883881 |
Revell, L.J., 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol., 3: 217-223. |
Ronquist, F., 1997. Dispersal-Vicariance Analysis: a new approach to the quantification of historical biogeography. Syst. Biol., 46: 195-203. |
Ronquist, F., Huelsenbeck, J.P., 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572-1574. |
Schluter, D., Price, T., Mooers, A.Ø., et al., 1997. Likelihood of ancestor states in adaptive radiation. Evolution, 51: 1699-1711. |
Sharma, S.K., Dkhar, J., Kumaria, S., et al., 2012. Assessment of phylogenetic inter-relationships in the genus Cymbidium (Orchidaceae) based on internal transcribed spacer region of rDNA. Gene, 495: 10-15. |
Shi, J.J., Rabosky, D.L., 2015. Speciation dynamics during the global radiation of extant bats. Evolution, 69: 1528-1545. DOI:10.1111/evo.12681 |
Shi, L.C., Chen, H.M., Jiang, M., et al., 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res., 47: W65-W73. DOI:10.1093/nar/gkz345 |
Sirichamorn, Y., Thomas, D.C., Adema, F.A.C.B., et al., 2014. Historical biogeography of Aganope, Brachypterum and Derris (Fabaceae, tribe Millettieae): insights into the origins of Palaeotropical intercontinental disjunctions and general biogeographical patterns in Southeast Asia. J. Biogeogr., 41: 882-893. DOI:10.1111/jbi.12262 |
Sosa, V., Cameron, K.M., Angulo, D.F., et al., 2016. Life form evolution in epidendroid orchids: ecological consequences of the shift from epiphytism to terrestrial habit in Hexalectris. Taxon, 65: 235-248. DOI:10.12705/652.2 |
Spicer, R.A., 2017. Tibet, the Himalaya, Asian monsoons and biodiversity - in what ways are they related?. Plant Divers., 39: 233-244. |
Stamatakis, A., Hoover, P., Rougemont, J., 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol., 57: 758-771. DOI:10.1080/10635150802429642 |
Tan, K., Pastor, L.M., Ren, M.X., 2020. Origin and evolution of biodiversity hotspots in Southeast Asia. Acta Ecol. Sin., 40: 3866-3877. |
Thomas, D.C., Hughes, M., Phutthai, T., et al., 2012. West to east dispersal and subsequent rapid diversification of the mega-diverse genus Begonia (Begoniaceae) in the Malesian archipelago. J. Biogeogr., 39: 98-113. DOI:10.1111/j.1365-2699.2011.02596.x |
Thomas, D.C., Tang, C.C., Saunders, R.M.K., 2017. Historical biogeography of Goniothalamus and Annonaceae tribe Annoneae: dispersal-vicariance patterns in tropical Asia and intercontinental tropical disjunctions revisited. J. Biogeogr., 44: 2862-2876. DOI:10.1111/jbi.13086 |
Turner, H., Hovenkamp, P., van Welzen, P.C., 2001. Biogeography of Southeast Asia and the west Pacific. J. Biogeogr., 28: 217-230. |
van den Berg, C., Ryan, A., Cribb, P.J., et al., 2002. Molecular phylogenetics of Cymbidium (Orchidaceae: Maxillarieae) sequence data from internal transcribed spacers (ITS) of ribosomal nuclearDNA and plastid matK. Lindleyana, 17: 102-111. |
van Welzen, P.C., Turner, H., Hovenkamp, P., 2003. Historical biogeography of Southeast Asia and the West Pacific, or the generality of unrooted area networks as historical biogeographic hypotheses. J. Biogeogr., 30: 181-192. |
Wang, B., LinHo, 2002. Rainy season of the Asian-Pacific summer monsoon. J. Clim., 15: 386-398. |
Westerhold, T., Marwan, N., Drury, A.J., et al., 2020. An astronomically dated record of Earth's climate and its predictability over the last 66 million years. Science, 369: 1383-1387. DOI:10.1126/science.aba6853 |
Wick, R.R., Schultz, M.B., Zobel, J., et al., 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics, 31: 3350-3352. DOI:10.1093/bioinformatics/btv383 |
Williams, E.W., Gardner, E.M., Harris, R., et al., 2017. Out of Borneo: biogeography, phylogeny and divergence date estimates of Artocarpus (Moraceae). Ann. Bot., 119: 611-627. |
Xie, J., Chen, Y., Cai, G., et al., 2023. Tree Visualization by One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res., 51: W587-W592. DOI:10.1093/nar/gkad359 |
Yang, Z.H., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol., 24: 1586-1591. DOI:10.1093/molbev/msm088 |
Ye, J.W., Tian, B., Li, D.Z., 2022. Monsoon intensification in East Asia triggered the evolution of its flora. Front. Plant Sci., 13: 1046538. |
Yu, W.B., Huang, P.H., Li, D.Z., et al., 2013. Incongruence between nuclear and chloroplast DNA phylogenies in Pedicularis section Cyathophora (Orobanchaceae). PLoS One, 8: e74828. DOI:10.1371/journal.pone.0074828 |
Yukawa, T., Miyoshi, K., Yokoyama, J., 2002. Molecular phylogeny and character evolution of Cymbidium (Orchidaceae). Bull. Natl. Mus. Nat. Sci., 28: 129-139. |
Zachos, J.C., Dickens, G.R., Zeebe, R.E., 2008. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature, 451: 279-283. DOI:10.1038/nature06588 |
Zeng, C.X., Hollingsworth, P.M., Yang, J., et al., 2018. Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods, 14: 43. |
Zhang, G.J., Hu, Y., Huang, M.Z., et al., 2023a. Comprehensive phylogenetic analyses of Orchidaceae using nuclear genes and evolutionary insights into epiphytism. J. Integr. Plant Biol., 65: 1204-1225. DOI:10.1111/jipb.13462 |
Zhang, G.Q., Chen, G.Z., Chen, L.J., et al., 2021. Phylogenetic incongruence in Cymbidium orchids. Plant Divers., 43: 452-461. |
Zhang, L., Huang, Y.W., Huang, J.L., et al., 2023b. DNA barcoding of Cymbidium by genome skimming: call for next-generation nuclear barcodes. Mol. Ecol. Resour., 23: 424-439. DOI:10.1111/1755-0998.13719 |
Zhang, L.G., Li, X.Q., Jin, W.T., et al., 2023c. Asymmetric migration dynamics of the tropical Asian and Australasian floras. Plant Divers, 45: 20-26. |
Zhang, M.Y., Sun, C.Y., Hao, G., et al., 2002. A preliminary analysis of phylogenetic relationships in Cymbidium (Orchidaceae) based on nrITS sequence data. Acta Bot. Sin., 44: 588. |
Zhou, M.Y., Liu, J.X., Ma, P.F., et al., 2022. Plastid phylogenomics shed light on intergeneric relationships and spatiotemporal evolutionary history of Melocanninae (Poaceae: Bambusoideae). J. Syst. Evol., 60: 640-652. DOI:10.1111/jse.12843 |
Zotz, G., Weigelt, P., Kessler, M., et al., 2021. EpiList 1.0: a global checklist of vascular epiphytes. Ecology, 102: e03326. |



