b. University of Chinese Academy of Sciences, Beijing 100093, China;
c. China National Botanical Garden, Beijing 100093, China
Orchidaceae are among the largest families of angiosperms, encompassing over 700 genera and more than 30, 000 species worldwide according to the Catalogue of Life (https://www.catalogueoflife.org/). The family is distributed across all continents except Antarctica and in extremely dry deserts (Fig. 1; Pridgeon et al., 2005; Chase et al., 2003; POWO, 2024). Many orchid taxa, such as Cattleya, Cymbidium, Dendrobium, and Paphiopedilum, play significant roles in horticulture, while species, such as Cremastra appendicula, Dendrobium officinale, and Gastrodia elata, possess vital medicinal value. All orchid species are listed in the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) appendices I or II (https://cites.org/), highlighting their conservation importance. Orchidaceae exhibit a variety of habits and ecological preferences, including terrestrial, epiphytic, climbing, and mycoheterotrophic lifestyles. This diverse array of characteristics represents an ideal case for the study of evolution and diversification (Jersakova et al., 2006; Lin et al., 2015; Kikuchi et al., 2020; Thompson et al., 2023).
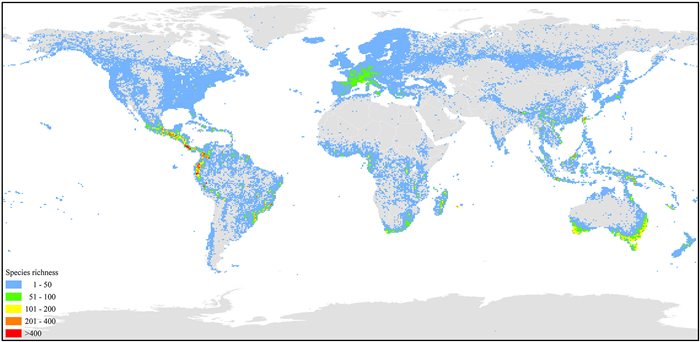
|
| Fig. 1 Species richness of georeferenced records in Orchidaceae, based on data of GBIF (GBIF.org, 2024). |
Historically, orchid taxonomy has heavily relied on the morphology of reproductive organs, such as pollinia and the lip (Dressler, 1993). Numerous frameworks have attempted to trace orchid evolution from the "three fertile anthers group" of apostasioids (Apostasia and Neuwiedia) to the "two fertile anthers group" of cypripedioids (Cypripedium, Mexipedium, Paphiopedilum, Phragmipedium, and Selenipedium), followed by the monandrous orchids (Epidendroideae, Orchidoideae, and Spiranthoideae) (Pfitzer, 1887; Schlecter, 1926; Dressler and Dodson, 1960; Dressler, 1993). However, significant discrepancies often exist among various classification systems proposed on the basis of morphology (Pridgeon et al., 2005; Chase et al., 2003, 2015).
Molecular systematics has greatly revised the phylogenetic placement of many tribes and genera within Orchidaceae (Judd et al., 1993; Kocyan et al., 2004; Sosa, 2008; Raskoti et al., 2016). Phylogenetic analysis based on rbcL gene sequences subdivide Orchidaceae into five major clades, subsequently recognized as five subfamilies in the widely-accepted classification system: Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae, and Epidendroideae (Cameron et al., 1999; Chase et al., 2003). Further advances in sequencing technology have resulted in a comprehensive phylogenetic framework for Orchidaceae (Chase et al., 2015), although unresolved issues persist, particularly concerning relationships within and between tribes such as Cymbidieae, Epidendreae, and Vandeae (Deng et al., 2015; Li et al., 2016, 2019b; Serna-Sanchez et al., 2021; Perez-Escobar et al., 2024; Zhang et al., 2023). Genomic data have generated numerous phylogenetic hypotheses and biogeographical inferences for Orchidaceae (Deng et al., 2015; Freudenstein and Chase, 2015; Givnish et al., 2015; Fernandez et al., 2019; Li et al., 2019b; Perez-Escobar et al., 2021, 2024; Serna-Sanchez et al., 2021; Zhang et al., 2023a, 2024), many of which have significantly clarified phylogenetic relationships within Orchidaceae. However, many previously recognized genera have been found to be non-monophyletic, leading to proposals for alternative generic delimitations (Zhang et al., 2015).
Orchid phylogenetics has also benefited from high-throughput sequencing technologies. Today, plant phylogenetic investigations routinely utilize organellar (plastid and mitochondria) genomic data and transcriptomic data (Wickett et al., 2014; Johnson et al., 2016; Bazinet et al., 2017; Unruh et al., 2018; Jin et al., 2021; Xia et al., 2022; Liu et al., 2023; Perez-Escobar et al., 2024; Zhang et al., 2023). For example, studies have shown that mitochondrial data is suitable for populations with diverse evolutionary rates, i.e., orchids (Ran et al., 2018; Guo et al., 2020; Stull et al., 2020). Although there are concerns that the transcriptomes of different tissues raise biases in phylogenetic inference, recent studies have demonstrated that lineage transcriptomics can be effectively employed for phylogenetic analysis as the results are only slightly affected by tissue type (Cheon et al., 2020).
The mechanisms underlying orchid diversity have long fascinated researchers. Previous work has examined net diversification rates and biogeography of genera, subtribes, and tribes within Ochidaceae (Micheneau et al., 2008; Guo et al., 2012; Givnish et al., 2016; Xiang et al., 2016; Perez-Escobar et al., 2017; Gamisch et al., 2021; Lai et al., 2021). Comprehensive studies of the biogeography and diversification of Orchidaceae require a robust phylogenetic framework, preferably resolved at the species level. However, efforts to resolve a phylogenetic framework of Ochidaceae at the genus level have been stymied by the pan-global distribution and exceptional diversity of the family (Givnish et al., 2016; Perez-Escobar et al., 2024).
In this review, we present a comprehensive synthesis of recent advances in phylogenetics, biogeography, and the mechanisms of diversification in Orchidaceae.
2. Phylogenetic researchPhylogenetic analyses based on plastid and nuclear genomes have advanced our understanding of orchid relationships at the subfamilial and tribal levels (Table 1; Unruh et al., 2018; Fernandez et al., 2019; Li et al., 2019b; Guo et al., 2020; Serna-Sanchez et al., 2021). Phylogenetic reconstructions based on chloroplast coding sequences (ptCDS) and mitochondrial coding sequences (mtCDS) have contrasted with previous studies based on morphology (Dressler and Dodson, 1960; Vermeulen, 1966; Garay, 1972; Dressler, 1981; Burns-Balogh and Funk, 1986; Berg et al., 2000; Chase et al., 2003), repositioning subfamilies Vanilloideae and Cypripedioideae (Li et al., 2019b), as well as designating Thaieae as a sister to the higher epidendroids (Xiang et al., 2012). However, recent research has also noted that phylogenetic trees based on nuclear data differ from those based on plastid data (Perez-Escobar et al., 2021).
| References | Subfamilies | Tribes | Subtribes | Species/genera | Datasets |
| Givnish et al. (2015) | 5 | 18 | 22 | 39/39 | 75 plastid genes |
| Freudenstein and Chase (2015) | 5 | 16 | 26 | 312/312 | 2 nuclear markers, 5 plastid markers, and 3 mitochondrial markers |
| Givnish et al. (2018) | 5 | 14 | 27 | 54/43 | 77 plastid genes |
| Li et al. (2019b) | 5 | 19 | 18 | 76/66 | 76 plastid genes and 38 mitochondrial genes |
| Kim et al. (2020) | 5 | 15 | 13 | 124/61 | 79 plastid genes |
| Serna-Sanchez et al. (2021) | 5 | 18 | 28 | 264/117 | 78 plastid genes |
| Perez-Escobar et al. (2021) | 5 | 16 | 28 | 75/68 | 292 low-copy nuclear genes and 78 plastid genes |
| Wong and Peakall (2022) | 5 | 13 | 21 | 69/48 | 633 nuclear genes |
| Zhang et al. (2023a) | 5 | 19 | 42 | 437/297 | 1450 low-copy nuclear genes |
| Perez-Escobar et al. (2024) | 5 | 17 | 40 | 448/285 | 610 to 1195 nuclear genes |
Transcriptomic data have emerged as a source of information that can confirm fundamental phylogenetic relationships in Orchidaceae, but perhaps more importantly elucidate the evolution of traits within the family (Cheon et al., 2020; Guo et al., 2020, 2023). For example, studies have used transcription data of 315 single-copy orthologous genes to confirm that Ochidaceae consists of five subfamilies (Deng et al., 2015). In addition, transcriptomes have been used to explore the phylogenetic relationships of European Ophrys and Gymnadenia, focusing on investigating the deceptive pollination mechanism in orchids (Fernandez et al., 2019). Transcriptomic data has also been used to reconstruct orchid phylogeny in a study on the evolution of epiphytism across 610 orchid species (Zhang et al., 2023).
Here we provide a classification Orchidaceae based on research since 2015 (Fig. 2). At the family level, the classification aligns with the main taxonomic systems of Orchidaceae as outlined by Chase et al. (2015). No revisions have been made to subfamilies Apostasioideae, Vanilloideae, or Cypripedioideae. However, the placement of several groups remains controversial, e.g., Orchidinae, Goodyerinae, Angraecinae, Aeridinae, and Podochileae. Thus, in the following two sections, we focus only on progress in understanding subfamilies, tribes, and subtribes of Orchidoideae and Epidendroideae. Our summary highlights several controversial taxa proposed by Chase et al. (2015), and includes updated information on orchids genera and species numbers for referring the species number provided by POWO (2024; Appendix A).
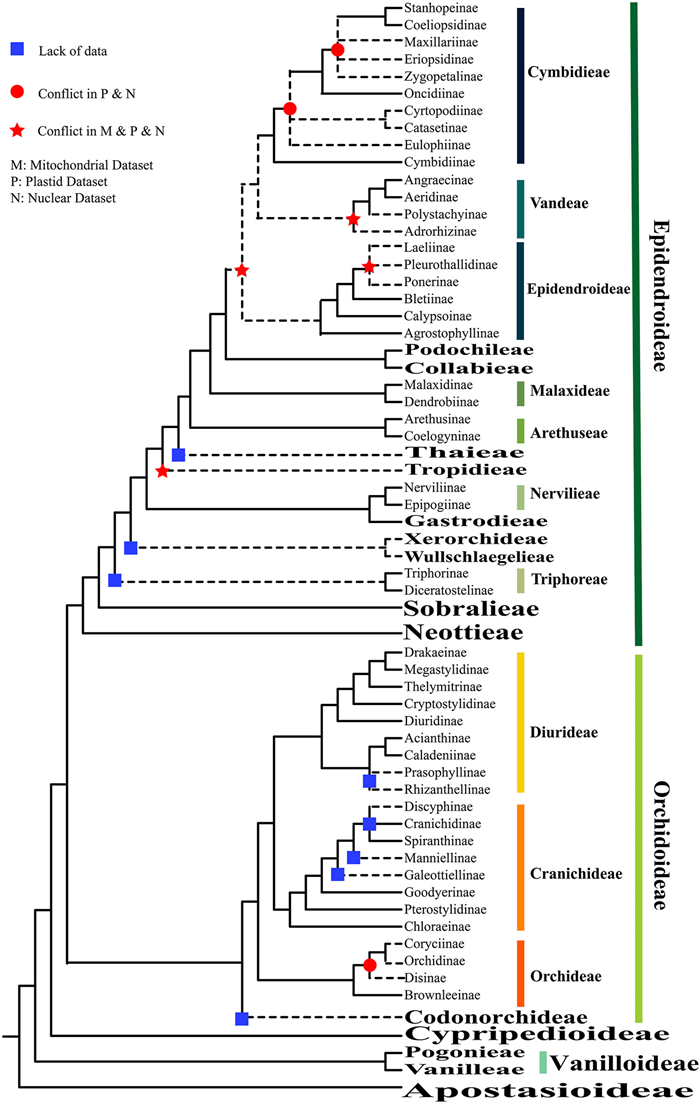
|
| Fig. 2 A 'classification summary' tree for subfamilies, tribes, and subtribes of Orchidaceae. Dashed lines denote taxa with ambiguous phylogenetic positions. Conflicts of phylogenetic position in different datasets are marked in red color. |
Orchidoideae comprises four tribes: Codonorchideae, Cranichideae, Diurideae, and Orchideae (Chase et al., 2015). Although few studies have been able to adequately sample Codonorchideae (Codonorchis as the only constituent genus), we tentatively place the tribe as a sister group to the remaining tribes (Fig. 2) (Singer et al., 2018, Serna-Sanchez et al., 2021; Zhang et al., 2023).
The tribe Orchideae has been divided into four sub-tribes: Brownleeinae, Disinae, Coryciinae, and Orchidinae (Fig. 2). Brownleeinae is recognized as the first-diverging group, however, the relationships among Disinae, Coryciinae, and Orchidinae remain unclear due to limited sampling of Coryciinae (Serna-Sanchez et al., 2021; Zhang et al., 2023). In their phylogenetic analysis of the subtribe Orchidinae, Jin et al. (2017) recognized five genera in the Ponerorhchis alliance — Hemipilia, Ponerorchis s.l., Sirindhornia, Shizhenia and Tsaiorchis — and proposed conserving one, Satyrium. Here, we recognize Ponerorchis s.l. and Hemipilia. Herminium as currently delimited is expanded to include Androcorys, Bhutanthera, Frigidorchis and Porolabium (Raskoti et al., 2016; Raskoti, 2017). The generic delimitation of Habenaria, one of the largest genera in the Orchidaceae, remains intensely debated (Kurzweil and Weber, 1992; Batista et al., 2013; Chase et al., 2015; Jin et al., 2017; Ngugi et al., 2020). Limited sampling has stymied efforts to resolve this issue.
In tribe Cranichideae, subtribes Chloraeinae, Pterostylidinae and Goodyerinae have been confidently placed as successive sister groups to the branch consisting of the remaining Cranichideae (Zhang et al., 2023; Perez-Escobar et al., 2024). All genera within subtribe Goodyerinae have been proposed to be monophyletic (Tu et al., 2021), except for Goodyera and Hetaeria (Chen et al., 2019; Smidt et al., 2021). Kim and Kim (2022) used 79 plastid-coding genes to determine the phylogenetic relationships of 18 Goodyera species. However, the phylogenetic relationships of Goodyera and Hetaeria remain unresolved (Chase et al., 2015; Hu et al., 2016; Chen et al., 2019; Smidt et al., 2021; Tu et al., 2021; Kim and Kim, 2022). Other subtribes within Cranichideae have also been limited by inadequate sampling, e.g., Galeottiellinae, Manniellinae, and Discyphinae. This lack of sampling has made it challenging to verify the precise phylogenetic positions of these subtribes and has caused ambiguity in understanding Spiranthinae, Cranichidinae and Discyphinae (Zhang et al., 2023; Perez-Escobar et al., 2024).
In tribe Diurideae, the branches of subtribes Caladeniinae and Acianthinae have been confidently identified as sister groups to each other. Prasophyllinae is the sister group to the clade comprising Caladeniinae and Acianthinae (Perez-Escobar et al., 2021; Zhang et al., 2023). The previously undetermined positions of Megastylidinae, Thelymitrinae, and Drakaeinae have been resolved, and Thelymitrinae was revealed as sister to (Megastylidinae + Drakaeinae) (Perez-Escobar et al., 2021; Serna-Sanchez et al., 2021; Zhang et al., 2023). Diuridinae and Cryptostylidinae are successive sisters to the branch of Thelymitrinae + (Megastylidinae + Drakaeinae) (Wong and Peakall, 2022; Zhang et al., 2023). To date, subtribe Rhizanthellinae has been inadequately sampled.
2.2. Phylogeny within subfamily EpidendroideaeThere are 16 tribes in subfamily Epidendroideae: Neottieae, Sobralieae, Triphoreae, Xerorchideae, Wullschlaegelieae, Gastrodieae, Nervilieae, Tropidieae, Thaieae, Arethuseae, Malaxideae, Podochileae, Collabieae, Vandeae, Cymbidieae and Epidendreae. Neottieae, Sobralieae + Triphoreae, and Nervilieae + Tropidieae are successive sisters to the remaining Epidendroideae (Givnish et al., 2015; Zhang et al., 2023). Freudenstein and Chase (2015) suggested that Tropidieae is the sister group of Wullschlaegelieae + Xerorchideae, and the Neottieae, Sobralieae, Tropidieae + (Wullschlaegelieae + Xerorchideae), Triphoreae, and Nervilieae are successive sisters to the higher epidendroids. Givnish et al. (2018) placed Neottieae, Sobralieae, and Nervilieae + Tropidieae as successive sister groups of the remaining Epidendroideae. With the same placements of Neottieae and Sobralieae, Li et al. (2019b) positioned Nervilieae as the sister group to Gastrodieae using mitochondrial genes. Neottieae and Sobralieae are successive sisters to the remaining Epidendroideae (Kim et al., 2020; Perez-Escobar et al., 2024; Zhang et al., 2023), Gastrodieae and Nervilieae stably placed as sister groups to each other (Kim et al., 2020; Wong and Peakall, 2022; Perez-Escobar et al., 2024; Zhang et al., 2023), hence the interrelationships of these tribes are stable. The positions of Triphoreae, Tropidieae, and Thaieae have been debated due to sampling and datasets. It seems that Triphoreae, Gastrodieae + Nervilieae, Tropidieae, and Thaieae successively diverged along the backbone of Orchidaceae (Kim et al., 2020; Wong and Peakall, 2022; Zhang et al., 2023; Perez-Escobar et al., 2024). The phylogenetic positions of Xerorchideae and Wullschlaegelieae, which both consist of difficult-to-sample single genera, are highly controversial (Freudenstein and Chase, 2015; Perez-Escobar et al., 2021, 2024; Zhang et al., 2023).
In the higher epidendroids, the relative phylogenetic positions of Arethuseae, Malaxideae, Podochileae, and Collabieae have been resolved (Givnish et al., 2015; Li et al., 2019b; Zhang et al., 2023; Perez-Escobar et al., 2024), although two alternative topologies for Vandeae, Epidendreae, and Cymbidieae have emerged, even between similar datasets. One topology, based on mitochondrial and nuclear data, is Vandeae + (Epidendreae + Cymbidieae) (Freudenstein and Chase, 2015; Li et al., 2019b; Perez-Escobar et al., 2021; Perez-Escobar et al., 2024; Zhang et al., 2023). An alternative topology based on transcriptome data or plastid genome data, and nuclear markers is Epidendreae + (Vandeae + Cymbidieae) (Givnish et al., 2015, 2018; Serna-Sanchez et al., 2021; Wong and Peakall, 2022). Both topologies are strongly supported. In this paper, we adopt the Epidendreae + (Vandeae + Cymbidieae) topology (Chase et al., 2015; Fig. 2). One reason why the relationships between these three tribes have been contentious is the complexity of phylogenetic relationships between subtribes within each tribe (Li et al., 2019b; Zhang et al., 2023; Perez-Escobar et al., 2024).
Generic delimitation has varied considerably in the tribe Arethuseae. Studies have determined that Arethuseae is a polyphyletic group (Goldman et al., 2001; Gravendeel et al., 2001; Chase et al., 2015, 2021; Huang et al., 2022a). Chase et al. (2021) combined 14 genera within the tribe (e.g., Dendrochilum, Pholidota and Bulleyia and others) into Coelogyne. Huang et al. (2022a) proposed a new genus, Mengzia to accommodate Bletilla sinensis based on molecular systematics and morphological characters. Nevertheless, owing to the low bootstrap values of some phylogenetic studies, relationships of the subtribe Coelogyninae require further investigation (Huang et al., 2022a).
The tribe Malaxideae consists of a large number of species and complex diversity of taxa. Although the phylogenetic relationships within the tribe have yet to be completely determined (Cameron, 2005; Chase et al., 2015), recent studies have made critical revisions. For example, Geiger (2023) placed the Hippeophyllum as the synonym of Oberonia. In addition, Li et al. (2020) proposed a new genus Blepharoglossum, subdivided from Liparis, based on morphological characters and molecular analysis.
In tribe Podochileae, Eria s.l. has been subdivided into 21 genera, including two newly recognized genera (Chase et al., 2015; Ng et al., 2018). In tribe Collabieae, morphological evidence and phylogenetic relationships within Calanthe have been used to expand Calanthe to include the species of Cephalantheropsis, Gastrorchis, Phaius, Cyanorkis and Gastorkis (Chase et al., 2020a, 2020b). This expansion has subsequently been supported by Zhou et al. (2023).
The relative positions of most of subtribes in Cymbidieae, such as Catasetinae, Cyrtopodiinae, Eulophiinae, Zygopetalinae, Eriopsidinae, and Maxillariinae, remain ambiguous. Freudenstein and Chase (2015) indicated that Eulophiinae is not monophyletic and that Catasetinae is nested within Eulophiinae. Givnish et al. (2015) suggested that Eulophiinae is sister to group containing Catasetinae and Cyrtopodiinae (Perez-Escobar et al., 2021; Zhang et al., 2023), whereas Serna-Sanchez et al. (2021) proposed that Cyrtopodiinae diverged earlier than Eulophiinae and Cyrtopodiinae was embedded within Catasetinae with low support. Molecular data support the placement of Eulophiinae as the common sister group of Catasetinae + Cyrtopodiinae (Perez-Escobar et al., 2024).
Previous studies suggested that subtribes Zygopetalinae, Eriopsidinae, and Maxillariinae diverged successively, and Maxillariinae diverged early in the Cymbidieae (Freudenstein and Chase, 2015; Givnish et al., 2015; Zhang et al., 2023). However, other studies suggested Zygopetalinae diverged early in the Cymbidieae (Perez-Escobar et al., 2021), and that Eriopsidinae is a younger group (Perez-Escobar et al., 2024). The phylogenetic placements of Cymbidiinae, Stanhopeinae, Coeliopsidinae, and Oncidiinae are not in dispute (Fig. 2).
Angraecinae and Aeridinae are monophyletic in Vandeae, respectively, but the delimitation of Adrorhizinae and Polystachyinae is currently speculative (Chase et al., 2015; Perez-Escobar et al., 2021; Serna-Sanchez et al., 2021; Zhang et al., 2023). Perez-Escobar et al. (2021) showed Polystachyinae diverged earlier, followed by Adrorhizinae (Freudenstein and Chase, 2015; Perez-Escobar et al., 2024). Zhang et al. (2023) showed that the Adrorhizinae diverged earlier, followed by Polystachyinae. In subtribe Aeridinae, Liu et al. (2020) proposed to combine Vandopsis, Diploprora, Cleisostoma and Schoenorchis into the Cleisostoma, and combine Trichoglottis into the Gastrochilus, and establish a new genus, Cymbilabia. In subtribe Angraecinae, Andriananjamanantsoa et al. (2016) suggested that Oeoniella and Sobennikoffia are nested within Angraecum, however, there are many structural differences among them; therefore, we followed the earlier classification.
For tribe Epidendreae, the phylogenetic placements are less disputed, with the exception of Ponerinae, Laeliinae, and Pleurothallidinae. Pleurothallidinae is sister group to Laeliinae and Ponerinae with high support (Zhang et al., 2023; Perez-Escobar et al., 2024). Agrostophyllinae, Calypsoinae, and Bletiinae successively diverged in Epidendreae with high support (Fig. 2) (Perez-Escobar et al., 2021, 2024; Serna-Sanchez et al., 2021; Zhang et al., 2023). Chysis is temporarily treated as a member of Bletiinae, and Coelia was incorporated into Calypsoinae (Chase et al., 2015). However, more recently, Coelia and Chysis were recognized as distinct clades with relatively strong support (Zhang et al., 2023). Risleya falls within Calypsoinae on the basis of ptCDS and mtCDS, instead of within Collabieae (Li et al., 2019b). We recommend broader sampling in the future to provide more robust phylogenetic outcomes.
3. Historical biogeography and molecular datingThe earliest divergence of monocots occurred between 185 and 139 Mya (million years ago, hereafter Mya; Kumar et al., 2017, 2022; Givnish et al., 2018; Li et al., 2019a; Shi et al., 2022). The Order Asparagales originated between 140 and 125 Mya (Givnish et al., 2018; Li et al., 2019a; Kumar et al., 2022). One early-diverging group within Asparagales included the Orchidaceae (Kumar et al., 2022). However, macrofossils of orchids remain scarce. Four fossils have been discovered, including two pollen fossils of Epidendroideae and subtribe Goodyerinae, as well as two leaf fossils of Earina and Dendrobium (Ramirez et al., 2007; Conran et al., 2009; Poinar and Rasmussen, 2017). The paucity of orchid fossils poses a further challenge to the study of temporal orchid evolution following the reconstruction of a phylogenetic framework (Givnish et al., 2016; Li et al., 2019b; Zhang et al., 2023).
Ramirez et al. (2007) inferred orchids originated during the Early Cretaceous between 76 and 84 Mya. More recent studies estimate that Orchidaceae originated between 76 and 132 Mya (Givnish et al., 2018; Li et al., 2019a; Shi et al., 2022; Zhang et al., 2023; Perez-Escobar et al., 2024). Although estimates of the age of Orchidaceae vary, these studies all suggest that orchids originated in the Cretaceous period and first diverged in the Late Cretaceous (Givnish et al., 2015, 2018; Serna-Sanchez et al., 2021), which supports the "ancient origin" of orchids. Some researchers have suggested that rapid diversification of orchids did not occur until the Cenozoic era (Givnish et al., 2015, 2018; Li et al., 2019b; Zhang et al., 2023). For example, the higher epidendroids have been shown to have diverged rapidly between 37.9 Mya and 30.8 Mya (Givnish et al., 2015). Zhang et al. (2023) indicated that most genera and species of orchids had recent origins less than 20 Mya. An additional study indicated that the majority of present-day orchid species diversity originated over the last 5 million years (Perez-Escobar et al., 2024).
Givnish et al. (2016) has proposed that the ancestor of extant orchids appeared in Australia ca. 120–102 Mya, and that repeated long- and short-distance dispersals have occurred throughout orchid history. Specifically, from Australia, Orchids spread to the Neotropics via Antarctica ca. 90 Mya, when vanilloid orchids diverged from the most recent common ancestor of orchids. Within vanilloids, both the Pogonieae and Vanilleae underwent long-distance dispersals, crossing the Pacific Ocean to New Caledonia between 64 and 59 Mya (Givnish et al., 2016). This hypothesis proposes that ancestors of Epidendroideae arose in the Neotropics ca. 64 Mya and the lower epidendroids then spread to Eurasia and Southeast Asia (Givnish et al., 2016).
An alternative biogeographical history of Orchidaceae proposes that Orchidaceae originated in Laurasia (Perez-Escobar et al., 2024). This proposal suggests that the most rapidly speciating lineages of Orchidaceae occurred in south-eastern Central America, e.g., Epidendrum and Maxillaria. In this scenario, Orchidoideae originated between 55 and 40 Mya as these plants dispersed from the Neotropics + Nearctic + Antarctic. Epidendroids were present in the Palearctic around 55–50 Mya, suggesting their earlier origin (Perez-Escobar et al., 2024).
The discrepancies between these two hypotheses can be attributed to two primary factors. One factor is that these studies used different datasets to construct phylogenetic frameworks of Orchidaceae. The phylogenetic framework of Australian origin of orchids is based on chloroplast genes (Givnish et al. (2016), whereas the phylogenetic framework of the Laurasia origin is based on a nuclear dataset (Perez-Escobar et al., 2024). Evolutionary rates between organellar data and nuclear data are known to differ, potentially introducing biases into phylogenetic frameworks (Cheon et al., 2020; Guo et al., 2023). Another factor is the scope and density of the sampling, which affects the accuracy of geographical history estimations (Givnish et al., 2016; Perez-Escobar et al., 2024).
Despite discrepancies between these two potential theories, both agree that the Neotropics (e.g., the Andes) have had profound effects on the spread and diversification of orchids (Givnish et al., 2016; Zizka, 2019; Perez-Escobar et al., 2022, 2024). Studies have shown that the Andes serve as both a key source and sink of Neotropical plant diversity (Perez-Escobar et al., 2022) and that the diversification of numerous Neotropical lineages has been significantly influenced by Andean uplift events (Perez-Escobar et al., 2017, 2022; Vitt et al., 2023). These uplift events rapidly alter topography within tropical climates, typically creating multiple microhabitats, potentially enhancing speciation rates and allowing islands within tropical mainland regions to foster higher evolutionary distinctness (Perez-Escobar et al., 2024; Vitt et al., 2023).
Phylogenetic studies of Orchidaceae genera have focused on biodiversity hotspots, elucidating orchid dispersal patterns and addressing challenges posed by species-rich taxa, e.g., Bulbophyllum, Dendrobium, and Paphiopedilum (Micheneau et al., 2008; Guo et al., 2012; Xiang et al., 2016; Perez-Escobar et al., 2017; Fernandez et al., 2019; Gamisch et al., 2021; Lai et al., 2021; Liu et al., 2023; Zhang et al., 2024). Xiang et al. (2016) indicated that Asian Dendrobium has been present in mainland Asia since the Oligocene and then spread to Malesia. Gamisch et al. (2021) proposed that Bulbophyllum dispersed from the Asian-Pacific region into Madagascar via a single long-distance dispersal event at ca. 12.9–7.0 Mya, or probably originated from similar habitats in the Asian-Pacific region. Tsai et al. (2020) explained that the taxa of Paphiopedilum in the Sundaland and Wallacea underwent repeated migration and isolation events between mainland south-eastern Asia and the Sunda Super Islands.
Several studies have examined how the biogeography of orchids in biodiversity hotspots is affected by mountain uplift events, e.g., Calochilus (Nargar et al., 2018), Cymbidium (Chen et al., 2024), Cypripedium (Liu et al., 2021; Szlachetko et al., 2021), Holcoglossum (Zhao et al., 2020), Ornithocephalus (Smidt et al., 2018), Pabstiella (Morales et al., 2021), Pleione (Wu et al., 2023). These studies have demonstrated that orchids have undergone multiple diversification and dispersal events (Lai et al., 2021; Thompson et al., 2023).
4. Diversification of OrchidaceaeOrchidaceae species, particularly their flowers and habitats, exhibit highly idiosyncratic features that appear to drive diversity. These features, which are either unique to this family or seldom found in other angiosperms, include epiphytism, CAM photosynthesis, and pollinia (Givnish et al., 2015; Zhang et al., 2023).
Epiphytic plants are integral components of numerous ecosystems, contributing greatly to global plant diversity (Ricogray and Thien, 1989; Spicer and Woods, 2022). Epiphytic orchid genera have been shown to contain more species than terrestrial genera (Gravendeel et al., 2004), and have higher speciation and extinction rates than do terrestrial orchids (Givnish et al., 2016; Zhang et al., 2023). The evolution of epiphytism in orchids coincides with rapid expansions of modern rainforests, posing the possibility that rainforests facilitated the transition from terrestrial to epiphytic habits (Zhang et al., 2023). Closed-canopy rainforest ecosystems provide ample ecological niches, e.g., trunks and branches of trees, with varying levels of sunlight and moisture (Nakamura et al., 2017; Spicer and Woods, 2022) that likely fostered epiphyte diversification. In addition, the higher fungal abundance and diversity that often characterize such habitats may have further contributed to high rates of epiphytic orchid diversity (Vance and Nadkarni, 1990; Cardelus et al., 2009).
More than 95% of epiphytic orchids descended from a single ancestor that became epiphytic at approximately 48.1–55.8 Mya, near the Paleocene-Eocene Thermal Maximum (Zhang et al., 2023). However, epiphytism may have evolved independently across several genera of Orchidaceae, underscoring its intricate evolutionary trajectory (Givnish et al., 2016; Zhang et al., 2023). Notably, epiphytic species are rare in Cypripedioideae and Orchidoideae. Epiphytes are entirely absent in Apostasioideae, and true epiphytes are absent in Vanilloideae, although there is debate regarding whether climbers such as most Vanilla species (Vanilloideae) should be considered epiphytes or hemiepiphytic (Chen et al., 2009; De Lima and Franco Pinheiro Moreira, 2022). Epiphytism was acquired no later than 35 Mya in Epidendroideae, but was subsequently lost at least three times (Givnish et al., 2015; Zhang et al., 2023). In Arethuseae, there were at least two independent acquisitions of epiphytism in Coelogyninae (Zhang et al., 2023).
Crassulacean acid metabolism (CAM) is a photosynthetic pathway that has evolved convergently in multiple plant lineages, especially in species inhabiting water-limited environments, such as hot semi-arid areas and tropical forests (Winter et al., 2021; Xue et al., 2023). Recent studies have indicated that the majority of epiphyte plants use the CAM pathway (Holtum et al., 2007), including bromeliads (Hermida-Carrera et al., 2020), pteridophytes (Silvera et al., 2010), and orchid species (Givnish et al., 2015; Zou et al., 2018; Gamisch et al., 2021; Hu et al., 2022; Zhang et al., 2022b). CAM photosynthesis, which has evolved at least four times within Orchidaceae, is thought to be a key innovation that has driven orchid diversity. Specifically, CAM photosynthesis has been linked to increased speciation and extinction in the higher epidendroids (Givnish et al., 2015, 2016).
Even in high-rainfall forests, CAM photosynthesis may confer selective advantages to tropical epiphytes without significantly affecting species diversity or diversification rates (Pierce et al., 2002; Gravendeel et al., 2004; Bone et al., 2015; Gamisch et al., 2021). Some researchers have argued CAM photosynthesis may lead to evolutionary dead-ends, noting that in certain lineages CAM photosynthesis may initially provide a short-term evolutionary advantage via speciation, but subsequently decrease net diversification rates (Igic and Busch, 2013; Gamisch et al., 2015; Hu et al., 2022). Although support for this hypothesis remains ambiguous, it challenges our prevailing assumption that CAM photosynthesis is a key innovation that drives diversity in tropical orchids.
Another innovation that has driven orchid diversity is pollinia. Pollinia is a single, cohesive mass of pollen, typically contained in a waxy body or other discrete compact unit found in members of Orchidaceae and Asclepiadaceae (Newton, 1984; Dressler, 1993; Johnson and Edwards, 2000). Pollinia evolved at least 64 Mya in Orchidoideae and Epidendroideae, transitioning from powdery to waxy. Research indicates that the evolution of pollinia significantly accelerated both speciation rates and extinction rates in Orchidaceae (Givnish et al., 2016).
The rate of diversification and morphological evolution varies in different groups of orchids. For example, previous research has identified increased rates of diversification in the orchidoids, including a second increase in the higher epidendroids, a further nested increase in Laeliinae + Pleurothallidinae + Ponerinae, and decreased rates of diversification in Agrostophyllinae + Calypsoinae (Givnish et al., 2015). An additional study has detected five increases and one decrease in diversification rate within Epidendroideae (Zhang et al., 2023). Although one increase in diversification is consistent between these studies, they differ on whether higher epidendroid diversification decreases at the node. Inconsistencies between these studies may be related to sampling sizes and the results of divergence time estimations (Zhang et al., 2023).
It is crucial to acknowledge that shifts in net diversification are scale-dependent. Taxon sampling in these studies is well suited to detect diversification shifts at tribal or subtribal levels within orchids, but may fail to identify such shifts within genera (Givnish et al., 2016; Zhang et al., 2023). A similar situation can be observed in families such as Asteraceae, Poaceae, and other families with a large number of species (Soreng et al., 2017; Zhang et al., 2021a; Huang et al., 2022b). Increased sampling density with a greater number of species is essential for a deeper understanding of the parallel evolution of life habits of orchids, notably at the species level (Givnish et al., 2016; Xiang et al., 2016; Guo et al., 2023). Given the challenges associated with sampling, it is critical to separately list species enrichment groups and hotspots for discussions on the mechanisms of orchid diversity. In recent years, numerous studies have contributed valuable information at the genus level, such as Angraecum (Andriananjamanantsoa et al., 2016), Bulbophyllum (Gamisch and Comes, 2019), Cymbidium (Chen et al., 2024), Holcoglossum (Zhao et al., 2020), Pleione (Wu et al., 2023), which to some extent support conclusions regarding the diversification of Orchidaceae.
5. Phylogenetic discrepancies between different datasetsMounting genomic data has empowered scientists with an unprecedented ability to unravel the phylogenetic relationships between orchids and examine the mechanisms underlying their massive diversity. Each study consistently supports a similar backbone phylogeny for Orchidaceae, although some notable "hard conflicts" regarding specific groups persist. These topological discrepancies may arise from factors such as taxon sampling, idiosyncratic rates of evolution in some groups, or characteristics inherent to the datasets, such as variations in substitution rate. The major contributing factor to these conflicts in topologies lies in the conflicting sequencing techniques employed, comprising nuclear data, chloroplast data, and mitochondrial data (Niu et al., 2017; Li et al., 2019b; Serna-Sanchez et al., 2021; Zhang et al., 2023). Significant conflicts between plastid and mitochondrial trees can be attributed to several factors. These include the different substitution rates of these two genomes, intragenomic substitution rate heterogeneity of plastid substitution rate, and RNA editing in mitochondrial genomes (Petersen et al., 2006; Wicke et al., 2016; Edera et al., 2018; Dong et al., 2023). While RNA editing in mitochondrial genes is a common occurrence and has been considered a potential issue in phylogenetic reconstructions, extensive studies have concluded that it has no direct effect on reconstructions of phylogeny. Instead, RNA editing may increase variability at edited sites (Petersen et al., 2006; Qiu et al., 2010; Liu et al., 2014; Bell et al., 2020; Dong et al., 2023). Incongruities can also arise from sampling paralogous sequences and highly divergent substitution rates, which can result in long-branch attraction and biased results (Petersen et al., 2006; Edera et al., 2018).
Compared to plastid genes, mitochondrial genes are characterized by lower substitution rates and homoplasy levels, which render them particularly valuable for reconstructing relationships of fast-evolving or ancient groups (e.g., Nymphaeales; Qiu et al., 2010; Richardson et al., 2013; Zhu et al., 2014; Qu et al., 2022).
Transcriptome and plastid sequences produce fewer conflicts, with strong support across trees with multiple species and supermatrix analyses (Givnish et al., 2015; Kim et al., 2020; Perez-Escobar et al., 2024; Zhang et al., 2023). Discordances between plastid and nuclear gene trees at a few nodes highlight the complexity of plant genome evolution, including events such as hybridization, polyploidization, impulses of rapid speciation, and extinction (Guo et al., 2023). Importantly, transcriptomes are mainly a collection of coding gene sequences, therefore, the informativeness of these sequences may be limited in rapidly differentiating species (Cheon et al., 2020; Guo et al., 2023, Leebens-Mack et al., 2019).
To date, 16 different complete orchid genomes have been published (Zhang et al., 2017, 2022a; Xu et al., 2021). These genomes indicate the orchids have undergone two whole-genome duplication (WGD) events, the most recent of which was shared by all orchids, whereas the older event was shared by most monocots (Van de Peer et al., 2017; Zhang et al., 2017). Changes within MADS-box gene classes, identified through these genomes, might have contributed to variations in labellum and pollinium morphology and accessory structures (Chao et al., 2018; Ai et al., 2021; Sun et al., 2021; Zhang et al., 2021b, 2021c).
These studies have elaborated on the evolutionary trajectory and divergence of orchids, leading to applications in various fields. For instance, they have facilitated the use of orchids in traditional Chinese medicine, and explored the heterotrophy of mycoheterotrophy and parasitic plants, or provided molecular evidence for partial endoreplication (Yuan et al., 2018; Hu et al., 2022; Jiang et al., 2022a, 2022b; Li et al., 2022).
6. ProspectsGenomic data and technological advances have profoundly enhanced our understanding of the phylogenetic relationships of orchids and facilitated more robust and reliable reconstructions of phylogenies. The phylogenetic relationships within taxa such as Xerorchideae and Wullschlaegelieae are expected to be resolved soon through comprehensive sampling. Furthermore, the application of single-molecular sequencing and other advanced methodologies promise to unlock additional layers of complexity and refinement, albeit with accompanying challenges, thus propelling Orchidaceae phylogenetic reconstructions to unprecedented scientific rigor and depth.
Important challenges need to be addressed before a clearer view of orchid phylogeny emerges. Several groups exhibit discordant positions across different datasets (e.g., Epidendreae, Vandeae, and Cymbidieae). Discrepancies in the phylogenetic positions of these groups will not be resolved by updating sequencing methods. Instead, the particularity of these taxa requires a more suitable approach and consideration of alternative interpretations of evolutionary relationships. Furthermore, despite notable progress, challenges still persist in topological analyses of clades where generic delimitation remains speculative (Aeridinae, Angraecinae, Arethuseae, Bletiinae, Goodyerinae, Malaxideae, Orchidinae, and Podochileae). The uneven sampling of biodiversity hotspots within Orchidaceae can be attributed to the extreme diversity and global distribution, coupled with practical limitations. This imbalance in sampling has led to gaps in the phylogenetic framework of Orchidaceae, particularly in regions with rich plant diversification. However, this scenario also underscores the immense potential value of orchids, highlighting the importance of further exploration and study, to better understand and harness the diverse and valuable traits within this botanical treasure trove.
The current state of phylogenomic inferences in Orchidaceae reveals a notable bias towards regions such as Europe, North America, and East Asia, with limited results from regions such as tropical Asia, Africa, and Latin America (Guo et al., 2023). This deficiency is particularly pronounced in narrow-ranged species in tropical countries, despite high species richness (Rudbeck et al., 2022). Additionally, many studies exhibit a restricted geographic and taxonomic scope, which can lead to excessive imbalanced sampling. Perez-Escobar et al. (2024) presented a novel hypothesis of biogeography of orchid plants, proving that taxon sampling depth and outgroups influence results. There is thus an urgent need for global collaboration to undertake large-scale phylogenomic work in other plant lineages, as in the case of Fabaceae (Azani et al., 2017), Lamiaceae (Boachon et al., 2018), and Poaceae (Soreng et al., 2017, 2022); and Carex (Roalson et al., 2021). Achieving large-scale sampling efforts are vital to enhance scientific comprehension of lineage diversification with a large number of species, leveraging existing genomic data and increasing the scope of cooperation in research (Givnish et al., 2016; Hendriks et al., 2023, Thompson et al., 2023).
AcknowledgmentsThis research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA0420203), the National Natural Science Foundation of China (32270214 to XJ), and China's National Basic Science and Technology Program (2018FY100801).
CRediT authorship contribution statement
Yajun Wang: Writing – original draft, Software, Project administration, Methodology, Formal analysis, Data curation, Conceptualization. Hanchen Wang: Writing – review & editing. Chao Ye: Writing – review & editing, Visualization. Zhiping Wang: Data curation. Chongbo Ma: Writing – review & editing. Dongliang Lin: Writing – review & editing. Xiaohua Jin: Writing – review & editing, Supervision, Resources, Funding acquisition, Data curation, Conceptualization.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.05.002.
Ai, Y., Li, Z., Sun, W.H., et al., 2021. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res., 8: 255. DOI:10.1038/s41438-021-00683-z |
Andriananjamanantsoa, H.N., Engberg, S., Louis Jr., E.E., et al., 2016. Diversification of Angraecum (Orchidaceae, Vandeae) in Madagascar: revised phylogeny reveals species accumulation through time rather than rapid radiation. PLoS One, 11: e0163194. DOI:10.1371/journal.pone.0163194 |
Azani, N., Babineau, M., Bailey, C.D., et al., 2017. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon, 66: 44-77. DOI:10.12705/661.3 |
Batista, J.A.N., Borges, K.S., de Faria, M.W.F., et al., 2013. Molecular phylogenetics of the species-rich genus Habenaria (Orchidaceae) in the New World based on nuclear and plastid DNA sequences. Mol. Phylogenet. Evol., 67: 95-109. DOI:10.1016/j.ympev.2013.01.008 |
Bazinet, A.L., Mitter, K.T., Davis, D.R., et al., 2017. Phylotranscriptomics resolves ancient divergences in the Lepidoptera. Syst. Entomol., 42: 305-316. DOI:10.1111/syen.12217 |
Bell, D., Lin, Q., Gerelle, W.K., et al., 2020. Organellomic data sets confirm a cryptic consensus on (unrooted) land-plant relationships and provide new insights into bryophyte molecular evolution. Am. J. Bot., 107: 91-115. DOI:10.1002/ajb2.1397 |
Boachon, B., Buell, C.R., Crisovan, E., et al., 2018. Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol. Plant, 11: 1084-1096. DOI:10.1016/j.molp.2018.06.002 |
Bone, R.E., Smith, J.A.C., Arrigo, N., et al., 2015. A macro-ecological perspective on crassulacean acid metabolism (CAM) photosynthesis evolution in Afro-Madagascan drylands: Eulophiinae orchids as a case study. New Phytol., 208: 469-481. DOI:10.1111/nph.13572 |
Burns-Balogh, P., Funk, V., 1986. A phylogenetic analysis of the Orchidaceae. Smithsonian Contrib. Bot., 61: 1-79. DOI:10.5479/si.0081024X.61 |
Cameron, K.M., 2005. Leave it to the leaves: a molecular phylogenetic study of Malaxideae (Epidendroideae, Orchidaceae). Am. J. Bot., 92: 1025-1032. DOI:10.3732/ajb.92.6.1025 |
Cameron, K.M., Chase, M.W., Whitten, W.M., et al., 1999. A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Am. J. Bot., 86: 208-224. DOI:10.2307/2656938 |
Cardelus, C.L., Mack, M.C., Woods, C., et al., 2009. The influence of tree species on canopy soil nutrient status in a tropical lowland wet forest in Costa Rica. Plant Soil, 318: 47-61. DOI:10.1007/s11104-008-9816-9 |
Chao, Y.T., Chen, W.C., Chen, C.Y., et al., 2018. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J., 16: 2027-2041. DOI:10.1111/pbi.12936 |
Chase, M.W., Cameron, K.M., Barrett, R., et al., 2003. DNA Data and Orchidaceae Systematics: a New Phylogenetic Classification. Natural History Publications.
|
Chase, M.W., Cameron, K.M., Freudenstein, J.V., et al., 2015. An updated classification of Orchidaceae. Bot. J. Linn. Soc., 177: 151-174. DOI:10.1111/boj.12234 |
Chase, M.W., Christenhusz, M.J.M., Schuiteman, A., 2020a. Expansion of Calanthe to include the species of Cephalantheropsis, Gastrorchis and Phaius (Collabieae; Orchidaceae). Phytotaxa, 472: 159-168. DOI:10.11646/phytotaxa.472.2.6 |
Chase, M.W., Christenhusz, M.J.M., Schuiteman, A., 2020b. Proposal to conserve Calanthe, nom. Cons., against the additional names Phaius, Cyanorkis, and Gastorkis (Orchidaceae, Collabieae). Taxon, 69: 1364-1365. DOI:10.1002/tax.12396 |
Chase, M.W., Gravendeel, B., Sulistyo, B.P., et al., 2021. Expansion of the orchid genus Coelogyne (Arethuseae; Epidendroideae) to include Bracisepalum, Bulleyia, Chelonistele, Dendrochilum, Dickasonia, Entomophobia, Geesinkorchis, Gynoglottis, Ischnogyne, Nabaluia, Neogyna, Otochilus, Panisea and Pholidota. Phytotaxa, 510: 94-134. DOI:10.11646/phytotaxa.510.2.1 |
Chen, S.C., Liu, Z.J., Zhu, G.H., et al., 2009. Orchidaceae, In: Wu Z.Y., Raven, P.H., Hong, D.Y. (Eds.), Flora of China. Science Press, Beijing & Missouri Botanical Garden pp. 167.
|
Chen, S.P., Tian, H.Z., Guan, Q.X., et al., 2019. Molecular systematics of Goodyerinae (Cranichideae, Orchidoideae, Orchidaceae) based on multiple nuclear and plastid regions. Mol. Phylogenet. Evol., 139. DOI:10.1016/j.ympev.2019.106542 |
Chen, H.Y., Zhang, Z.R., Yao, X., et al., 2024. Plastid phylogenomics provides new insights into the systematics, diversification, and biogeography of Cymbidium (Orchidaceae). Plant Divers., 46: 448-461. DOI:10.1016/j.pld.2024.03.001 |
Cheon, S.M., Zhang, J.Z., Park, C.G., 2020. Is phylotranscriptomics as reliable as phylogenomics?. Mol. Biol. Evol., 37: 3672-3683. DOI:10.1093/molbev/msaa181 |
Conran, J.G., Bannister, J.M., Lee, D.E., 2009. Earliest orchid macrofossils: early miocene Dendrobium and Earina (Orchidaceae: Epidendroideae) from New Zealand. Am. J. Bot., 96: 466-474. DOI:10.3732/ajb.0800269 |
De Lima, J.F., Franco Pinheiro Moreira, A.S., 2022. Structural plasticity in roots of the hemiepiphyte Vanilla phaeantha Rchb. f. (Orchidaceae): a relationship between environment and function. Sci. Nat., 109: 46. DOI:10.1007/s00114-022-01816-7 |
Deng, H., Zhang, G.Q., Lin, M., et al., 2015. Mining from transcriptomes: 315 single-copy orthologous genes concatenated for the phylogenetic analyses of Orchidaceae. Ecol. Evol., 5: 3800-3807. DOI:10.1002/ece3.1642 |
Dong, S. -S., Zhou, X. -P., Peng, T., et al., 2023. Mitochondrial RNA editing sites affect the phylogenetic reconstruction of gymnosperms. Plant Divers., 45: 485-489. DOI:10.1016/j.pld.2023.02.004 |
Dressler, R.L., 1981. The orchids: natural history and classification. Taxon. DOI:10.2307/1219717 |
Dressler, R.L., 1993. Phylogeny and Classification of the Orchid Family. Cambridge University Press pp. 71-89.
|
Dressler, R.L., Dodson, C., 1960. Classification and phylogeny of the Orchidaceae. Ann. Mo. Bot. Gard., 47: 25-68. DOI:10.2307/2394615 |
Edera, A.A., Gandini, C.L., Sanchez-Puerta, M.V., 2018. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol., 97: 215-231. DOI:10.1007/s11103-018-0734-9 |
Fernandez, L.P., Byers, K., Cai, J., et al., 2019. A phylogenomic analysis of the floral transcriptomes of sexually deceptive and rewarding European orchids, Ophrys and Gymnadenia. Front. Plant Sci., 10: 01553. DOI:10.3389/fpls.2019.01553 |
Freudenstein, J.V., Chase, M.W., 2015. Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann. Bot., 115: 665-681. DOI:10.1093/aob/mcu253 |
Gamisch, A., Comes, H.P., 2019. Clade-age-dependent diversification under high species turnover shapes species richness disparities among tropical rainforest lineages of Bulbophyllum (Orchidaceae). BMC Evol. Biol., 19: 93. DOI:10.1186/s12862-019-1416-1 |
Gamisch, A., Fischer, G.A., Comes, H.P., 2015. Multiple independent origins of auto-pollination in tropical orchids (Bulbophyllum) in light of the hypothesis of selfing as an evolutionary dead end. BMC Evol. Biol., 15: 192. DOI:10.1186/s12862-015-0471-5 |
Gamisch, A., Winter, K., Fischer, G.A., et al., 2021. Evolution of crassulacean acid metabolism (CAM) as an escape from ecological niche conservatism in Malagasy Bulbophyllum (Orchidaceae). New Phytol., 231: 1236-1248. DOI:10.1111/nph.17437 |
Garay, L.A., 1972. On the origin of the Orchidaceae II. J. Arnold Arbor., 53: 202-215. DOI:10.5962/p.185781 |
GBIF. org, Citation Guidelines. 2024 (20 February). https://www.gbif.org/citation-guidelines.
|
Geiger, D.L., 2023. Studies in oberonia 11: the genus Hippeophyllum reduced to oberonia, with ten new synonyms of Oberonia scortechinii (Orchidaceae: Malaxideae). Gard. Bull. Singapore, 75: 129-148. DOI:10.26492/gbs75(1).2023-10 |
Givnish, T.J., Spalink, D., Ames, M., et al., 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. Roy. Soc. B-Biol. Sci., 282: 171-180. DOI:10.1098/rspb.2015.1553 |
Givnish, T.J., Spalink, D., Ames, M., et al., 2016. Orchid historical biogeography, diversification, Antarctica and the paradox of orchid dispersal. J. Biogeogr., 43: 1905-1916. DOI:10.1111/jbi.12854 |
Givnish, T.J., Zuluaga, A., Spalink, D., et al., 2018. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am. J. Bot., 105: 1888-1910. DOI:10.1002/ajb2.1178 |
Goldman, D.H., Freudenstein, J.V., Kores, P.J., et al., 2001. Phylogenetics of Arethuseae (Orchidaceae) based on plastid matK and rbcL sequences. Syst. Bot., 26: 670-695. DOI:10.1043/0363-6445-26.3.670 |
Gravendeel, B., Chase, M.W., De Vogel, E.F., et al., 2001. Molecular phylogeny of Coelogyne (Epidendroideae; Orchidaceae) based on plastid RFLPS, matK, and nuclear ribosomal its sequences: evidence for polyphyly. Am. J. Bot., 88: 1915-1927. DOI:10.2307/3558367 |
Gravendeel, B., Smithson, A., Slik, F.J.W., et al., 2004. Epiphytism and pollinator specialization: drivers for orchid diversity?. Proc. Roy. Soc. B-Biol. Sci., 359: 1523-1535. DOI:10.1098/rstb.2004.1529 |
Guo, Y.Y., Luo, Y.B., Liu, Z.J., et al., 2012. Evolution and biogeography of the slipper orchids: eocene vicariance of the conduplicate genera in the Old and New World Tropics. PLoS One, 7: e38788. DOI:10.1371/journal.pone.0038788 |
Guo, J., Xu, W.B., Hu, Y., et al., 2020. Phylotranscriptomics in Cucurbitaceae reveal multiple whole-genome duplications and key morphological and molecular innovations. Mol. Plant, 13: 1117-1133. DOI:10.1016/j.molp.2020.05.011 |
Guo, C., Luo, Y., Gao, L.M., et al., 2023. Phylogenomics and the flowering plant tree of life. J. Integr. Plant Biol., 65: 299-323. DOI:10.1111/jipb.13415 |
Hendriks, K.P., Kiefer, C., Al-Shehbaz, I.A., et al., 2023. Global Brassicaceae phylogeny based on filtering of 1, 000-gene dataset. Curr. Biol., 33: 4052-4068. DOI:10.1016/j.cub.2023.08.026 |
Hermida-Carrera, C., Fares, M.A., Font-Carrascosa, M., et al., 2020. Exploring molecular evolution of rubisco in C3 and CAM Orchidaceae and bromeliaceae. BMC Evol. Biol., 20: 11. DOI:10.1186/s12862-019-1551-8 |
Holtum, J.A.M., Winter, K., Weeks, M.A., et al., 2007. Crassulacean acid metabolism in the ZZ plant, Zamioculcas zamiifolia (Araceae). Am. J. Bot., 94: 1670-1676. DOI:10.3732/ajb.94.10.1670 |
Hu, C., Tian, H.Z., Li, H.Q., et al., 2016. Phylogenetic analysis of a 'Jewel Orchid' genus Goodyera (Orchidaceae) based on DNAsequence data from nuclear and plastid regions. PLoS One, 11: e0150366. DOI:10.1371/journal.pone.0150366 |
Hu, A.Q., Gale, S.W., Liu, Z.J., et al., 2022. Diversification slowdown in the Cirrhopetalum alliance (Bulbophyllum, Orchidaceae): insights from the evolutionary dynamics of crassulacean acid metabolism. Front. Plant Sci., 13: 794171. DOI:10.3389/fpls.2022.794171 |
Huang, W.C., Liu, Z.J., Jiang, K., et al., 2022a. Phylogenetic analysis and character evolution of tribe Arethuseae (Orchidaceae) reveal a new genus Mengzia. Mol. Phylogenet. Evol., 167: 107362. DOI:10.1016/j.ympev.2021.107362 |
Huang, W.C., Zhang, L., Columbus, J.T., et al., 2022b. A well-supported nuclear phylogeny of Poaceae and implications for the evolution of C-4 photosynthesis. Mol. Plant, 15: 755-777. DOI:10.1016/j.molp.2022.01.015 |
Igic, B., Busch, J.W., 2013. Is self-fertilization an evolutionary dead end?. New Phytol., 198: 386-397. DOI:10.1111/nph.12182 |
Jersakova, J., Johnson, S.D., Kindlmann, P., 2006. Mechanisms and evolution of deceptive pollination in orchids. Biol. Rev., 81: 219-235. DOI:10.1017/S1464793105006986 |
Jiang, L., Lin, M.F., Wang, H., et al., 2022a. Haplotype-resolved genome assembly of Bletilla striata (Thunb.) Reichb. f. to elucidate medicinal value. Plant J., 111: 1340-1353. DOI:10.1111/tpj.15892 |
Jiang, Y., Hu, X.D., Yuan, Y., et al., 2022b. The Gastrodia menghaiensis (Orchidaceae) genome provides new insights of orchid mycorrhizal interactions. BMC Plant Biol., 22: 179. DOI:10.1186/s12870-022-03573-1 |
Jin, W.T., Schuiteman, A., Chase, M.W., et al., 2017. Phylogenetics of subtribe Orchidinae s. l. (Orchidaceae; Orchidoideae) based on seven markers (plastid matK, psaB, rbcL, trnL-F, trnH-psbA, and nuclear nrITS, Xdh): implications for generic delimitation. BMC Plant Biol., 17: 222. DOI:10.1186/s12870-017-1160-x |
Jin, W.T., Gernandt, D.S., Wehenkel, C., et al., 2021. Phylogenomic and ecological analyses reveal the spatiotemporal evolution of global pines. Proc. Natl. Acad. Sci. U.S.A., 118: e2022302118. DOI:10.1073/pnas.2022302118 |
Johnson, S.D., Edwards, T.J., 2000. The structure and function of orchid pollinaria. Plant Syst. Evol., 222: 243-269. DOI:10.1007/bf00984105 |
Johnson, M.G., Malley, C., Goffinet, B., et al., 2016. A phylotranscriptomic analysis of gene family expansion and evolution in the largest order of pleurocarpous mosses (Hypnales, Bryophyta). Mol. Phylogenet. Evol., 98: 29-40. DOI:10.1016/j.ympev.2016.01.008 |
Judd, W.S., Stern, W.L., Cheadle, V.I., 1993. Phylogenetic position of apostasia and Neuwiedia (Orchidaceae). Bot. J. Linn. Soc., 113: 87-94. DOI:10.1111/j.1095-8339.1993.tb00331.x |
Kikuchi, I.A.B.S., Kessler, P.J.A., Schuiteman, A., et al., 2020. Molecular phylogenetic study of the tribe Tropidieae (Orchidaceae, Epidendroideae) with taxonomic and evolutionary implications. PhytoKeys, 140: 11-22. DOI:10.3897/phytokeys.140.46842 |
Kim, T.H., Kim, J.H., 2022. Molecular phylogeny and historical biogeography of Goodyera R. Br. (Orchidaceae): a case of the vicariance between East Asia and North America. Front. Plant Sci., 13: 850170. DOI:10.3389/fpls.2022.850170 |
Kim, Y.K., Jo, S., Cheon, S.H., et al., 2020. Plastome evolution and phylogeny of subtribe Aeridinae (Vandeae, Orchidaceae). Mol. Phylogenet. Evol., 144: 106721. DOI:10.1016/j.ympev.2019.106721 |
Kocyan, A., Qiu, Y.L., Endress, P.K., et al., 2004. A phylogenetic analysis of Apostasioideae (Orchidaceae) based on ITS, trnL-F and matK sequences. Plant Syst. Evol., 247: 203-213. DOI:10.1007/s00606-004-0133-3 |
Kumar, S., Stecher, G., Suleski, M., et al., 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol., 34: 1812-1819. DOI:10.1093/molbev/msx116 |
Kumar, S., Suleski, M., Craig, J.M., et al., 2022. TimeTree 5: an expanded resource for species divergence times. Mol. Biol. Evol., 39: msac174. DOI:10.1093/molbev/msac174 |
Kurzweil, H., Weber, A., 1992. Floral morphology of southern african Orchideae. II. Habenariinae. Nord. J. Bot., 12: 39-61. DOI:10.1111/j.1756-1051.1992.tb00200.x |
Lai, Y.J., Han, Y., Schuiteman, A., et al., 2021. Diversification in Qinghai-Tibet Plateau: Orchidinae (Orchidaceae) clades exhibiting pre-adaptations play critical role. Mol. Phylogenet. Evol., 157: 107062. DOI:10.1016/j.ympev.2020.107062 |
Leebens-Mack, J.H., Barker, M.S., Carpenter, E.J., et al., 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature, 574: 679. DOI:10.1038/s41586-019-1693-2 |
Li, M.H., Zhang, G.Q., Lan, S.R., et al., 2016. A molecular phylogeny of Chinese orchids. J. Syst. Evol., 54: 349-362. DOI:10.1111/jse.12187 |
Li, H.T., Yi, T.S., Gao, L.M., et al., 2019a. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants, 5: 461-470. DOI:10.1038/s41477-019-0421-0 |
Li, Y.X., Li, Z.H., Schuiteman, A., et al., 2019b. Phylogenomics of Orchidaceae based on plastid and mitochondrial genomes. Mol. Phylogenet. Evol., 139: 106540. DOI:10.1016/j.ympev.2019.106540 |
Li, L., Chung, S.W., Li, B., et al., 2020. New insight into the molecular phylogeny of the genus Liparis s. l. (Orchidaceae: Malaxideae) with a new generic segregate: Blepharoglossum. Plant Syst. Evol., 306: 54. DOI:10.1007/s00606-020-01679-3 |
Li, M.H., Liu, K.W., Li, Z., et al., 2022. Genomes of leafy and leafless Platanthera orchids illuminate the evolution of mycoheterotrophy. Nat. Plants, 8: 373. DOI:10.1038/s41477-022-01127-9 |
Lin, C.S., Chen, J.J.W., Huang, Y.T., et al., 2015. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep., 5: 9040. DOI:10.1038/srep09040 |
Liu, Y., Cox, C.J., Wang, W., et al., 2014. Mitochondrial phylogenomics of early land plants: mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Syst. Biol., 63: 862-878. DOI:10.1093/sysbio/syu049 |
Liu, D.K., Tu, X.D., Zhao, Z., et al., 2020. Plastid phylogenomic data yield new and robust insights into the phylogeny of Cleisostoma-Gastrochilus clades (Orchidaceae, Aeridinae). Mol. Phylogenet. Evol., 145: 106729. DOI:10.1016/j.ympev.2019.106729 |
Liu, H., Jacquemyn, H., Chen, W., et al., 2021. Niche evolution and historical biogeography of lady slipper orchids in North America and Eurasia. J. Biogeogr., 48: 2727-2741. DOI:10.1111/jbi.14224 |
Liu, L.K., Du, J.X., Liu, Z.H., et al., 2023. Comparative and phylogenetic analyses of nine complete chloroplast genomes of Orchidaceae. Sci. Rep., 13: 21403. DOI:10.1038/s41598-023-48043-2 |
Micheneau, C., Carlsward, B.S., Fay, M.F., et al., 2008. Phylogenetics and biogeography of mascarene angraecoid orchids (Vandeae, Orchidaceae). Mol. Phylogenet. Evol., 46: 908-922. DOI:10.1016/j.ympev.2007.12.001 |
Morales, N.G., Toscano De Brito, A.L.V., Silverio Righetto Mauad, A.V., et al., 2021. Molecular phylogeny and biogeography of Pabstiella (Pleurothallidinae: Orchidaceae) highlight the importance of the Atlantic Rainforest for speciation in the genus. Bot. J. Linn. Soc., 195: 568-587. DOI:10.1093/botlinnean/boaa092 |
Nakamura, A., Kitching, R.L., Cao, M., et al., 2017. Forests and their canopies: achievements and horizons in canopy science. Trends Ecol. Evol., 32: 438-451. DOI:10.1016/j.tree.2017.02.020 |
Nargar, K., Molina, S., Wagner, N., et al., 2018. Australasian orchid diversification in time and space: molecular phylogenetic insights from the beard orchids (Calochilus, Diurideae). Aust. Syst. Bot., 31: 389-408. DOI:10.1071/sb18027 |
Newton, L.E., 1984. Tterminology of structures associated with pollinia of the Asclepiadaceae. Taxon, 33: 619-621. DOI:10.2307/1220779 |
Ng, Y.P., Schuiteman, A., Pedersen, H.A., et al., 2018. Phylogenetics and systematics of Eria and related genera (Orchidaceae: Podochileae). Bot. J. Linn. Soc., 186: 179-201. DOI:10.1093/botlinnean/box088 |
Ngugi, G., Le Péchon, T., Martos, F., et al., 2020. Phylogenetic relationships amongst the African genera of subtribe Orchidinae s. l. (Orchidaceae; Orchideae): implications for subtribal and generic delimitations. Mol. Phylogenet. Evol., 153: 106946. DOI:10.1016/j.ympev.2020.106946 |
Niu, Z.T., Zhu, S.Y., Pan, J.J., et al., 2017. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci. Rep., 7: 2073. DOI:10.1038/s41598-017-02252-8 |
Perez-Escobar, O.A., Gottschling, M., Chomicki, G., et al., 2017. Andean mountain building did not preclude dispersal of lowland epiphytic orchids in the Neotropics. Sci. Rep., 7: 4919. DOI:10.1038/s41598-017-04261-z |
Perez-Escobar, O.A., Dodsworth, S., Bogarin, D., et al., 2021. Hundreds of nuclear and plastid loci yield novel insights into orchid relationships. Am. J. Bot., 108: 1166-1180. DOI:10.1002/ajb2.1702 |
Perez-Escobar, O.A., Zizka, A., Bermudez, M.A., et al., 2022. The Andes through time: evolution and distribution of Andean floras. Trends Plant Sci., 27: 364-378. DOI:10.1016/j.tplants.2021.09.010 |
Perez-Escobar, O.A., Bogarin, D., Przelomska, N.A., et al., 2024. The origin and speciation of Orchids. New Phytol., 242: 700-716. DOI:10.1111/nph.19580 |
Petersen, G., Seberg, O., Davis, J.I., et al., 2006. Mitochondrial data in monocot phylogenetics. Aliso, 22: 52-62. DOI:10.5642/aliso.20062201.05 |
Pfitzer, E., 1887. Entwurf einer natürlichen Anordnung der Orchideen. Carl Winter's
Universitätsbuchhandlung, Heidelburg. https://doi.org/10.5962/bhl.title.166408.
|
Pierce, S., Winter, K., Griffiths, H., 2002. The role of CAM in high rainfall cloud forests: an in situ comparison of photosynthetic pathways in Bromeliaceae. Plant Cell Environ., 25: 1181-1189. DOI:10.1046/j.1365-3040.2002.00900.x |
Poinar Jr., G., Rasmussen, F.N., 2017. Orchids from the past, with a new species in Baltic amber. Bot. J. Linn. Soc., 183: 327-333. DOI:10.1093/botlinnean/bow018 |
POWO, Plants of the World Online. 2024 (20 February) https://powo.science.kew.org/.
|
Pridgeon, A.M., Cribb, P.J., Chase, M.W., et al., 2005. Genera Orchidacearum. Oxford University Press, New York.
|
Qiu, Y.L., Li, L.B., Wang, B., et al., 2010. Angiosperm phylogeny inferred from sequences of four mitochondrial genes. J. Syst. Evol., 48: 391-425. DOI:10.1111/j.1759-6831.2010.00097.x |
Qu, X.J., Zhang, X.J., Cao, D.L., et al., 2022. Plastid and mitochondrial phylogenomics reveal correlated substitution rate variation in Koenigia (Polygonoideae, Polygonaceae) and a reduced plastome for Koenigia delicatula including loss of all ndh genes. Mol. Phylogenet. Evol., 174: 107544. DOI:10.1016/j.ympev.2022.107544 |
Ramirez, S.R., Gravendeel, B., Singer, R.B., et al., 2007. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature, 448: 1042-1045. DOI:10.1038/nature06039 |
Ran, J.H., Shen, T.T., Wang, M.M., et al., 2018. Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc. Roy. Soc. B-Biol. Sci., 285: 1012. DOI:10.1098/rspb.2018.1012 |
Raskoti, B.B., 2017. A taxonomic revision of Herminium L. (Orchidoideae, Orchidaceae). PhytoKeys, 79: 1-74. DOI:10.3897/phytokeys.79.11215 |
Raskoti, B.B., Jin, W.T., Xiang, X.G., et al., 2016. A phylogenetic analysis of molecular and morphological characters of Herminium (Orchidaceae, Orchideae): evolutionary relationships, taxonomy, and patterns of character evolution. Cladistics, 32: 198-210. DOI:10.1111/cla.12125 |
Richardson, A.O., Rice, D.W., Young, G.J., et al., 2013. The "fossilized" mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol., 11: 29. DOI:10.1186/1741-7007-11-29 |
Ricogray, V., Thien, L.B., 1989. Effect of different ant species on reproductive fitness of Schomburgkia tibicinis (Orchidaceae). Oecologia, 81: 487-489. DOI:10.1007/bf00378956 |
Roalson, E.H., Jimenez-Mejias, P., Hipp, A.L., et al., 2021. A framework infrageneric classification of Carex (Cyperaceae) and its organizing principles. J. Syst. Evol., 59: 726-762. DOI:10.1111/jse.12722 |
Rudbeck, A.V., Sun, M., Tietje, M., et al., 2022. The Darwinian shortfall in plants: phylogenetic knowledge is driven by range size. Ecography, 8: e06142. DOI:10.1111/ecog.06142 |
Schlecter, R., 1926. Das system der Orchidaceen. Notizbl. Bot. Gart. U. Mus. BerlinDahlem., 9: 563-591. |
Serna-Sanchez, M.A., Perez-Escobar, O.A., Bogarin, D., et al., 2021. Plastid phylogenomics resolves ambiguous relationships within the orchid family and provides a solid timeframe for biogeography and macroevolution. Sci. Rep., 11: 6858. DOI:10.1038/s41598-021-83664-5 |
Shi, T., Huneau, C., Zhang, Y., et al., 2022. The slow-evolving Acorus tatarinowii genome sheds light on ancestral monocot evolution. Nat. Plants, 8: 764. DOI:10.1038/s41477-022-01187-x |
Silvera, K., Neubig, K.M., Whitten, W.M., et al., 2010. Evolution along the crassulacean acid metabolism continuum. Funct. Plant Biol., 37: 995-1010. DOI:10.1071/fp10084 |
Singer, R.B., Buzatto, C.R., Sanguinetti, A., et al., 2018. Found again: the extremely rare Codonorchis canisioi (Orchidaceae: Codonorchideae) reappears after being missing for 78 years. Plant Syst. Evol., 304: 1157-1163. DOI:10.1007/s00606-018-1538-8 |
Smidt, E.C., Toscano De Brito, A.L.V., Martins, A.C., et al., 2018. Phylogenetics, biogeography and character evolution in the Ornithocephalus clade (Orchidaceae, Oncidiinae). Bot. J. Linn. Soc., 188: 339-354. DOI:10.1093/botlinnean/boy067 |
Smidt, E.C., Salazar, G.A., Silverio Righetto Mauad, A.V., et al., 2021. An Indomalesian origin in the Miocene for the diphyletic New World jewel orchids (Goodyerinae, Orchidoideae): molecular dating and biogeographic analyses document non-monophyly of the Neotropical genera. Bot. J. Linn. Soc., 197: 322-349. DOI:10.1093/botlinnean/boab028 |
Soreng, R.J., Peterson, P.M., Romaschenko, K., et al., 2017. A worldwide phylogenetic classification of the Poaceae (Gramineae) II: an update and a comparison of two 2015 classifications. J. Syst. Evol., 55: 259-290. DOI:10.1111/jse.12262 |
Soreng, R.J., Peterson, P.M., Zuloaga, F.O., et al., 2022. A worldwide phylogenetic classification of the Poaceae (Gramineae) III: an update. J. Syst. Evol., 60: 476-521. DOI:10.1111/jse.12847 |
Sosa, V., 2008. A molecular and morphological phylogenetic study of subtribe Bletiinae (Epidendreae, Orchidaceae). Cladistics, 24: 103-104. DOI:10.1600/036364407780360175 |
Spicer, M.E., Woods, C.L., 2022. A case for studying biotic interactions in epiphyte ecology and evolution. Perspect. Plant Ecol. Evol. Syst., 54: 125658. DOI:10.1016/j.ppees.2021.125658 |
Stull, G.W., Soltis, P.S., Soltis, D.E., et al., 2020. Nuclear phylogenomic analyses of asterids conflict with plastome trees and support novel relationships among major lineages. Am. J. Bot., 107: 790-805. DOI:10.1002/ajb2.1468 |
Sun, Y., Chen, G.Z., Huang, J., et al., 2021. The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornam. Plant Res., 1: 1-13. DOI:10.48130/OPR-2021-0010 |
Szlachetko, D.L., Gorniak, M., Kowalkowska, A.K., et al., 2021. The natural history of the genus Cypripedium (Orchidaceae). Plant Biosyst., 155: 772-796. DOI:10.1080/11263504.2020.1785963 |
Thompson, J.B., Davis, K.E., Dodd, H.O., et al., 2023. Speciation across the Earth driven by global cooling in terrestrial orchids. Proc. Natl. Acad. Sci. U.S.A., 120: e2102408120. DOI:10.1073/pnas.2102408120 |
Tsai, C.C., Liao, P.C., Ko, Y.Z., et al., 2020. Phylogeny and historical biogeography of Paphiopedilum Pfitzer (Orchidaceae) based on nuclear and plastid DNA. Front. Plant Sci., 11: 00126. DOI:10.3389/fpls.2020.00126 |
Tu, X.D., Liu, D.K., Xu, S.W., et al., 2021. Plastid phylogenomics improves resolution of phylogenetic relationship in the Cheirostylis and Goodyera clades of Goodyerinae (Orchidoideae, Orchidaceae). Mol. Phylogenet. Evol., 164: 107269. DOI:10.1016/j.ympev.2021.107269 |
Unruh, S.A., McKain, M.R., Lee, Y.I., et al., 2018. Phylotranscriptomic analysis and genome evolution of the Cypripedioideae (Orchidaceae). Am. J. Bot., 105: 631-640. DOI:10.1002/ajb2.1047 |
Van de Peer, Y., Mizrachi, E., Marchal, K., 2017. The evolutionary significance of polyploidy. Nat. Rev. Genet., 18: 411-424. DOI:10.1038/nrg.2017.26 |
van den Berg, C., Higgins, W.E., Dressler, R.L., et al., 2000. A phylogenetic analysis of Laeliinae (Orchidaceae) based on sequence data from internal transcribed spacers (ITS) of nuclear ribosomal DNA. Lindleyana 15, 96-114. https://api.semanticscholar.org/CorpusID:55324941.
|
Vance, E.D., Nadkarni, N.M., 1990. Microbial biomass and activity in canopy organic matter and the forest floor of a tropical cloud forest. Soil Biol. Biochem., 22: 677-684. DOI:10.1016/0038-0717(90)90015-r |
Vermeulen, P., 1966. The system of the Orchidales. Acta Bot. Neerl., 15: 224-253. DOI:10.1111/J.1438-8677.1966.TB00228.X |
Vitt, P., Taylor, A., Rakosy, D., et al., 2023. Global conservation prioritization for the Orchidaceae. Sci. Rep., 13: 6718. DOI:10.1038/s41598-023-30177-y |
Wicke, S., Mueller, K.F., DePamphilis, C.W., et al., 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. U.S.A., 113: 9045-9050. DOI:10.1073/pnas.1607576113 |
Wickett, N.J., Mirarab, S., Nam, N., et al., 2014. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U.S.A., 111: E4859-E4868. DOI:10.1073/pnas.1323926111 |
Winter, K., Virgo, A., Garcia, M., et al., 2021. Constitutive and facultative crassulacean acid metabolism (CAM) in Cuban oregano, Coleus amboinicus (Lamiaceae). Funct. Plant Biol., 48: 647-654. DOI:10.1071/fp20127 |
Wong, D.C.J., Peakall, R., 2022. Orchid phylotranscriptomics: the prospects of repurposing multi-tissue transcriptomes for phylogenetic analysis and beyond. Front. Plant Sci., 13: 910362. DOI:10.3389/fpls.2022.910362 |
Wu, S.S., Jiang, M.T., Miao, J. l., et al., 2023. Origin and diversification of a Himalayan orchid genus Pleione. Mol. Phylogenet. Evol., 184: 107797. DOI:10.1016/j.ympev.2023.107797 |
Xia, X.M., Yang, M.Q., Li, C.L., et al., 2022. Spatiotemporal evolution of the global species diversity of Rhododendron. Mol. Biol. Evol., 39: msab314. DOI:10.1093/molbev/msab314 |
Xiang, X.G., Li, D.Z., Jin, W.T., et al., 2012. Phylogenetic placement of the enigmatic orchid genera Thaia and Tangtsinia: evidence from molecular and morphological characters. Taxon, 61: 45-54. DOI:10.1002/tax.611003 |
Xiang, X.G., Mi, X.C., Zhou, H.L., et al., 2016. Biogeographical diversification of mainland Asian Dendrobium (Orchidaceae) and its implications for the historical dynamics of evergreen broad-leaved forests. J. Biogeogr., 43: 1310-1323. DOI:10.1111/jbi.12726 |
Xu, Y.X., Lei, Y.T., Su, Z.X., et al., 2021. A chromosome-scale Gastrodia elata genome and large-scale comparative genomic analysis indicate convergent evolution by gene loss in mycoheterotrophic and parasitic plants. Plant J., 108: 1609-1623. DOI:10.1111/tpj.15528 |
Xue, Q.Q., Yang, J.P., Yu, W.H., et al., 2023. The climate changes promoted the chloroplast genomic evolution of Dendrobium orchids among multiple photosynthetic pathways. BMC Plant Biol., 23: 189. DOI:10.1186/s12870-023-04186-y |
Yuan, Y., Jin, X.H., Liu, J., et al., 2018. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun., 9: 1615. DOI:10.1038/s41467-018-03423-5 |
Zhang, Y.B., Du, H.H., Jin, X.H., et al., 2015. Species diversity and geographic distribution of wild Orchidaceae in China. Chin. Sci. Bull., 60: 179-188. DOI:10.1360/N972014-00480 |
Zhang, G.Q., Liu, K.W., Li, Z., et al., 2017. The Apostasia genome and the evolution of orchids. Nature, 549: 379. DOI:10.1038/nature23897 |
Zhang, C.F., Huang, C.H., Liu, M., et al., 2021a. Phylotranscriptomic insights into Asteraceae diversity, polyploidy, and morphological innovation. J. Integr. Plant Biol., 63: 1273-1293. DOI:10.1111/jipb.13078 |
Zhang, W.X., Zhang, G.Q., Zeng, P., et al., 2021b. Genome sequence of Apostasia ramifera provides insights into the adaptive evolution in orchids. BMC Genom., 22: 536. DOI:10.1186/s12864-021-07852-3 |
Zhang, Y.X., Zhang, G.Q., Zhang, D.Y., et al., 2021c. Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic. Res., 8: 183. DOI:10.1038/s41438-021-00621-z |
Zhang, D.Y., Zhao, X.W., Li, Y.Y., et al., 2022a. Advances and prospects of orchid research and industrialization. Hortic. Res., 28: 9. DOI:10.1093/hr/uhac220 |
Zhang, M.Z., Liu, N., Da Silva, J.T.A., et al., 2022b. Physiological and transcriptomic analysis uncovers salinity stress mechanisms in a facultative crassulacean acid metabolism plant Dendrobium officinale. Front. Plant Sci., 13: 1028245. DOI:10.3389/fpls.2022.1028245 |
Zhang, G.J., Hu, Y., Huang, M.Z., et al., 2023. Comprehensive phylogenetic analyses of Orchidaceae using nuclear genes and evolutionary insights into epiphytism. J. Integr. Plant Biol., 65: 1204-1225. DOI:10.1111/jipb.13462 |
Zhang, J.Y., Cheng, Y.H., Liao, M., et al., 2024. A new infrageneric classification of Gastrochilus (Orchidaceae: Epidendroideae) based on molecular and morphological data. Plant Divers, 46: 435-447. DOI:10.1016/j.pld.2023.08.001 |
Zhao, J.H., Zhou, P., Li, X.Q., et al., 2020. Temporal and spatial pattern of Holcoglossum Schltr. (Orchidaceae), an east Asian endemic genus, based on nuclear and chloroplast genes. Front. Ecol. Evol., 8: 00245. DOI:10.3389/fevo.2020.00245 |
Zhou, P., Li, J.H., Liu, Y.Z., et al., 2023. Species richness disparity in tropical terrestrial herbaceous floras: evolutionary insight from Collabieae (Orchidaceae). Mol. Phylogenet. Evol., 186: 107860. DOI:10.1016/j.ympev.2023.107860 |
Zhu, A.D., Guo, W.H., Jain, K., et al., 2014. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol. Biol. Evol., 31: 1228-1236. DOI:10.1093/molbev/msu079 |
Zizka, A., 2019. Big data suggest migration and bioregion connectivity as crucial for the evolution of Neotropical biodiversity. Front. Biogeogr., 11: e40617. DOI:10.21425/F5FBG40617 |
Zou, L.H., Wan, X., Deng, H., et al., 2018. RNA-seq transcriptomic profiling of crassulacean acid metabolism pathway in Dendrobium catenatum. Sci. Data, 5: 180252. DOI:10.1038/sdata.2018.252 |



