b. University of Chinese Academy of Sciences, Beijing, China;
c. Tibet Plateau Institute of Biology, Lhasa 850001, Tibet, China
Trichomes, epidermal protuberances situated on above-ground plant tissues, function as a physical barrier to safeguard plants against biotic threats such as fungal infections and insect herbivory. They also provide protection against abiotic stressors such as water loss, freezing, and UV radiation (Hauser, 2014). The morphology of trichomes exhibits a diverse range, with glandular trichomes capable of synthesizing metabolites for the medicinal purposes. Given their simple structure, the development of trichomes serves as an ideal model for studying the cell differentiation in plants (Ishida et al., 2008).
Trichome development has been extensively studied in model plants over the past decades, and numerous transcription factors that regulate trichome development have been identified in Arabidopsis. Among them, the R2R3 MYB-related transcription factor GLABRA1 (GL1), the Basic Helix–Loop–Helix transcription factor GL3, or its homolog ENHANCER OF GL3 (EGL3), and the WD40-repeat protein TRANSPARENT TESTA GLABRA1 (TTG1) collectively form a trimeric activator complex known as MYB-bHLH-WD40 (MBW) (Oppenheimer et al., 1991; Payne et al., 2000; Walker et al., 1999). Lose-of-function in GL1, GL3 or EGL3 leads to an obvious lack of trichomes on the leaves. Complete function mutations in TTG1 result in a loss of trichome initiation, indicating that TTG1 is crucial in regulating trichome development (Wang et al., 2021). The MBW complex can promote the trichome formation by directly activating the expression of GLABRA2 (GL2), which encodes a conserved homeodomain-leucine zipper (HZ-Zip) regulator (Rerie et al., 1994). Lan et al. (2021) found that TCP4 directly interacts with GL3 to interfere with the transactivation activity of MBW complex, thereby inhibits the formation of trichomes. Our recently published study also showed that the trichome number was significantly increased in CRISPR/Cas9-edited BrrTCP4b turnip (Brassica rapa var. rapa) plants (Liu et al., 2022). This observation led us to speculate about a potential interaction between BrrTCP4b and BrrGL3 in turnip. The yeast two-hybrid (Y2H) and bimolecular luciferase complementation (BiLC) results showed that there was no interaction between BrrTCP4b and BrrGL3 (Fig. S1a and b). These data prompted us to investigate the molecular mechanism of BrrTCP4b in regulating trichome formation.
2. Materials and methods 2.1. Yeast two-hybrid analysisThe coding sequences of BrrTCP4b was fused in-frame with the GAL4 DNA-binding domain of the bait vector pGBKT7 (Takara Bio, Beijing, China) and transformed into yeast strain Y2HGold yeast strain. The library of turnip seedling cDNA fused with the GAL4 activation domain of the prey vector pGADT7-Rec was in the Y187 yeast strain (Taraka Bio). Y2HGold yeast strains were mated with Y187 yeast strains, suspended in synthetic medium dropout of SD-Leu-Trp-His-ADE (SD/-LTHA) and incubated at 30 ℃. The positive interactions were further tested through cloning the full-length CDS of BrrGL3 and BrrTTG1 were cloned into pGADT7 vectors, respectively. The respective combinations of vectors were co-transformed into the yeast strain Y2HGold. pGADT7 and pGBKT7 vectors were cotransformed as negative controls. And selection medium supplemented with SD-LTH containing 5 mM 3-AT was used. The interactions were observed after 3 days of incubation at 30 ℃. Primers used are listed in Table S1.
2.2. Co-immunoprecipitation assayFor Co-immunoprecipitation (Co-IP) assay, the coding sequences of BrrTCP4b and BrrTTG1 were cloned into the pRI101-GFP and pRI101-6flag, respectively. The resulting constructs were transformed into Agrobacterium strain GV3101 and subsequently co-infiltrated into Nicotiana benthamiana leaves. The leaves were harvested after 3 days infiltration and ground to powder in liquid nitrogen. Total protein was extracted with lysis buffer [25 mM Tris–HCl (pH7.5), 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5% (v/v) NP-40, 1 mM PMSF] and incubated with GFP-Trap (ChromoTek, Germany) at 4 ℃ for 3 h. The magnetic beads were washed three times with washing buffer, and then eluted into a volume of 30 μL using loading buffer before immunoblot analysis.
2.3. Bimolecular fluorescence complementation assay and bimolecular luciferase complementation assayFor Bimolecular fluorescence complementation (BiFC) assay, BrrTCP4b or BrrGL3 were fused to the N or C terminus of GFP respectively, and BrrTTG1 was fused with mCherry. The constructs were separately transformed into Agrobacterium strain GV3101. Nicotiana benthamiana leaves were observed at 3 d after co-infiltration. Confocal laser scanning microscopy was performed using epi-fluorescence microscopy (Olympus 80i, Olympus Corporation, Japan).
For Bimolecular luciferase complementation (BiLC) assay, BrrTCP4b was fused to the N-terminus of LUC, BrrGL3 and BrrTTG1 were fused to the C-terminus of LUC respectively. The resulting constructs were transformed into Agrobacterium strain GV3101, respectively. Luciferase activity was measured using Tanon 5200S Luminescent Imaging Workstation at 3 d after co-infiltration in Nicotianabenthamiana leaves. Three biological replicates were analyzed. Primers used are listed in Table S1.
2.4. Chromatin immunoprecipitation (ChIP) assayA ChIP assay was performed according to Meng et al. (2018). Two-week-old BrrTCP4b-GFP transgenic plants were used. Immunoprecipitation was carried out using an Anti-GFP antibody (ab290, Abcam, Shanghai, China). The enriched DNA fragments were analyzed through qRT-PCR. The primers used were: P1–F: 5′ TAGCTGACGATGAGGATCGGA 3′, P1-R: 5′ TGATCCTCCTCACCCTCCAAA 3′, P2–F: 5′ ACATACATATACGCTAATGAG 3′ and P2-R: 5′ GTGTAAGCATGTGCAAACAT 3′.
2.5. Quantitative real-time PCRFor quantitative real-time PCR (qRT-PCR), total RNA was extracted using the Eastep® Super Total RNA Extraction Kit (Promega). 2 μg total RNA was used to reverse transcription using GoScript™ Reverse Transcriptase (Promega) according to the manufacturer's instructions. qRT-PCR was performed using FastStart Universal SYBR Green Master Mix (ROX). The data analysis was performed using Step One Plus Real-Time PCR System (Applied Biosystems). Three biological replicates were carried out for each sample. Primers used are listed in Table S1.
2.6. Transient transcriptional activation assayFor generating the reporter constructs, 2 Kb of BrrGL2 promoter was amplified from the genomic DNA of turnip and cloned into the pRI101-LUC. ProGL2:LUC was transformed into Agrobacterium strain GV3101, and co-injected with 35S: BrrTCP4b, 35S: BrrTTG1 and 35S: BrrGL3 were used as effectors. The infiltrated leaves of Nicotiana benthamiana plants were sprayed with luciferin solution at 3 d after infiltration, and imaged using a Tanon 5200S Luminescent Imaging Workstation. Three biological replicates were analyzed. Primers used are listed in Table S1.
3. Results and discussionIn order to investigate the molecular mechanism by which BrrTCP4b regulates trichome development in turnip, we performed a Y2H screen using BrrTCP4b as the bait. Our finding revealed that BrrTTG1, a key WD40-repeat protein transcription factor in the MBW complex, interacts with BrrTCP4b in yeast (Fig. 1a).

|
| Fig. 1 BrrTCP4b physically interacts with BrrTTG1. a, Y2H assays of BrrTCP4b and BrrTTG1 proteins. The transformed yeasts were spotted on selection medium (SD-Leu-Trp-His) containing 5mM 3-AT (3-amino-1, 2, 4-triazole). 3-AT was used to inhibit the self-activation for the co-transformed yeast. AD, activation domain. BD, DNA-binding protein. b, Firefly luciferase complementation assay to test the interaction between BrrTCB4b and BrrTTG1 in Nicotiana benthamiana. BrrTTG1 was fused with cLUC at its N-terminus. c, CoIP assay to confirm the interaction between BrrTCP4b and BrrTTG1 in vivo. d, Bimolecular fluorescence complementation assay to test the interaction between BrrTCP4b and BrrGL3 in Nicotiana benthamiana. BrrTCP4b was fused with nGFP at its C-terminus and BrrGL3 was fused with cGFP at its N-terminus. e, BrrTCP4b interacts with MBW complex in Nicotiana benthamiana. BrrTTG1 was fused with mCherry. BrrTCP4b, BrrGL3 and BrrTTG1 co-localized in nucleus and formed a complex, Scale Bar = 10 μm. All experiments repeated three times, yielding consistent results. |
To determine whether BrrTCP4b can interact with TTG1 in planta, BiLC was performed. The luminescence was obviously detected in the co-injection of BrrTCP4b and BrrTTG1 in Nicotiana benthamiana, but not in the control (Fig. 1b), suggesting that BrrTCP4b interacted with BrrTTG1 in vivo. To further confirm the interaction, the co-immunoprecipitation (Co-IP) assay was performed by co-expressing the GFP-tagged BrrTCP4b and Flag-tagged BrrTTG1 protein using the transient expression system in N. benthamiana. The results showed that BrrTTG1 was pulled down by BrrTCP4b (Fig. 1c). These results indicated that BrrTCP4b physically interact with BrrTTG1. And BrrTTG1 could interact with BrrGL3, it suggested that BrrTTG1 and BrrGL3 might form the MBW complex in turnip (Fig S1c). To test whether BrrTCP4b affected the transactivation activity of MBW complex, we firstly detected the interaction between BrrTCP4b and MBW complex. We found that there was no fluorescence signal when the constructs of nGFP-BrrTCP4b and cGFP-BrrGL3, driven by the CaMV 35S promoter, were co-injected in leaves of N. benthamiana (Fig. 1d). However, co-injection of 35S: nGFP-BrrTCP4b, 35S: cGFP-BrrGL3 and 35S: mCherry-BrrTTG1 into the leaves of N. benthamiana resulted in the detection of a fluorescence signal. The signal was also observed to be co-localized within the nucleus (Fig. 1e). These results implied that BrrTCP4b could interact with MBW complex. In the MBW complex, TTG1 functions as a scaffold, on which the DNA-binding MYB and bHLH proteins interact to generate the transcriptional complex (Airoldi et al., 2019). In this study, BrrTCP4b cannot interact with BrrGL3. When BrrTTG1 is present, BrrTCP4b can interact with BrrGL3. These results implied that BrrTCP4b might regulate trichome development through BrrTTG1 in turnip.
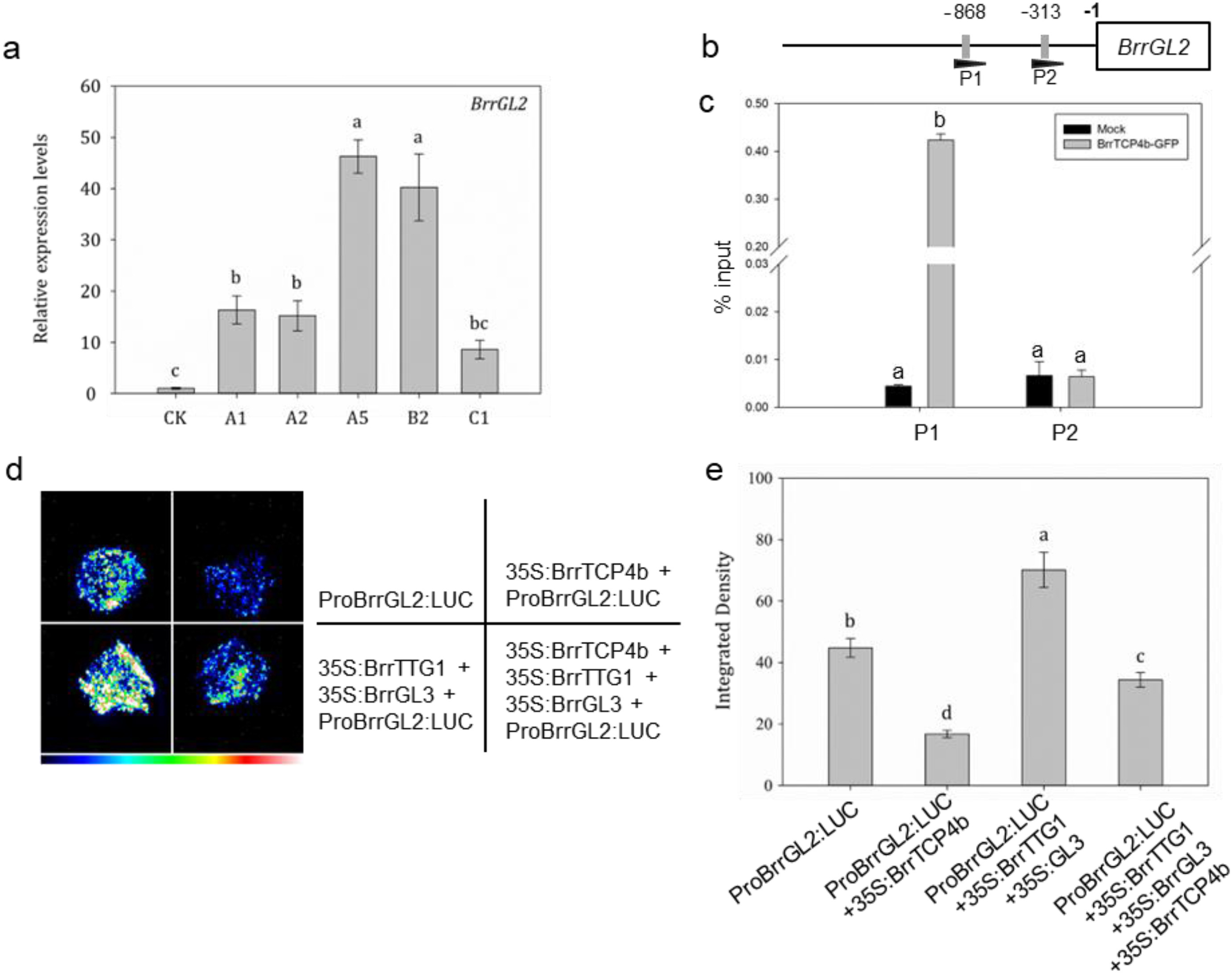
GL2 is a direct target of MBW in regulation of trichome formation (Zhao et al., 2008). The expression level of BrrGL2 was detected in the CRISPR/Cas9-edited BrrTCP4b turnip, the result showed that the expression level of BrrGL2 CRISPR//Cas9-edited BrrTCP4b turnip was obviously increased (Fig. 2a). ChIP-qPCR analysis revealed that BrrTCP4b directly binds to the promoter of BrrGL2 (Fig. 2b). This finding suggests that BrrTCP4b may directly bind to the GL2 promoter, thereby inhibiting its expression. Subsequently, a transient expression assay was performed to further validate this interference. The data demonstrated that co-injection of 35S: BrrTTG1 and 35S: BrrGL3 with ProBrrGL2:LUC could significantly enhanced the LUC activity. Whereas the LUC activity of the co-injection of 35S: BrrTCP4b, 35S: BrrTTG1, and 35S: BrrGL3 with ProBrrGL2:LUC was significantly reduced (Fig. 2c and d). BrrTTG1 and BrrGL3 could active the expression of BrrGL2 in turnip (Fig. S2). These results suggested that BrrTCP4b inhibited the activity of MBW complex by directly interacting with the WD-40 repeat protein BrrTTG1 and was a negative regulator in the development of trichomes in turnip.
|
| Fig. 2 BrrTCP4b disrupts the transactivation activity of MBW complex. a, Relative expression level of BrrGL2 in CRISPR-Cas9-BrrTCP4b turnip. A1, A2, A5, B2 and C1 represent five BrrTCP4b gene-edited lines. The expression level of BrrGL2 in wild-type turnip was set as one. BrrTUB was used as the reference gene. The data are means (±SE) of three biological replicates. b, Schematic of the location of BrrTCP4b-binding motif in the promoter of BrrGL2 gene. P1 represents BrrTCP4b-binding site. P2 represents the non-binding site of BrrTCP4b, as a negative control. c, Chromatin immunoprecipitation assay of BrrTPC4b binding to the promoter region of BrrGL2. Relative enrichment was calculated as the value of the amplified signal normalized against that of the input DNA. Data presented are the means ± SD of three biological replicates. d, Transient expression assay showing that the activation of GL2 expression by BrrTTG1/BrrGL3 was repressed by BrrTCP4b. The ProBrrGL2:LUC reporter was co-injected with corresponding constructs. e, Quantification of relative luminescence. The data are the means (±SE) of four biological replicates. Letters indicate statistically significant difference in the means (P < 0.05). Statistical analyses were performed by one-way ANOVA analysis. |
In summary, our findings revealed that BrrTCP4b physically interacts with BrrTTG1 to suppress trichome formation in turnip, which is different from the model plant Arabidopsis. The Brassica lineage underwent an additional whole genome triplication (WGT) event after its divergence from the Arabidopsis lineage (Cheng et al., 2014). Multiple copies of genes can arise from WGT and these copies must provide a function to the organism to be preserved in the genome (Birchler and Yang, 2022). This led us to hypothesize that BrrTCP4b might acquire a new function following the genome triplication in turnip. These findings contribute to our understanding of the regulatory mechanism of trichome development in the non-model plants turnip.
AcknowledgementsThis work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, Pan-Third Pole Environment Study for a Green Silk Road (Pan-TPE) (XDA2004010306) and the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502), and Science and Technology Program of Xizang Autonomous Region (XZ202001ZY0003G).
CRediT authorship contribution statement
Cheng Li: Writing – original draft, Data curation. Li Zhang: Formal analysis, Data curation. Hefan Li: Resources. Yuanwen Duan: Resources, Data curation. Xuemei Wen: Writing – review & editing. Yongping Yang: Writing – review & editing, Funding acquisition. Xudong Sun: Writing – review & editing, Funding acquisition, Formal analysis, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.03.003.
Airoldi, C.A., Hearn, T.J., Brockington, S.F., et al., 2019. TTG1 proteins regulate circadian activity as well as epidermal cell fate and pigmentation. Nat. Plants, 5: 1145-1153. DOI:10.1038/s41477-019-0544-3 |
Birchler, J.A., Yang, H., 2022. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell, 34: 2466-2474. DOI:10.1093/plcell/koac076 |
Cheng, F., Wu, J., Wang, X., 2014. Genome triplication drove the diversification of Brassica plants. Horticulture Research, 1: 14024. DOI:10.1038/hortres.2014.24 |
Hauser, M.T., 2014. Molecular basis of natural variation and environmental control of trichome patterning. Front. Plant Sci., 5: 320. |
Ishida, T., Kurata, T., Okada, K., et al., 2008. A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol., 59: 365-386. DOI:10.1146/annurev.arplant.59.032607.092949 |
Lan, J., Zhang, J., Yuan, R., et al., 2021. TCP transcription factors suppress cotyledon trichomes by impeding a cell differentiation-regulating complex. Plant Physiol., 186: 434-451. DOI:10.1093/plphys/kiab053 |
Liu, Y., Zhang, L., Li, C., et al., 2022. Establishment of Agrobacterium-mediated genetic transformation and application of CRISPR/Cas9 genome-editing system to Brassica rapa var. rapa. Plant Methods, 18: 98. DOI:10.1186/s13007-022-00931-w |
Meng, L.S., Li, C., Xu, M.K., et al., 2018. Arabidopsis ANGUSTIFOLIA3 (AN3) is associated with the promoter of CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) to regulate light-mediated stomatal development. Plant Cell Environ., 41: 1645-1656. DOI:10.1111/pce.13212 |
Oppenheimer, D.G., Herman, P.L., Sivakumaran, S., et al., 1991. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell, 1: 483-493. |
Payne, C.T., Zhang, F., Lloyd, A.M., 2000. GL3 encodes a bHLH protein that regulates trichome development inI through interaction with GL1 and TTG1. Genetics, 156: 1349-1362. DOI:10.1093/genetics/156.3.1349 |
Rerie, W.G., Feldmann, K.A., Marks, M.D., 1994. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev., 8: 1388-1399. DOI:10.1101/gad.8.12.1388 |
Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., et al., 1999. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell, 11: 1337-1350. DOI:10.2307/3870753 |
Wang, X., Shen, C., Meng, P., et al., 2021. Analysis and review of trichomes in plants. BMC Plant Biol., 21: 70. DOI:10.18282/l-e.v10i3.2394 |
Zhao, M., Morohashi, K., Hatlestad, G., et al., 2008. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development, 135: 1991-1999. DOI:10.1242/dev.016873 |



