Wheat is usually sown in fall and then passes through the winter to flower in warm spring. This cooling of winter wheat seeds, which accelerates flowering, is called vernalization. The genetic basis of vernalization response in winter wheat has been extensively studied. To date, four genes have been identified that directly participate in the wheat vernalization response, namely VRN1, VRN2, VRN3 and VRN4 (Yan et al., 2003, 2004, 2006; Kippes et al., 2015). However, it is unclear whether these same genes play a role in responding to vernalization in spring wheat varieties.
VRN1 (also known as VRT1), which is homologous to APTALA1 in Arabidopsis thaliana, encodes a MADS-box family transcription factor (Yan et al., 2003). VRN1 is expressed at low levels before vernalization, but after vernalization induction, VRN1 is highly expressed in the apex and leaves, and the expression level increases with the increasing duration under low temperature (Danyluk et al., 2003; Distelfeld et al., 2009). In hexaploid wheat, there are three homologous copies of TaVRN1 gene (Trevaskis, 2010; Konopatskaia et al., 2016). VRN2 is a flowering inhibitor that includes two closely located genes encoding CCT domain containing proteins, ZCCT1 and ZCCT2 (Yan et al., 2004). In hexaploid wheat, there are six homologous copies of TaVRN2. Mutations in the CCT domain of TaVRN2 or the deletion of the entire TaVRN2 gene can weaken the requirement of vernalization (Yan et al., 2004). In addition, TaVRT2 has been shown to interact with TaVRN1 and overexpression of TaVRT2 in some winter wheat varieties (e.g., Kenong199) promotes heading even under incomplete vernalization conditions (Xie et al., 2021). VRN3 is an ortholog of the Flowering T locus (FT) gene in Arabidopsis and the Heading date 3a (Hd3a) gene in rice. It encodes "florigen" in wheat. VRN3 expression levels are higher in varieties with retrotransposon insertions in the VRN3 promoter region. Studies have shown that TaVRN3 interacts with TaFDL2 and 14-3-3 proteins to form a flowering activation complex, which then binds to the ACGT cis-acting element in the TaVRN1 promoter region to activate the expression of TaVRN1, thus promoting flowering in wheat (Li and Dubcovsky, 2008; Li et al., 2015). VRN3 can integrate vernalization and photoperiod signals to accelerate flowering. Under long day conditions, VRN2 suppresses the expression of VRN3 by inhibiting the expression of VRN1 to delay wheat flowering (Distelfeld and Dubcovsky, 2010; Chen and Dubcovsky, 2012). VRN1 binds to the promoter regions of VRN2 and VRN3 to suppress the expression of VRN2 and to promote the expression of VRN3 (Deng et al., 2015). The photoperiod genes PPD1 and CO also affect the expression of VRN3. Phytochrome C (PHYC) mediates the transcriptional activation of PPD1 and CO, thereby upregulating the expression of VRN3 (Chen et al., 2014; Pearce et al., 2016).
Changes in the expression of vernalization-responsive genes are often coupled with changes in histone modification. Studies have shown that histone modifications are extensive in genes of the flowering pathway, photoperiod, and circadian clock (Huan et al., 2018). Previous studies have also reported that the overall levels of H3K27me3 and H3K4me3 modifications in winter (Norstar) and spring wheat (Manitou) increase after vernalization (Diallo et al., 2012). Specifically, Chromatin Immunoprecipitation followed by Quantitative Real-Time PCR (ChIP-qPCR) has been used to show that the H3K4me3 modification level on TaVRN1A increased in these two varieties, whereas there was no significant difference in H3K27me3 modification level. Similarly, in an additional wheat variety (i.e., Kenong199), vernalization increased H3K4me3 modification levels of TaVRN1A, but decreased H3K27me3 modification levels of the same gene (Xie et al., 2021). Interestingly, in both a wheat variety that requires vernalization (Sonja) and one that does not (Morex), H3K4me3 modification levels on HvVRN1 were found to increase, whereas the H3K27me3 modification levels on HvVRN1 decreased (Oliver et al., 2009). However, more data is required to further confirm the dynamic modifications of H3K4me3 and H3K27me3 on the VRN1 gene before and after vernalization.
In this study, we examined the molecular mechanisms that regulate vernalization response in winter and spring wheat varieties. For this purpose, we determined how major vernalization genes (VRN1, VRN2, and VRN3) respond to vernalization in these varieties and whether modifications to histones play a role in changes in gene expression. We also identified genes that are differentially expressed in response to vernalization in winter and spring wheat varieties.
2. Materials and methods 2.1. Plant materials and vernalization treatmentsWheat varieties involved in this study included Chinese Spring (CS), Fielder, Kenong199 (KN199), and Aikang 58 (AK58). The five vernalization treatment conditions were as follows: (a) plants were grown at 22 ℃ for 2 weeks (referred to as V0); (b) plants were grown at 22 ℃ for 2 weeks and then transferred to 4 ℃ for 2 weeks (referred to as V14); (c) plants were grown at 22 ℃ for 2 weeks and then transferred to 4 ℃ for 2 weeks before being returned to 22 ℃ for 1 week (referred to as V14N7); (d) plants were grown at 22 ℃ for 2 weeks and then transferred to 4 ℃ for 4 weeks (referred to as V28); (e) plants were grown at 22 ℃ for 2 weeks then transferred to 4 ℃ for 4 weeks before being returned to 22 ℃ for 1 week (referred to as V28N7). All vernalization treatments were carried out in a climate-controlled growth chamber.
2.2. RNA extraction and reverse transcriptionEntire above-ground parts of wheat plants were sampled for RNA extraction by TRIzol. Reverse transcription was performed using the TransGen Reverse Transcription kit (ET101-01). Two micrograms of RNA were used for cDNA synthesis, and after reverse transcription, the cDNA was diluted 4-fold for subsequent experiments.
2.3. RNA-seq data processingAK58 seedlings grown under different conditions (i.e., V0, V28, and V28N7) were collected for RNA extraction and library preparation. Barcoded cDNA libraries were constructed using the VAHTS Universal V8 RNA-seq Library Prep Kit (NRM605-02) and were sequenced on MGI DNBseq-T7 platforms at 150 bp. Trimmomatic (version 0.32; Bolger et al., 2014) was used to remove adaptors from raw sequencing reads following the following parameter: -q 25 –paired –length 36 –stringency 3 -e 0.1. Clean data were aligned to the Chinese Spring reference sequence from the International Wheat Genome Sequencing Consortium (IWGSC) (version 1.1) using the STAR software (v.2.7.0c; Dobin et al., 2013). RSEM software (v.1.3.3; Li and Dewey, 2011) was used for quantification analysis. Differentially expressed genes were identified using DESeq2 software (Love et al., 2014) with P value < 0.05 and |log2 fold changes| ≥ 1, 2, or 3. STEM analysis was performed using the OmicStudio online software (Lyu et al., 2023). The TGT Online website (http://wheat.cau.edu.cn/TGT/m3/?navbar=GOEnrichment) was used for GO analysis on differentially expressed genes (Chen et al., 2020).
2.4. Chromatin immunoprecipitation (ChIP) assayChIP assay was performed according to a previous report (Zhao et al., 2020) with minor modifications. For each ChIP experiment about 0.2 g of tissue was used. Fresh seedlings were cut into small pieces and placed in MC buffer (10 mM sodium phosphate, pH 7, 50 mM NaCl and 0.1 M sucrose) that contained 1% formaldehyde for cross-linking. The samples were vacuum-fixed twice (10 min for each time). Then 0.125 M glycine was added to terminate the crosslinking. After vacuuming for 5 min, the samples were washed with MC buffer and stored at -80 ℃ for later use. Samples were ground into powder using liquid nitrogen and chromatin was fragmented into 200–600 bp by sonication (Covaris S220) at 200 cycles per burst, peak power for 10 min. Chromatin was immunoprecipitated using anti-H3K27me3 (9733S, CST) or anti-H3K4me3 (ab8580, Abcam). After de-crosslinking and proteinase K treatment, the precipitated DNA was purified by QIAquick® PCR Purification Kit (28106, QIAGEN) for later qPCR analysis.
2.5. Quantitative polymerase chain reaction (qPCR) and ChIP-qPCRGene expression analysis was carried out with the 2X SYBR Green Fast qPCR Mix Kit (RK21204, ABclonal Corporation). qPCR reactions included 1 μL cDNA, 10 μL 2X Supermix, 0.8 μL Primer mix, and 8.2 μL ddH2O. Amplification was performed on a CFX96 Real-Time System PCR instrument (Bio-Rad) and fluorescence values were collected. The relative expression levels of genes were calculated using the 2^(-ΔΔCt) method with TaActin as the internal reference gene. For ChIP-qPCR assay, input DNA and immunoprecipitated DNA were used simultaneously for qPCR detection, with input DNA serving as control. The primers used for qPCR and ChIP-qPCR are listed in Table S6.
3. Results 3.1. The effect of vernalization on the expression of major vernalization genes in both winter and spring wheatSpring wheat varieties can be planted in autumn in areas where winter is not too harsh. In this case, plants would experience low temperature in winter. To study the effect of vernalization on spring wheat varieties, we compared the expression of VRNs in four wheat varieties, including two spring varieties (i.e., Chinese Spring, CS and Fielder) and two winter varieties (i.e., Aikang58, AK58 and Kenong199, KN199), that were grown under five vernalization conditions: no vernalization; two- or four-week vernalization; two- or four-week vernalization followed by 7 days at room temperature (Fig. 1A). We found that in both spring and winter wheat varieties the homologs of VRN1 (VRN1A, VRN1B, and VRN1D) were expressed at higher levels following vernalization treatment. In spring wheat varieties, the expression of all the three VRN1s continued to increase after plants were returned to normal temperatures, except in Chinese Spring wheat following the two-week vernalization treatment (Fig. 1B, C and D). In winter wheat varieties, expression of VRN1s decreased greatly after plants were returned to normal temperatures, with the largest decrease in expression observed in plants vernalized for four weeks. These results indicate that vernalization induces expression of VRN1 genes in both spring and winter wheat. However, in winter wheat varieties, VRN1 expression must be maintained by low temperatures.

|
| Fig. 1 The effect of vernalization on the expression of VRN1A in different wheat varieties. (A) A schematic diagram illustrating five vernalization treatment conditions. The red color represents plant growth at 22 ℃, the green color represents plant grown at 4 ℃. The expression of VRN1A (B), VRN1B (C) and VRN1D (D) in four wheat varieties under five vernalization conditions are shown. The internal reference is TaActin. Results represent data from three biological replicates. Bar = mean ± sd. |
In both spring and winter wheat varieties, VRN1 and VRN2 expression patterns were negatively correlated. Specifically, in winter wheat, when plants were returned to normal temperatures after two- and four-week vernalization treatments, VRN1 expression decreased, whereas VRN2 expression increased (Fig. S1A). Our findings suggest that VRN1 acts as a major upstream negative regulator of VRN2 in both winter wheat and spring wheat, which is consistent with models put forth in previous studies (Xu and Chong, 2018). VRN3 was also expressed at higher levels following a four-week vernalization treatment (Fig. S1B). However, VRN3 expression did not decrease in plants returned to normal temperatures following either a two- or four-week vernalization treatment. These results indicate that factors other than VRN1 and VRN2 regulate VRN3 expression.
3.2. Common and also divergent histone modification dynamics on VRN1 gene in spring and winter wheat regulated by vernalizationGene transcription is highly associated with histone modification. Furthermore, in barley and wheat vernalization has been shown to induce VRN1 expression by altering chromatin state (Oliver et al., 2009). To determine whether histone modifications underlie differences in VRN1 expression between winter and spring wheat, we evaluated dynamic changes of H3K27me3 and H3K4me3 on the VRN1 gene in Fielder and KN199 wheat varieties grown with no vernalization, a four-week vernalization, and a four-week vernalization followed by a return to normal temperatures (Fig. 2A). We found that H3K4me3 modification levels at the VRN1 gene increased following vernalization in both spring and winter wheat varieties (Fig. 2B). However, H3K4me3 modification levels decreased in KN199 but not in Fielder wheat following the return to a normal temperature. In Fielder wheat, levels of H3K27me3 modification at the VRN1A gene decreased after a four-week vernalization and a four-week vernalization followed by a return to normal temperatures. After returning plants from vernalization to normal temperatures, levels of H3K27me3 modification at the VRN1A gene decreased in KN199, but not as much as in Fielder wheat (Fig. 2C). This indicates that although H3K27me3 and H3K4me3 modifications are both involved in the vernalization response in wheat and have an impact on VRN1 expression, the catabolism of H3K4me3 and H3K27me3 on VRN1 induced by vernalization may differ between winter and spring wheat.
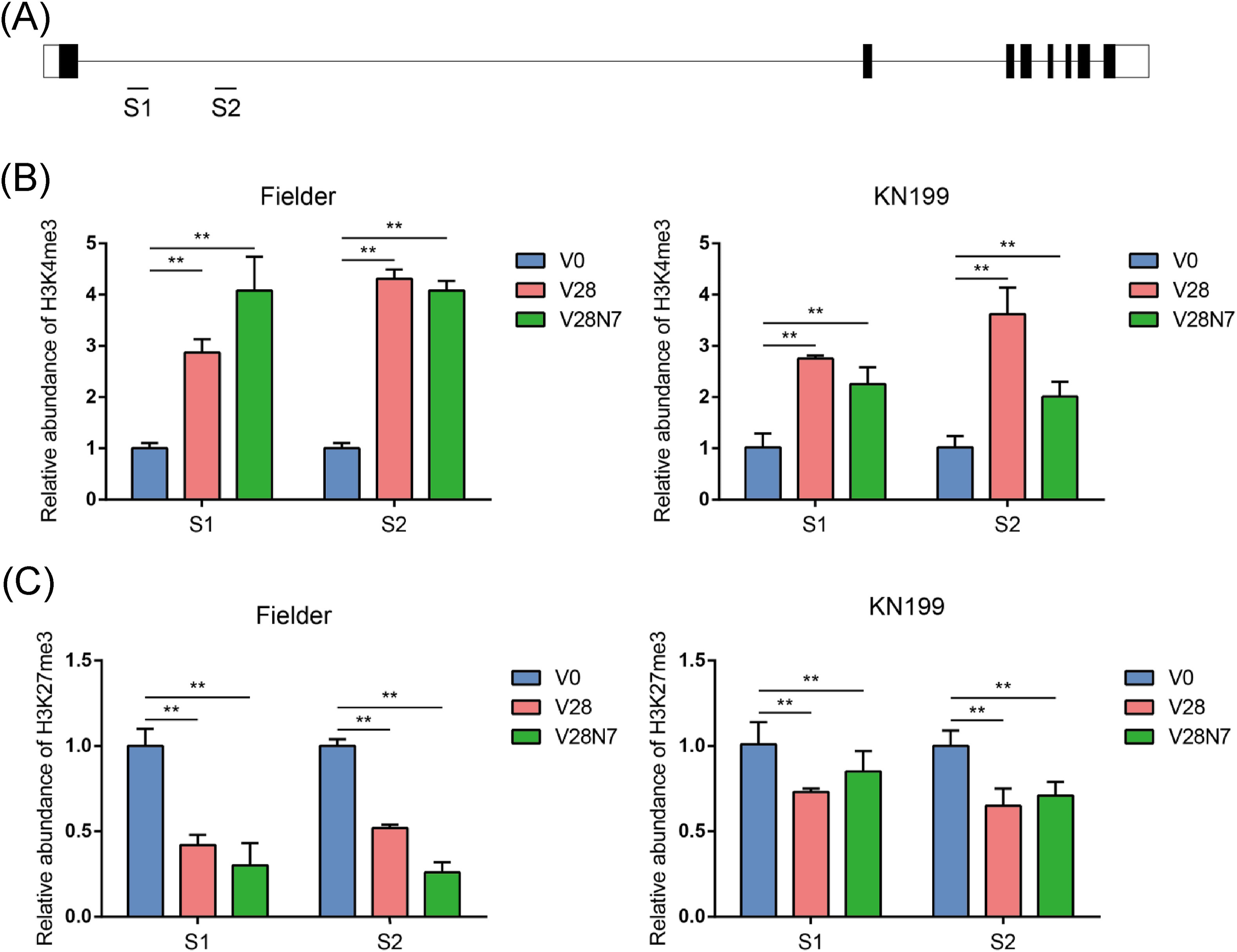
|
| Fig. 2 The effect of vernalization on the levels of H3K27me3 and H3K4me3 modifications at the VRN1A locus. (A) Gene structure diagram of VRN1A. The black lines represent introns, black rectangles represent exons, and white regions represent non-coding regions. S1 and S2 indicate the positions of primers for ChIP-qPCR detection. Changes in H3K4me3 modification levels (B) and H3K27me3 modification levels (C) of VRN1A in Fielder and KN199 varieties grown with no vernalization (V0), a four-week vernalization (V28), and a four-week vernalization followed by a return to room temperature (V28N7). The calculated values represent relative enrichment levels of ChIP-DNA compared to input DNA with three replicates, and was normalized to histone modification levels in plants that were not vernalized (V0). Significance was obtained from the student's t-test. **p < 0.01. |
We also examined the genome-wide effect of vernalization on wheat gene expression in a winter variety (AK58) grown with no vernalization, a four-week vernalization, and a four-week vernalization followed by 1 week at normal temperatures. For these treatments, RNA sequencing (RNA-seq) generated 106, 154, 530, 1, 203, 515, 301, and 125, 884, 336 reads on average, respectively. To assess the quality of the transcriptome data, we confirmed that the expression profiles of six randomly selected genes were consistent with those of the transcriptome (Fig. S2). In addition, PCA analysis clustered replicates from the same treatment together (Fig. 3A), indicating the reproducibility of the data.

|
| Fig. 3 Variation in gene expression profiles in a winter wheat variety (AK58) under different vernalization treatments. (A) PCA analysis of differentially expressed genes in AK58 grown with no vernalization (V0), a four-week vernalization (V28), and a four-week vernalization followed by a return to normal temperatures (V28N7). Comparisons were performed with AK58 grown after a four-week vernalization (V28) and those returned to room temperature following a four-week vernalization (V28N7). The number of downregulated genes (B), the number of upregulated genes (C) and the total number of differentially expressed genes (D) are shown in comparison with the V0 condition. (E) Differentially expressed genes between V28 and V0 conditions and between V28N7 and V0 conditions are also shown. |
To determine which genes respond to vernalization, we identified genes that were differentially expressed (DEGs) between different vernalization treatments. Genes with the highest probability of being regulated by vernalization were those that were differentially expressed between the following two treatments: no vernalization vs. a four-week vernalization; and no vernalization vs. a four-week vernalization followed by a return to normal temperatures. In total, 13, 256 and 6776 genes (fold change greater than 2) were differentially expressed between these treatments. Moreover, 5673 and 1719 DEGs with fold changes greater than 4 and 2991 and 671 DEGs with fold changes greater than 8 were also identified in each treatment (Fig. 3B, C and D).
For the complementary comparisons (i.e., a four-week vernalization vs. no vernalization; a four-week vernalization followed by return to normal temperature vs. no vernalization), there were a total of 3560 DEGs (fold change greater than 2) in common (Fig. 3E). Among these genes, 1919 genes were downregulated during the four-week vernalization and 1906 genes were downregulated in plants that were returned to normal temperatures following the vernalization. These DEGs are enriched for genes that play roles in the flavonoid biosynthesis, protein autoprocessing and the positive regulation of anthocyanin metabolism (Fig. 4A and C). Specifically, these DEGs include members of the cytochrome P450 superfamily, chalcone and stilbene synthase gene family, N-terminal nucleophile aminohydrolases (Ntn hydrolases) superfamily and basic-leucine zipper (bZIP) transcription factor family (Table S1). In addition, 1641 genes were upregulated in in response to a four-week vernalization treatment and 1654 genes were upregulated in plants that were returned to normal temperatures after vernalization. These genes are mainly enriched for the leucine catabolic process pathway (Fig. 4B and D). These findings suggest that vernalization activates flavonoid biosynthesis, protein autoprocessing, anthocyanin metabolism, and leucine catabolic processes.

|
| Fig. 4 GO analysis of genes differentially expressed under different vernalization conditions. |
VRN1 is a key regulator during vernalization. Our transcriptome data show that VRN1 expression is strongly induced by vernalization and significantly decreased after being returned to normal temperatures (Figs. 1 and S1A). To identify genes involved in the dynamic expression of VRN1 following a change in temperature, we used Short Time-series Expression Miner (STEM) analysis of 3560 genes to trace their dynamic expression during the vernalization process. These 3560 genes were divided into 16 clusters (Cluster 0–15; Fig. 5A, Table S2). Expression of genes in Cluster0, Cluster1, Cluster3, Cluster4, Cluster11, and Cluster14 changed significantly across three pseudo-time points. These clusters contain a total of 2531 genes. The expression patterns of genes in Cluster11 and Cluster14 were similar to that of VRN1, whereas genes in Cluster1 showed the opposite pattern. Cluster11 and Cluster14 are enriched for genes that are involved in the negative regulation of red or far-red light signaling pathway and leucine catabolic process.

|
| Fig. 5 Identification and functional analysis of vernalization-responsive genes in wheat. (A) STEM analysis of vernalization-responsive genes in wheat. A total of 16 clusters were identified with Cluster 0, Cluster 1, Cluster 3, Cluster 4, Cluster 11, and Cluster 14 showing significant gene expression dynamics. (B) Heatmap of expression dynamics of 110 transcription factors under V0, V28, and V28N7 treatments. (C) Regulatory network based on transcription factors differentially expressed in response to vernalization. |
Cluster1 is enriched for genes in the flavonoid biosynthetic process (Fig. S3; Table S3). Clusters1, 11, and 14 collectively contain 1575 genes, including 110 transcription factors (Table S4). Transcription factor expression patterns differed among plants treated with no vernalization, a four-week vernalization, and a four-week vernalization followed by a return to normal temperatures (Fig. 5B). These transcription factors include MADS-box, MYB, TCP, and WRKY genes. Previous studies that identified DNA elements that bind to these transcription factors (Inukai et al., 2017) allowed us to predict the regulatory relationships among these transcription factors. Accordingly, we speculate that VRN1 regulates VRN2 and FPF, which is in line with previous reports (Greenup et al., 2010; Deng et al., 2015). In addition, four cold-regulated genes (TaCORs), a class of genes believed to respond to low-temperatures (Park et al., 2015), appear to be targets of VRN1. Moreover, VRN1 has an indirect effect on Wheat PHYTOCLOCK 1 (WPCL1) mediated by VRN2. WPCL1 is a homologous to LUX ARRHYTHMO (LUX)/PHYTOCLOCK 1 (PCL1) in Arabidopsis, which participates in the regulation of the circadian clock and flowering time (Mizuno et al., 2012, 2016). VRN1, VRT2, HSFB, ERF, and ZHD are all involved in the regulation of WPCL1, indicating the link of vernalization and circadian clock. In addition, VRN2 is appears to be regulated by multiple transcription factors, including TCP, ARF, HSFB and ABI. TaFT-5A, a homolog of VRN3, is also regulated by TCP.
To identify the possible regulators that mediate changes of histone modifications on the VRN1 gene during vernalization, we analyzed the expression of genes encoding histone writers or erasers in wheat (Wang et al., 2021). We found that the expression of some H3K4me3 methyltransferases in wheat was induced by vernalization, whereas the expression of other H3K4me3 demethylases was inhibited by vernalization. The expression of some H3K27me3 methyltransferases was inhibited by vernalization, whereas the expression of some H3K27me3 demethylases was induced by vernalization. However, the expression of H3K4me3 demethylase, JMJ14 and large proportion of H3K27me3 methyltransferases, including MEA, LHP1 and FIS, was induced also by vernalization (Fig. S4). This indicates that only particular histone writers or erasers are involved in the vernalization-induced transcription of VRN1A in wheat. Further investigation on these vernalization-related transcription factors will provide new insights into wheat vernalization.
4. Discussion 4.1. The impact of vernalization time on wheatWheat can be cultivated in a wide range of regions and high yield is greatly enhanced by appropriate flowering time (Gororo et al., 2001; Kamran et al., 2014). As a major gene controlling the flowering of wheat, VRN1 mainly gives wheat its spring or winter characteristics depending on different genotypic alleles (Yan et al., 2003). The combination of VRN1 genotype alleles and other genes controlling flowering time determines the regions where wheat can be grown. Winter wheat cannot complete its reproductive transition without vernalization. The expression level of VRN1 in winter wheat is strongly induced by vernalization and maintained at a high level after vernalization, preparing wheat for reproductive growth.
In this study, the expression levels of VRN1, VRN2, and VRN3 were examined in four wheat varieties under five vernalization treatment conditions. It was found that sufficient vernalization time (28 days of vernalization followed by 7 days of recovery at room temperature) could promote the expression of VRN1, while insufficient vernalization time (14 days of vernalization followed by 7 days of recovery at room temperature) could not fully induce the expression of VRN1. This indicates that both spring and winter wheat varieties respond to vernalization, but the degree of response varies among different wheat varieties. Additional vernalization treatments on more wheat varieties will lay a foundation for determining the time of response to vernalization for different wheat varieties and provide more insight into wheat breeding.
4.2. The effect of epigenetic factors on the expression of vernalization gene VRN1The expression of the vernalization gene VRN1 is affected by epigenetic factors, such as DNA methylation, non-coding RNA, and histone modifications (Khan et al., 2013; Huan et al., 2018; Xu et al., 2021). Studies have shown that DNA methylation and demethylation are also involved in the vernalization process in plants. CG methylation exists throughout the VRN1 gene region, while non-CG methylation mainly exists in the first intron of VRN1. Low temperature specifically induces an increase in non-CG methylation in the first intron of VRN1, and this change in methylation is reset in the next generation (Khan et al., 2013). In addition to DNA methylation, the expression of TaVRN1A is also regulated by non-coding RNA. VAS is a non-coding RNA generated from the antisense strand of TaVRN1A that only exists in winter wheat. During early vernalization, VAS expression is induced, which promotes wheat flowering by positively regulating the expression of TaVRN1A. During mid-vernalization, VAS helps the complex formed by TaRF2b and TaRF2a anchor in the Sp1 site of TaVRN1A, activating the expression of TaVRN1A (Xu et al., 2021).
Both H3K27me3 and H3K4me3 modifications are involved in the wheat vernalization process. In this study, the H3K27me3 and H3K4me3 modification levels at the VRN1A locus in Fielder and KN199 varieties were tested before vernalization (V0) and after vernalization (V28N7). The results showed that in these two varieties, vernalization could reduce the H3K27me3 modification level at the VRN1A locus, and it could increase the H3K4me3 modification level at the VRN1A locus. Transcription data indicated that specific histone writers or/and erasers are involved in the vernalization-induced transcription of VRN1A in wheat (Fig. S4). This may help us to discover the upstream regulator of vernalization response in wheat. In rice, distinct forms and levels of histone modification vary across the genome and among varieties (Zhao et al., 2020), and may also occur among wheat varieties.
4.3. Regulation of vernalization in cereal cropsThe distribution of H3K4me3 and H3K27me3 modifications in the genome before and after vernalization in Brachypodium distachyon revealed that H3K27me3 modification is selectively regulated in multiple regions during vernalization and Chromosome 4 may be the main target of this selective regulation, whereas H3K4me3 modification does not have this feature (Huan et al., 2018). During vernalization, the levels of H3K4me3 and H3K27me3 modification on thousands of genes change. Cluster analysis of gene function classifies these genes into several categories, including maintenance of vernalization memory, low temperature response, and flowering and stress response. Vernalization changes the overall distribution of H3K27me3 and H3K4me3 modifications in B. distachyon. However, the effect of vernalization on the overall distribution of H3K27me3 and H3K4me3 modifications in hexaploid wheat are not clear yet and requires further study.
By comparing the transcriptome data of non-vernalized, vernalized and post-vernalized Brachypodium distachyon, 674 genes that continuously respond to vernalization were defined as vernalization-memory-related genes (Huan et al., 2013). There were conserved and specific vernalization-memory-related genes found in B. distachyon and barley. In this study, transcriptome analysis of wheat seedlings grown under different vernalization conditions identified seven clusters of 2531 genes that continuously respond to vernalization. Many of these genes play roles in leucine catabolism, the biosynthesis of unsaturated fatty acid, monoterpenoid biosynthesis, and flavonoid biosynthesis; in addition, several of these genes are known to act as negative regulators in the red or far-red light signaling pathway and positive regulators during anthocyanin metabolism. Twelve of these genes overlap with orthologues in Brachypodium and barley (Table S5), indicating that vernalization response is both conserved, although with some species specificity, in temperate plants.
Furthermore, we discovered a large number of transcription factors that may be involved in the vernalization response in wheat. We have used data on differentially expressed transcription factors to propose a regulatory network that highlights novel regulatory relationships in the vernalization response, e.g., VRNs regulate WPCL1. Our findings improve our understanding of vernalization based on the response to environmental stimulates.
AcknowledgementsThis work was supported by Project 2662020ZKPY002 supported by the Fundamental Research Funds for the Central Universities.
Data availability
The RNA-Seq data generated in this study have been deposited in the NCBI/Sequence Read Archive (SRA) database under accession code PRJNA1019635.
CRediT authorship contribution statement
Yunzhen Li: Investigation, Formal analysis, Data curation, Writing – original draft. Liujie Jin: Resources, Validation. Xinyu Liu: Resources. Chao He: Software, Formal analysis. Siteng Bi: Software. Sulaiman Saeed: Writing – review & editing. Wenhao Yan: Conceptualization, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.02.005.
Bolger, A.M., Lohse, M., Usadel, B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30: 2114-2120. DOI:10.1093/bioinformatics/btu170 |
Chen, A., Dubcovsky, J., 2012. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genetics, 8: e1003134. DOI:10.1371/journal.pgen.1003134 |
Chen, A., Li, C.X., Hu, W., et al., 2014. Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc. Natl. Acad. Sci. U.S.A., 111: 10037-10044. DOI:10.1073/pnas.1409795111 |
Chen, Y.M., Song, W.J., Xie, X.M., et al., 2020. A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the Triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant, 13: 1694-1708. DOI:10.1016/j.molp.2020.09.019 |
Danyluk, J., Kane, N.A., Breton, G., et al., 2003. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol., 132: 1849-1860. DOI:10.1104/pp.103.023523 |
Deng, W., Casao, M.C., Wang, P., et al., 2015. Direct links between the vernalization response and other key traits of cereal crops. Nat. Commun., 6: 5882. DOI:10.1038/ncomms6882 |
Diallo, A.O., Ali-Benali, M.A., Badawi, M., et al., 2012. Expression of vernalization responsive genes in wheat is associated with histone H3 trimethylation. Mol. Genet. Genom., 287: 575-590. DOI:10.1007/s00438-012-0701-0 |
Distelfeld, A., Dubcovsky, J., 2010. Characterization of the maintained vegetative phase deletions from diploid wheat and their effect on VRN2 and FT transcript levels. Mol. Genet. Genom., 283: 223-232. DOI:10.1007/s00438-009-0510-2 |
Distelfeld, A., Tranquilli, G., Li, C.X., et al., 2009. Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol., 149: 245-257. DOI:10.1104/pp.108.129353 |
Dobin, A., Davis, C.A., Schlesinger, F., et al., 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29: 15-21. DOI:10.1093/bioinformatics/bts635 |
Gororo, N.N., Flood, R.G., Eastwood, R.F., et al., 2001. Photoperiod and vernalization responses in Triticum turgidum×T. Tauschii synthetic hexaploid wheats. Ann. Bot., 88: 947-952. DOI:10.1006/anbo.2001.1531 |
Greenup, A.G., Sasani, S., Oliver, S.N., et al., 2010. ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol., 153: 1062-1073. DOI:10.1104/pp.109.152488 |
Huan, Q., Mao, Z.W., Chong, K., et al., 2018. Global analysis of H3K4me3/H3K27me3 in Brachypodium distachyon reveals VRN3 as critical epigenetic regulation point in vernalization and provides insights into epigenetic memory. New Phytol., 219: 1373-1387. DOI:10.1111/nph.15288 |
Huan, Q., Mao, Z.W., Zhang, J.Y., et al., 2013. Transcriptome-wide analysis of vernalization reveals conserved and species-specific mechanisms in Brachypodium. J. Integr. Plant Biol., 55: 696-709. DOI:10.1111/jipb.12050 |
Inukai, S., Kock, K.H., Bulyk, M.L., 2017. Transcription factor-DNA binding: beyond binding site motifs. Curr. Opin. Genet. Dev., 43: 110-119. DOI:10.1016/j.gde.2017.02.007 |
Kamran, A., Iqbal, M., Spaner, D., 2014. Flowering time in wheat (Triticum aestivum L.): a key factor for global adaptability. Euphytica, 197: 1-26. DOI:10.1007/s10681-014-1075-7 |
Khan, A.R., Enjalbert, J., Marsollier, A.C., et al., 2013. Vernalization treatment induces site-specific DNA hypermethylation at the VERNALIZATION-A1 (VRN-A1) locus in hexaploid winter wheat. BMC Plant Biol., 13: 209. DOI:10.1186/1471-2229-13-209 |
Kippes, N., Debernardi, J.M., Vasquez-Gross, H.A., et al., 2015. Identification of the VERNALIZATION 4 gene reveals the origin of spring growth habit in ancient wheats from South Asia. Proc. Natl. Acad. Sci. U.S.A., 112: E5401-E5410. DOI:10.1073/pnas.1514883112 |
Konopatskaia, I., Vavilova, V., Kondratenko, E.Y., et al., 2016. VRN1 genes variability in tetraploid wheat species with a spring growth habit. BMC Plant Biol., 16: 244. DOI:10.1186/s12870-016-0924-z |
Li, B., Dewey, C.N., 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12: 323. DOI:10.1186/1471-2105-12-323 |
Li, C.X., Dubcovsky, J., 2008. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J., 55: 543-554. DOI:10.1111/j.1365-313X.2008.03526.x |
Li, C.X., Lin, H.Q., Dubcovsky, J., 2015. Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J., 84: 70-82. DOI:10.1111/tpj.12960 |
Love, M.I., Huber, W., Anders, S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 550. DOI:10.1186/s13059-014-0550-8 |
Lyu, F.Y., Han, F.R., Ge, C.L., et al., 2023. OmicStudio: a composable bioinformatics cloud platform with real-time feedback that can generate high-quality graphs for publication. iMeta, 2: e85. DOI:10.1002/imt2.85 |
Mizuno, N., Kinoshita, M., Kinoshita, S., et al., 2016. Loss-of-Function mutations in three homoeologous PHYTOCLOCK 1 genes in common wheat are associated with the extra-early flowering phenotype. PLoS One, 11: e0165618. DOI:10.1371/journal.pone.0165618 |
Mizuno, N., Nitta, M., Sato, K., et al., 2012. A wheat homologue of PHYTOCLOCK 1 is a candidate gene conferring the early heading phenotype to einkorn wheat. Genes Genet. Syst., 87: 357-367. DOI:10.1266/ggs.87.357 |
Oliver, S.N., Finnegan, E.J., Dennis, E.S., et al., 2009. Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proc. Natl. Acad. Sci. U.S.A., 106: 8386-8391. DOI:10.1073/pnas.0903566106 |
Park, S., Lee, C.M., Doherty, C.J., et al., 2015. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J., 82: 193-207. DOI:10.1111/tpj.12796 |
Pearce, S., Kippes, N., Chen, A., et al., 2016. RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways. BMC Plant Biol., 16: 141. DOI:10.1186/s12870-016-0831-3 |
Trevaskis, B., 2010. The central role of the VERNALIZATION1 gene in the vernalization response of cereals. Funct. Plant Biol., 37: 479-487. DOI:10.1071/FP10056 |
Wang, M., Li, Z., Zhang, Y., et al., 2021. An atlas of wheat epigenetic regulatory elements reveals subgenome divergence in the regulation of development and stress responses. Plant Cell, 33: 865-881. DOI:10.1093/plcell/koab028 |
Xie, L., Zhang, Y., Wang, K., et al., 2021. TaVrt2, an SVP-like gene, cooperates with TaVrn1 to regulate vernalization-induced flowering in wheat. New Phytol., 231: 834-848. DOI:10.1111/nph.16339 |
Xu, S.J., Chong, K., 2018. Remembering winter through vernalisation. Nat. Plants, 4: 997-1009. DOI:10.1038/s41477-018-0301-z |
Xu, S.J., Dong, Q., Deng, M., et al., 2021. The vernalization-induced long non-coding RNA VAS functions with the transcription factor TaRF2b to promote TaVRN1 expression for flowering in hexaploid wheat. Mol. Plant, 14: 1525-1538. DOI:10.1016/j.molp.2021.05.026 |
Yan, L., Fu, D., Li, C., et al., 2006. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A., 3: 19581-19586. DOI:10.1073/pnas.0607142103 |
Yan, L., Loukoianov, A., Blechl, A., et al., 2004. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science, 303: 1640-1644. DOI:10.1126/science.1094305 |
Yan, L., Loukoianov, A., Tranquilli, G., et al., 2003. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A., 100: 6263-6268. DOI:10.1073/pnas.0937399100 |
Zhao, L., Xie, L., Zhang, Q., et al., 2020. Integrative analysis of reference epigenomes in 20 rice varieties. Nat. Commun., 11: 2658. DOI:10.1038/s41467-020-16457-5 |



