b. National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China
Plant organ development involves continuing cell expansion and dynamics cell differentiation, leading to the acquisition of new functions (De Rybel et al., 2016; Guiziou et al., 2021; Ramirez-Parra et al., 2005). This intricate process relies on precise gene expression programs at the proper time and position. A plant organ is comprised of multiple cell types with distinct features, which are orchestrated by transcription factors (TFs) in intricate regulatory networks in a cell-type specific manner (Kaufmann and Airoldi, 2018). Triggered by internal and external signals, TFs bind to cis-elements of downstream genes to regulate gene expression to further fulfil cell identity establishment and maintenance. Gene regulatory networks (GRNs) containing TFs and their target genes shed light on how genes co-ordinately orchestrate cell fate specification. Therefore, network-based approaches can help elucidate the mechanisms that link genes, cells, tissues and organs in a systemic manner (Wu et al., 2022).
GRN analyses that integrate multiple data types at increased resolution have significantly improved our understanding of the complex molecular mechanisms controlling the development of flowers (Chen et al., 2018; Yan et al., 2016), roots (Brady et al., 2011; Moreno-Risueno et al., 2015; Reynoso et al., 2022), seeds (Santos-Mendoza et al., 2008), and secondary cell walls (Taylor-Teeples et al., 2015). The rapid development of single cell sequencing techniques which have been fully embraced by the plant community (Seyfferth et al., 2021), has greatly promoted in-depth knowledge for plant organ morphogenesis at single-cell resolution. In fact, GRNs constructed at single-cell level revealed novel mechanisms in the root morphogenesis process in Arabidopsis (Denyer et al., 2019; Roszak et al., 2021; Yang et al., 2021). This suggests that single-cell GRN (scGRN) analysis can recapitulate the complex and heterogeneous biological processes in the plant (Qian and Huang, 2020; Tripathi and Wilkins, 2021). Although progress has been made on constructing cell transcriptomic atlases in plants (Seyfferth et al., 2021), our current view of cellular taxonomy is restricted to specific organs or tissues, hampering our holistic understanding of plant cell fate specification. In fact, efforts from human studies have demonstrated that integration multiple single-cell transcriptomic datasets can boost the statistical power for discovery of rare cell phenotypes and potentially novel markers (Butler et al., 2018; Liu et al., 2021; Ryu et al., 2023; Stuart et al., 2019), highlighting the potential value of large-scale single-cell data integration.
In the current study, we presented for the first time a reference single-cell transcriptomic atlas of the whole Arabidopsis seedling by integration of 63 single cell transcriptomic datasets from representative tissues. In total, 47 distinct cell types have been identified from 382,724 cells, and several potential new cell types have been annotated. Additionally, we have established a cell marker gallery featuring numerous potentially new markers. Those markers can precisely reflect to the unique characteristics of their respective cell types since all the analysis is contacted in a holistic context. Furthermore, we systematically constructed cell-type specific GRNs and uncovered key regulators that act in a coordinated manner to control cell-type specific gene expression. Therefore, our integrative analysis will not only be a valuable resource for future plant single cell atlas construction, but also provides new insights for developmental regulatory mechanisms for specific cell types.
2. Materials and methods 2.1. scRNA-seq data collectionscRNA-seq datasets (n = 45; Table S1) generated in Arabidopsis juvenile seedlings by 10X Genomics or Drop-seq technologies were collected from previous studies (Denyer et al., 2019; Farmer et al., 2021; Jean-Baptiste et al., 2019; Liu et al., 2020; Long et al., 2021; Lopez-Anido et al., 2021; Ryu et al., 2019; Shulse et al., 2019; Wendrich et al., 2020; Zhang et al., 2019). We reanalysed these scRNA-seq data using the raw sequencing data in the fastq format, which were downloaded from the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/).
2.2. Pre-processing of raw sequencing dataThe raw 10X Genomics and Drop-seq fastq files were aligned, filtered and counted using the Cell Ranger pipeline v.3.1.0 (10X Genomics) and the Drop-seq tool v.1.13 (https://github.com/broadinstitute/Drop-seq) respectively. The Arabidopsis reference genome (v.TAIR10) and the corresponding GTF annotation file (v.Araport11) were downloaded from the Arabidopsis Information Resource (TAIR) database (https://www.arabidopsis.org/), followed by genome index built and filtered reads alignment using STAR (v.2.7.3a), which used as an aligner in Cell Ranger and Drop-seq tools. Specifically, for the alignment of reads generated from single nucleus, in order to accommodate the expression of precursor RNAs that contain introns, the intron regions of each read were removed and realigned to the reference genome. After barcodes and UMIs counting, the feature-barcode matrixes generated from Cell Ranger and the digital expression matrixes returned by Drop-seq tools, all of which with each unique molecular identifiers (UMIs) for every detected gene as a row and per valid cell barcode as a column, were used for subsequent analyses.
2.3. Integration of single cell dataFeature-barcode count matrices for each sample were processed with Seurat package (v.4.0.0) (Hao et al., 2021). Three types of cells were removed from the analysis: the ones expressed less than 200 genes, the ones detected as potential doublets by R package scater (v.1.20.1), or the ones have more than 10% of mitochondrial gene expression in UMI counts. “vst” in “FindVariableFeatures” function was utilized to determine the Top 3000 most variably expressed genes, “mt.percent” in “ScaleData” was further used with regression on the proportion of mitochondrial UMIs. “Two-stepwise strategy” was applied for scRNA-seq data integration. Firstly, scRNA-seq datasets from a specific study were aligned with canonical correlation analysis (CCA), then Reciprocal Principal Component Analysis (RPCA) method in Seurat was used for integration of scRNA-seq data from different studies (Büttner et al., 2019; Luecken et al., 2022). The datasets were normalized individually with SCTransform prior to integration (Hafemeister and Satija, 2019). We evaluated the performance of seven integration methods on the collected 63 scRNA-seq datasets using a benchmarking pipeline (https://github.com/theislab/scib-pipeline). We employed ten evaluation metrics, categorized into two groups: (1) batch effect removal and (2) preservation of biological variance. The first category includes principal component regression (batch), ASW (batch), graph connectivity, graph iLISI, and kBET. The second category comprises NMI, ARI, ASW (cell-type), graph cLISI, isolated label F1, and isolated label silhouette. The metrics were aggregated to generate an overall score and rank the methods. Seurat RPCA achieved the highest overall score, indicating its effectiveness in eliminating batch effects while preserving biological variation.
2.4. Cell clustering and annotation“RunPCA” function was used to compute the top 30 principal components (PCs) using top variably expressed genes on visualization purposes. Clustering was performed with integrated expression values based on shared-nearest-neighbor (SNN) graph clustering (Louvain community detection-based method) using “FindClusters” with a resolution of 0.8. Cell clusters were visualized using UMAP (Uniform Manifold Approximation and Projection).
The gene expression in the integrated RNA assay have been normalized, differential expressed genes (DEGs) in each cluster were identified using “FindAllMarkers” with non-parametric Wilcoxon rank sum test method. For cell annotation, we firstly constructed a cell type-specific marker genes database via collected annotated marker genes from previous published dataset, including single-cell studies as well as abundant gene expression studies (Table S3). Then, the defined cell types of our detected conserved cluster-specific marker genes were retrieved from the cell type-specific marker genes database. Finally, we performed function confirmation by reviewing literature and verified the expression pattern of each cell-specific marker gene. The expression pattern of cell known and expected cell type-specific marker genes across clusters were visualized to further confirm the cell type of each cluster.
2.5. Cell type deconvolutionWe first generated a cell-type expression matrix of top 100 marker genes from scRNA-seq data. Using this gene expression matrix, we then applied CIBERSORTx (Newman et al., 2019) with default parameters to deconvolute cell-type abundance from 96 bulk mRNA-seq datasets collected from the Plant Public RNA-seq Database (PPRD) database (Yu et al., 2022) (Table S5).
2.6. Gene regulatory network inference and regulons discoveryTF DNA binding motifs of Arabidopsis were downloaded from the JASPAR (Fornes et al., 2020) and CIS-BP (Weirauch et al., 2014) databases and subjected to redundancy filtering. The cisTarget database was constructed according to the SCENIC (Aibar et al., 2017) protocol (https://github.com/aertslab/create_cisTarget_databases). We used pySCENIC 0.11.2 (https://github.com/aertslab/pySCENIC) to infer gene regulatory network (Aibar et al., 2017). Briefly, SCENIC contains three steps: (1) identify co-expression modules between TF and the potential target genes; (2) for each co-expression module, infer direct target genes based on motif enrichment of the corresponding TF. Each regulon is then defined as a TF and its direct target genes; (3) calculate the Regulon Activity Score (RAS) in each single cell via the area under the recovery curve. The cell-type specificity of a regulon was quantified using an entropy-based approach, building upon the previously reported methodology centered on RAS (Suo et al., 2018).
2.7. Regulon-regulon co-association analysisWe used Paired Motif Enrichment Tool (PMET; https://github.com/kate-wa/PMET-software) (Rich-Griffin et al., 2020) to detect the co-localization of pairs of TF binding motifs within the promoters (gene upstream 1 kb) of cell type-specific differentially expressed genes. Briefly, PMET contains two major steps: (1) For each motif in the provided motif set, PMET uses FIMO (Grant et al., 2011) to assess motif matches within all of the promoters; (2) Identify co-localized motif pairs using binomial test. Then the gene lists provided by the user are tested for enrichment of motif pairs using pairwise hypergeometric test.
We provided 1368 motifs corresponding to 708 TFs of Arabidopsis thaliana, collected from databases of CIS-BP (http://cisbp.ccbr.utoronto.ca), JASPAR (http://jaspar.genereg.net) and PlantTFDB (http://planttfdb.gao-lab.org/), as input of PMET program. Top 70 differentially expressed genes with the highest expression fold change of each cluster were provided as input of the second step of PMET program. DNA sequences of 1000 bp upstream of the transcription start site (TSS) were taken as promoters when running PMET program.
To reduce false positive rate and improve signal-to-noise ratio in the predicted result, experimentally verified protein-protein interaction (PPI) information of Arabidopsis from the databases of BIND (http://bind.ca), BIOGRID (https://thebiogrid.org/), IntAct (https://www.ebi.ac.uk/intact/home) and MINT (https://mint.bio.uniroma2.it/) were collected and utilized to filter the predicted TF pairs. What's more, only key TF regulators in regulon were retained in the predicted TF pairs.
2.8. Gene set enrichment analysisGene ontology (GO) enrichment analysis for a specific gene set was performed using the clusterProfiler (Wu et al., 2021) package.
2.9. Topic modeling of cell-type specific gene expression programsIn order to identify cell-type specific gene expression programs (GEPs) with biological meanings, we used the CellFunTopic framework (https://github.com/compbioNJU/CellFunTopic) to infer enriched biological pathways of DEGs among different cell types. Specifically, CellFunTopic scores the activity of each functional gene set defined by GO vocabularies using the gene set-scoring method of Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005). The resulting enrichment scores in terms of clusters-by-gene-set scoring matrix is subjected to topic modeling. CellFunTopic applies latent Dirichlet allocation (LDA) (Blei et al., 2003) with variational expectation maximization (VEM) (Nasios and Bors, 2006) to factorize the scoring matrix into a clusters-by-topics matrix θ (the probability of a cluster belonging to a topic) and a topics-by-gene-set matrix φ (the contribution of a topic within a cell cluster). The results are highly interpretable: the top topics under each cell cluster directly reveal the specificity of biological programs belonging to a particular cell type, which is analogous cell-type GEPs.
2.10. Integration of scRNA-seq and scATAC-seq dataThe published scATAC-seq dataset generated in root of Arabidopsis thaliana (Dorrity et al., 2021) was downloaded from GEO under accession number GSE173834. The R package ArchR (Granja et al., 2021) v.1.0.1 was used to process the scATAC-seq dataset and perform the integration of the scATAC-seq data with scRNA-seq data, and to transfer cell type labels from the scRNA data to the scATAC data. Only cells derived from root and from clusters with root cells accounting for more than 65% in our integrated scRNA-seq data were used for integration with the scATAC-seq dataset.
ArchR was used to infer TF motif enrichments from the scATAC-seq data, which indicate the activity of regulatory factors in different cell types and are defined as motif deviations in the outputs of ArchR. To validate the regulon activity in scRNA-seq data inferred by SCENIC, motif deviations in scATAC-seq data and regulon activity in scRNA-seq data of representative TFs are shown in heatmaps.
2.11. Statistical analysisIf not specified, all statistical analyses and data visualization were done in R (v.4.0.0). R packages such as ggplot2, igraph and ComplexHeatmap (Gu, 2022) were used for graphics.
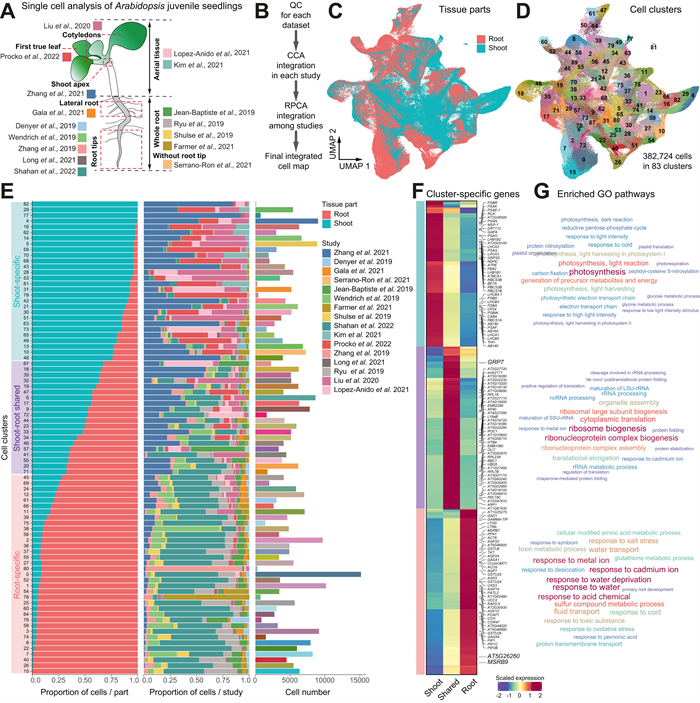
3. Results 3.1. Comprehensive integration of single-cell transcriptomic data in Arabidopsis juvenile seedlingsTo generate a comprehensive reference single-cell transcriptomic atlas of the whole Arabidopsis seedling and thus to reconstruct the cellular taxonomy across the plant body, we performed comprehensive integration analysis of single-cell RNA sequencing (scRNA-seq) data generated from representative tissues in Arabidopsis juvenile seedlings (Fig. 1A-B; see Methods). A total of 63 scRNA-seq datasets were collected from 16 different studies (Denyer et al., 2019; Farmer et al., 2021; Gala et al., 2021; Jean-Baptiste et al., 2019; Kim et al., 2021; Liu et al., 2020; Long et al., 2021; Lopez-Anido et al., 2021; Procko et al., 2022; Ryu et al., 2019; Serrano-Ron et al., 2021; Shahan et al., 2022; Shulse et al., 2019; Wendrich et al., 2020; Zhang et al., 2019, 2021) (Table S1). These data were generated from root tips, whole root, true leaves, aerial tissues and cotyledons of Arabidopsis seedling plants at 5–17 days after germination (DAG), representing both under- and above-ground part of the whole plant (Fig. 1A). After stringent quality control and eliminating low-quality cells and potential doublets, a total of 382,724 high-quality cells were retained for further analysis. Among these cells, 56.5% (216,193) were from the root, and 43.5% (166,531) were from the shoot (Table S2).
|
| Fig. 1 Mapping a reference single-cell transcriptomic atlas of Arabidopsis seedlings at the whole-organism level. (A) Single cell transcriptomic data used for constructing the reference single-cell transcriptomic atlas. In total, 63 scRNA-seq datasets were collected from ten different studies (in different color codes). The datasets were either generated in shoot or root tissues, as annotated according to the original studies. (B) Bioinformatic analysis pipeline for integrative analysis of single cell datasets. All scRNA-seq datasets were subjected to uniform processing, quality control and integration. The Uniform Manifold Approximation and Projection (UMAP) plot shows the integrated cell map where cells are colored by the tissues of origin. (C) UMAP plot displaying the effective of data integration from root and shoot. (D) UMAP plot displaying the integrated cell map, which consists of 83 distinct cell clusters. (E) Bar plots displaying the distribution of cells in each cluster based on the tissue of origin (left) or the original study (middle; color codes as in A), and the number of cells (right; color codes as in D). Cell clusters are assigned to three different categories based on the composition of cells from shoot or root tissues. (F) Heatmap showing the highly expressed genes in the three different categories of cell clusters. Representative genes are highlighted on the right. (G) Word cloud graphs indicate enriched gene ontology (GO) biological progresses for genes in the three categories in (F). |
In complex integration tasks, such as the one in our study, there exists a tradeoff between batch effect removal and conservation of biological variance (Luecken et al., 2022). We employed a two-step process for integrating single-cell RNA data, utilizing both Seurat CCA and RPCA methods (Fig. 1B). In the first step, we employed the Seurat CCA method to integrate datasets within the same study. This involved selecting highly variable genes and scaling the data based on log-transformation (Hafemeister and Satija, 2019). The choice of Seurat CCA was motivated by its proven effectiveness and consistent superior performance for small-scale data integration (Büttner et al., 2019; Luecken et al., 2022). Correcting batch effects between studies is particularly challenging, especially when there are biological variations in cell types across different studies. In the second step, we applied Seurat RPCA to merge the integrated data from the previous step, which gained the highest overall score after comparing the strength of integration with the other six integration methods (Fig. S1A). This approach can preserve distinct cell identities and better handle the challenges associated with between-study batch correction (Luecken et al., 2022). Therefore, by incorporating both Seurat CCA and RPCA methods, we aimed to strike a balance between batch effect removal and the conservation of biological variance in our integrated analysis.
Following dimensionality reduction using the Uniform Manifold Approximation and Projection (UMAP) algorithm, unsupervised clustering based on the Leiden algorithm reveals that shoot and root tissues have common and unique cell populations (Fig. 1C). Overall, 83 distinct cell clusters were identified in the integrated cell map (Fig. 1D). The number of cells in each cluster ranges from 115 (C82) to 15,162 (C0). It turns out that all the identified cell clusters are supported by different datasets from at least five different studies (Fig. 1E and Fig. S1B-C), suggesting a decent part of the batch effects have been removed. To be noted, although there are only two snRNA-seq studies with 7861 cells (2.053% of the total cells) integrated into the whole datasets, they can still be effectively integrated through our strategy (Fig. S1D-E).
We further categorized these 83 clusters into three groups based on the proportion of cell origin (shoot or root), resulting in three distinct groups: root-specific, shoot-specific, and root-shoot shared cell clusters (Fig. 1E). Conducting gene ontology (GO) enrichment analysis on the cluster-specific expressed genes in each group (Fig. 1F), we found significant (P < 0.05) enrichments in pathways associated with photosynthesis, salicylic acid signaling, and jasmonic acid signaling in the shoot-specific cell clusters; In contrast, root-specific clusters exhibited substantial enrichment in pathways related to nutrient absorption and transport (Fig. 1G). On the other hand, the shoot-root shared clusters demonstrated enrichment in fundamental cellular processes such as RNA modification and ATP biosynthesis (Fig. 1G). The consistency between the functional characteristics of cell clusters and their tissue origins underscores the robustness and reliability of our single-cell data integration strategy in capturing the inherent biological variations and characteristics of these cells.
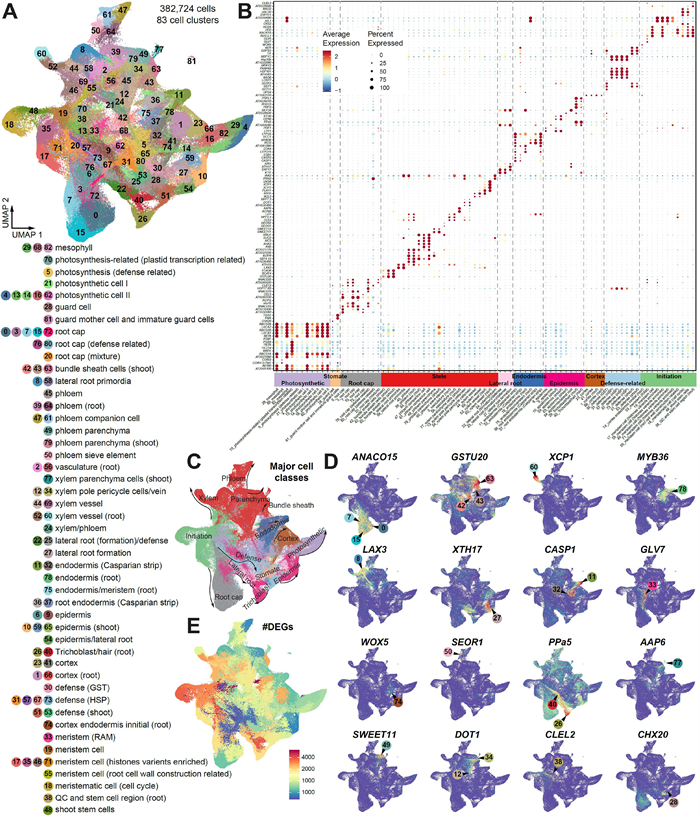
3.2. Systemic annotation of Arabidopsis cell types from integrated datasetsIn order to enhance the interpretation of the integrated single-cell transcriptomic atlas, we employed a rigorous approach to ascertain the cell type of each cluster. To confirm the identity of the clusters, we utilized two layers of information. Firstly, we compiled a comprehensive list of 1587 known markers from various sources(He et al., 2023). This compilation included markers that have been validated through techniques such as in situ hybridization or confocal images (Table S3). Additionally, we incorporated predicted marker genes derived from previous single-cell studies (DEGs; Table S3). By leveraging this wealth of marker information, we aimed to accurately assign cell types to each cluster within the integrated atlas (Fig. 2A-B). In total, 47 distinct cell types were identified and they can be grouped into ten different major types (cortex, endodermis, epidermis, defence-related cells, initiation cells, photosynthetic cells, root cap, stele cells, lateral root cells and stomata cells) according to their functions or spatial locations on the UMAP (Fig. 2C). To the best of our knowledge, our analysis here provides the most comprehensive cell type annotation so far at single cell resolution, largely expanding our current view of cell compositions in Arabidopsis (Farmer et al., 2021; Long et al., 2021; Ryu et al., 2019; Shahan et al., 2022; Zhang et al., 2019). In addition, we identified an extensive list of potential novel marker genes that are at least top 50 differentially expressed genes with high cell-type specificity (Shannon index > 0.4) and functional related to the corresponding cell clusters based on the integrated cell atlas (Table S4), which could enlarge the marker gene gallery of A. thaliana and contribute to build a benchmarking plant marker gene database.
|
| Fig. 2 Comprehensive annotation of cell types. (A) The annotation of the 83 cell clusters. (B) Dot plots showing the expression patterns of top marker genes (row) in each cell cluster (column). Cell types or states were annotated based on these marker genes and can be roughly assigned to ten different major cell classes according to their structure and/or function. (C) The ten major cell types (color codes as in the x-axis) distribution on the UMAP plot. (D) UMAP plots demonstrating the expression patterns of examples of marker genes. (E) UMAP plots displaying the number of DEGs among different cell clusters. |
Notably, several putative novel cell types were identified in our integrative analysis, including suberin lamellae (Clusters 11 and 78), myrosin cells (C81) and defense-related cells (C30, C31, C51, C53, C57, C67 and C73) (Fig. 2A). In Arabidopsis roots, suberin lamellae and casparin strips are both located in the endodermis and function as transmembrane barriers to limit the movement of apoplastic solutes into the endodermal cells (Franke and Schreiber, 2007). However, there are currently no available markers to distinguish these two cell types. We observed that specific expression of MYB39 (AT4G17785) and two GDSL family genes (AT2G23540 and AT5G37690) in clusters C11 and C78, which are known to be involved in suberin monomer biosynthesis (Vishwanath et al., 2015; Yadav et al., 2014) and degradation (Ursache et al., 2021) (Fig. 2B). Meanwhile, clusters C26 and C37 showed specific expression of MYB36 and CASP1-2 (Fig. 2D), which are two genes essential for casparian strip formation (Hosmani et al., 2013; Kamiya et al., 2015; Roppolo et al., 2011). Consequently, clusters C11 and C78 were annotated as putative suberin lamellae cells, while clusters C26 and C37 were identified as casparian strip cells. We also identified seven clusters enrich with genes annotated with responses to external stimuli (C30, C31, C51, C53, C57, C67 and C73), we annotated them as defence-related cell although the formation of a cohesive tissue remains to be ascertained. Those clusters originate from multiple data resources, but most of cells stem from shoot, especially the epidermal tissues (77.24%). Interestingly, we found two distinct groups within these clusters: one exhibiting an abundance of heat shock proteins (HSPs), while the other group is primarily comprised of cells studied in the context of stomatal cells with a good proportion of genes response to hypoxia, salicylic acid and bacterial origin molecules. The HSP-enriched cells mainly come from seedlings that had been subjecting to heat stress treatment (Jean-Baptiste et al., 2019). Therefore, it is possible that those are a collection of cells that have been influenced by heat stress during the scRNA-Seq experiment.
To assess the reliability of our functional annotation, we employed our in-house tool, CellFunTopic, to identify functional topics within each cell cluster (details in Methods; Fig. S2A-B). The results demonstrate a strong correlation between the enrichment of biological processes in each cluster, based on their DEGs, and their corresponding annotations. For instance, photosynthesis process and response to light stimulus were highly enriched in the photosynthetic-related cell clusters (including C4, C5, C14, C16, C29, C79 and C82). The topic functional roles of casparian strips and suberin lamellae are associated with lignin deposition and phenylpropanoid pathway, respectively, aligning with the composition of these two transmembrane barriers (Beisson et al., 2012; Doblas et al., 2017; Franke et al., 2005; Holbein et al., 2021) (Fig. S2B). This suggests that our manual annotation is robust. To further validate the confidence of cell type annotation, we used CIBERSORTx (Newman et al., 2019) to deconvolve cell-type specific gene expression from published bulk RNA-seq data that were generated from various tissues, organs or cell types in Arabidopsis seedlings (Table S5). Generally, the estimated cell-type specific gene expression patterns from bulk data are consistent with the samples of origin (Fig. S3). For instance, photosynthetic cells and the corresponding marker genes were overrepresented in aboveground samples. Samples enriched for quiescent center and stem cell niche showed dominant abundance of initiation cell clusters. The above analyses further confirm the reliability of cell-type annotation based on marker genes.
Furthermore, we evaluated the transcriptome similarity of different cell clusters using two calculation method: Jaccard index of DEGs and Pearson correlation coefficient of the overall expression in each cluster (Fig. S4A). In general, clusters in the same major types showed significantly (P < 0.05) higher similarity than clusters from different major types (Fig. S4B), suggesting that cells from same major types harbour similar active gene expression pattern. We observed that 11 initiation cell clusters (C33, C19, C17, C35, C48, C71, C56, C18, C43, C38 and C74) are significantly distinct from other clusters in terms of transcriptome similarity (Fig. S4A). Accordingly, these clusters have the greatest number of DEGs (Fig. 2E) and six of them were root-shoot shared cell clusters (Fig. 1E). Genes with roles in regulating cell fate, such as BBM (Burkart et al., 2022), GLV7 (RGF3) (Ou et al., 2022), PLT1 (Xiong et al., 2020) and DI21, were highly expressed in these cell clusters (Fig. 2B and D), aligning with the characteristics of meristem cells (Stahl and Simon, 2010). These findings strengthen the credibility of our cell annotation and the classification of major cell types.
3.3. The cell type specificity of regulonsSince our integrated cell atlas enables the comparison of gene expression across the entire plant at cell-type level, we aim to construct the regulatory relationships in a cell-type specific manner. SCENIC (Aibar et al., 2017) was used to predict active regulons based on co-expression analysis and TF motif enrichment. A regulon consists of a key TF regulator and its co-expressed target genes(Aibar et al., 2017). In total, we identified 638 regulons across all types of cells, the average target genes in each regulon are 691. All the key regulatory and their target genes knit together to form a network with 441019 interactions (Table S6). As expected, the regulon activity and the expression intensity of the key regulator in the corresponding regulon are highly correlated, confirming the reliability of regulon identification (Fig. 3A). The identified regulons are generally specific to cell types (Fig. 3A), and several TFs within these regulons have been previously demonstrated to play significant roles in their respective cell types, indicating the sensitivity and robustness of our analysis. For instance, we found that the regulon mediated by ANAC046 is notably active in the root cap (Fig. 3B). This observation aligns with recent finding that highlight ANAC046 as a crucial regulator of cell death and suberin biosynthesis in the Arabidopsis root cap (Huysmans et al., 2018; Mahmood et al., 2019). Similarly, the regulon containing JKD is highly active in the mature cortex in our analysis, consistent with its known role in cortex regulation. JKD has been reported to interact with cell fate determinants SCR and SHR (Ogasawara et al., 2011) and to restrict CYCIND6 expression in the cortex. Mutations in JKD lead to periclinal division in the cortex (Welch et al., 2007). Additionally, the DOF5.6-containing regulon demonstrated explicit roles in the phloem cells, in line with the established biological function of DOF5.6 in vascular tissue development (Guo et al., 2009; Haga et al., 2011). Besides, we also noticed that a number of regulons were shared among different cell types that are functionally correlated, as exemplified regulons mediated by CRF10 and MYB40 (Fig. 3A). These two regulons showed high activity in the initiation cells and stele cells, respectively, indicating their potential role in the development of specific cell types or in the transition between cell states.

|
| Fig. 3 Inference of cell-type specific regulons by SCENIC. (A) Heatmaps showing the activity of 628 regulons per cell type (left), and the expression specificity of the corresponding key TF regulator in regulon (right). Top representative regulons in each cell types are highlighted. (B) UMAP plots depicting the activity and expression specificity of selected regulons. (C) Correlation of regulon activity and TF expression in shoot for each cell types. Positive correlations indicate cell types enriched in shoot and otherwise in root. (D) A network view of top ten representative regulons in each cell type, revealing specific and shared regulons across cell classes. Small nodes in pie charts represent the relative expression of key regulatory TFs in regulon in shoot (blue-green) and root (red), the ratio of TF expression in above-ground tissues is calculated as the expression of TFs in above-ground tissues divided by their total expression in both above- and below-ground tissues, the same approach is used to calculate the relative expression in underground tissues. Large nodes indicate cell types which are colored according to the major cell classes. |
To evaluate the specificity of regulons thoroughly, we performed correlation analysis between the activity of regulon (both TF and its regulated targets) and the expression level of its corresponding key TF regulator in either above- or under-ground tissues for each cell type (Fig. 3C and Fig. S5A). As expected, the activity of regulons identified in the cell types in aboveground tissue, such as mesophyll cell, stomate cell and myrosin cells, showed positive correlations with the proportion of the corresponding key TF regulators expressed in above-ground cell types, suggesting the shoot specificity of those regulons. In contrast, the activity of regulons from typical under-ground tissue like lateral root and trichoblast is negatively correlated with the expression ratio of their key regulators in the above-ground cell types, indicating that they are root specific regulons. As a proof of concept, we visualized the top ten active regulons in each cell type in a network view, where the relative expression of key TF regulator in regulon in root and shoot tissues were shown in pie charts (Fig. 3D). In general, the TFs of identified regulons mainly expressed at the anatomy position of the plant (underground or aboveground) that correspond to the cell types they were assigned to. For instance, PLT1 and GLV7 mediated regulons were highly activated in root meristem (RAM) cells (Cluster 33); while in QC and stem cell niche cells (Cluster 38), BBM mediated regulons was one of the predominant regulators (Fig. 3D and Fig. S5B). All these three TFs can induce embryonic activity in plant cells (Kareem et al., 2015; Liu et al., 2014; Lowe et al., 2016). Specially, we have compared the cell-type specific GRNs between casparin strips and suberin lamellae. We found that ANAC038, MYB67 and MYB107 emerged as key regulators within suberin lamellae (Cluster 11 and 78) (Fig. 3D). Notably, the target genes under the influence of these three key transcription factors were significantly enriched in pathways associated with ion transport, the phenylpropanoid synthesis pathway, and suberin biosynthesis processes (Fig. S6A). On the other hand, ERF15, GRF6, and HB23 were identified as key regulators within Casparian strips (Fig. 3D), and the target genes regulated by these three key regulators showed enrichments in proton transport, water transmembrane transporter functions, and carbohydrate transmembrane transporter functions (Fig. S6B).
As a regulon inferred by SCENIC is based on the motif location and the gene expression pattern, to further validated the regulatory relations between key regulator and its targets in a certain cell type specific regulon, we used a published scATAC-seq dataset generated in root (Dorrity et al., 2021) to identify the TF motif of the key regulators in the corresponding regulons among different cell types. The motif deviations of the key regulators were highly positively correlated with the activity of their involved regulons, indicated a high accuracy of the regulons identified in root cells (Fig. S7).
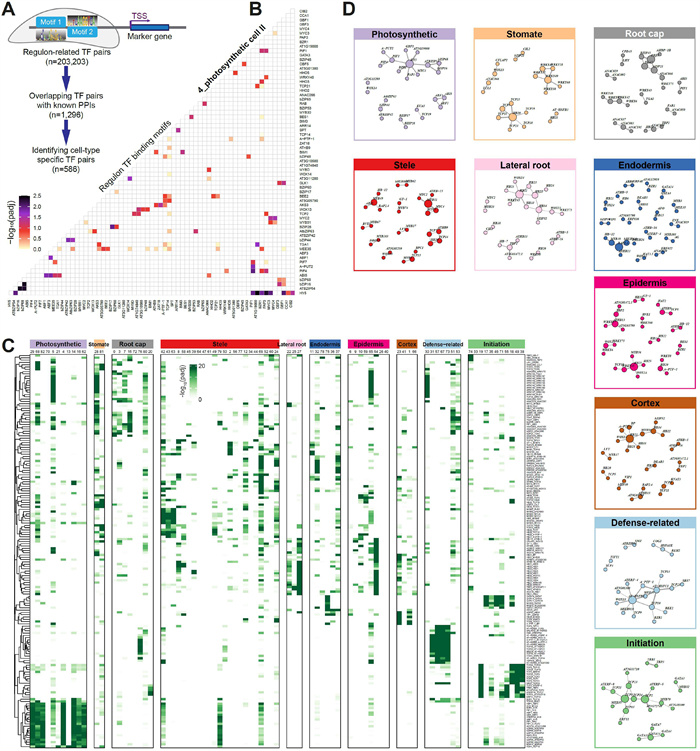
3.4. Dynamics of regulon-regulon associationsSince the dynamics of gene expression usually depend on cooperative binding of multiple TFs to the promoter regions (Jolma et al., 2015), we explored potential co-associations of regulons in different cell types. We adapted the Paired Motif Enrichment Tool (PMET) to detect pairs of TF binding motifs within the promoter regions of cell-type specific marker genes (Rich-Griffin et al., 2020). Only key TF regulators in regulons were used in the PMET analysis. The enriched TF pairs were subject to filtering using experimentally verified protein-protein interacted TFs. This filtering process does not exhibit any bias in terms of cell type specificity. However, However, it resulted in significantly closer motif location distances between interacting TFs (Fig. S8A-B). In total, 1296 reliable TF-TF pairs were obtained (Fig. 4A and Table S7). Taken the TF-TF pairs in photosynthetic cells as an example, TCP2, HY5, PTF1 and MEE35 (TCP4) were found to pair with a variety of other TFs to co-ordinately regulate photosynthesis related genes (Toledo-Ortiz et al., 2014; Zheng et al., 2022) (Fig. 4B).
|
| Fig. 4 Combinational regulon-regulon pairs specifying cell type gene regulatory networks. (A) Workflow for identification of cell-type specific motif pairs. The diagram illustrates the paired-motif enrichment analysis in cell type identity genes, i.e., identifying pairs of TF binding motifs within the promoters of cell-type specific marker genes. (B) Heatmap showing the top enriched co-localized TF pairs in photosynthetic cells. The row and column both represent the key TF regulators in the regulons. (C) Heatmap showing the enrichment of representative regulon pairs (rows) whose enrichment scores are cell-type specific (columns). (D) Network view of representative regulon-regulon interactions for each major cell classes. |
Consistent with the high cell-type specificity of the regulons mediated by these TFs (Fig. 3A), a substantial portion of the predicted co-associated TF-TF pairs is also cell-type specific (45.2%, 586 out of 1296), indicating differences in the co-regulation patterns of transcription factors among cell types. The representative modules of TF-TF pairs showed clear cell-type-specific patterns (Fig. 4C-D and Table S7). For instance, TF-TF pairs in WRKY family were specifically enriched in stomatal cells, epidermis, root cap cells and defense-related cells (Fig. 4D), in agreement with the biotic and abiotic stress related function of this TF family (Bakshi and Oelmüller, 2014). GATA-associated TF-TF pairs displayed a high degree of enrichment in initiation cells, and GATA factors have been reported to play critical roles in differentiation and control of cell proliferation (Fig. S9A). The TCPs-containing pairs have the largest number and presented in almost all cell types (Fig. S9B), in line with the hub function of the TCP family members in various regulatory networks (Bemer et al., 2017). We conducted a detailed examination of intricate TCP pairing patterns for gene regulation across different cell types. Notably, some TCPs, such as TCP19 and TCP22, exhibited simple co-associations with TFs in only two cell types. In contrast, TCP10, TCP14, TCP15, TCP23, and MEE35 (TCP4) tended to form multiple partnerships with TFs in various cell types (Fig. S9C-D), suggesting diverse roles of the TCP family members.
3.5. Construction of cell type-specific GRNsTo examine how the cell-type specific TF regulators control dynamics of gene expression in different tissues, we constructed GRNs based on cell-type specific regulons consisting of both the key TF regulators and their target genes with tissue-specific gene expression (Table S6). In the GRNs of photosynthetic cells, we found that six TF regulators, namely AGL3, ERF59, ESE3, GLK1, MEE35, and MYBS1, all of which have RSS specificity scores ranking in the top 1%, collectively regulate target genes associated with photosynthesis and tetrapyrrole metabolic processes (Fig. S10A-B). However, these six TF regulators appear to have distinct roles in photosynthetic cells. Specifically, AGL3 uniquely regulates genes involved in the flavonoid biosynthetic process and vitamin metabolism, while ERF59, ESE3, GLK1, MEE35, and MYBS1 uniquely regulate genes associated with S-glycoside biosynthesis, responses to light intensity, cellular responses to salicylic acid stimulation, leaf morphogenesis, and heme biosynthesis processes, respectively (Fig. S10C). To highlight the key information of the entire network, we only visualized the key regulators in representative regulons in each cell types (Fig. 3D) and their top five target genes with high specificity are displayed in the view of networks. The key regulators were coloured according to their enriched major cell types and the target genes were shown in pie charts based on their relative expression levels in above- and underground tissues (Fig. 5A). The result suggests that cell-type specific TFs tend to cluster together and their target genes are exclusively expressed in the corresponding tissue. For instance, the root cap regulators, represented by grey circles, exhibit a strong preference for targeting genes expressed in underground tissues. On the other hand, the photosynthesis regulators (indicated by purple circles) specifically target genes that are exclusively expressed in shoots. This observation suggests that the identified GRNs accurately reflect the regulatory relationships occurring in different parts of the plant body. Furthermore, a closer examination of specific TFs further confirms the robustness of our GRNs. For example, GLK1 (Fujii et al., 2022) and AGL20 (SOC1), which are known for their roles in chloroplast development and biogenesis, respectively, were identified as key regulators of photosynthetic cells in our analysis. Notably, they both target the gene CURT1A (Armbruster et al., 2013), which is highly enriched at the grana margin and involved in modifying thylakoid architecture. DEL1, identified as a regulator of meristem cells and specifically targeting histone proteins, is an important inhibitor of endocycles in plants (Vlieghe et al., 2005). In cases where TFs belong to multiple cell types, such as NLP4, which has important role in root morphological responses to rhizobia (Hernández-Reyes et al., 2022), is a key regulator in both root cap and defense-related cells. These findings, which reproduce the biology role of well-studied TFs and target genes in their respective cell types, together suggest the effectiveness and reliability of our GRNs which is constructed at the single-cell resolution. A detailed depiction of the GRNs of defense related cells is shown in Fig. 5B. Several TFs and genes participate in this process. Notably, ERF022, a TF previously associated with leaf senescence (Hinckley et al., 2019), is found to target NDR1 and CAF1A, both of which are genes involved in plant defense responses process (Samaradivakara et al., 2022; Walley et al., 2010). This suggests a novel role for ERF022 in the context of defence-related cellular processes. Some of the regulators identified in defence-related cells are also shared with photosynthesis cells and stomatal cells, implying the multifunctionality of these TFs and a potential interplay between stomatal and defence-related cellular functions. A-PTF-1 (TCP13), a regulator shared by defence and stomatal cells, plays a vital role during dehydration stress (Urano et al., 2022). It targets GPT2 and ARCK1, which have been reported to be induced by abiotic stress (Dyson et al., 2015; Tanaka et al., 2012). Another target gene, AT5G43260, remains functionally unknown but is upregulated during abiotic stress (Song et al., 2013). The regulatory relationship and functions of these defense-related regulator and target genes further verified the existence of this cell type.
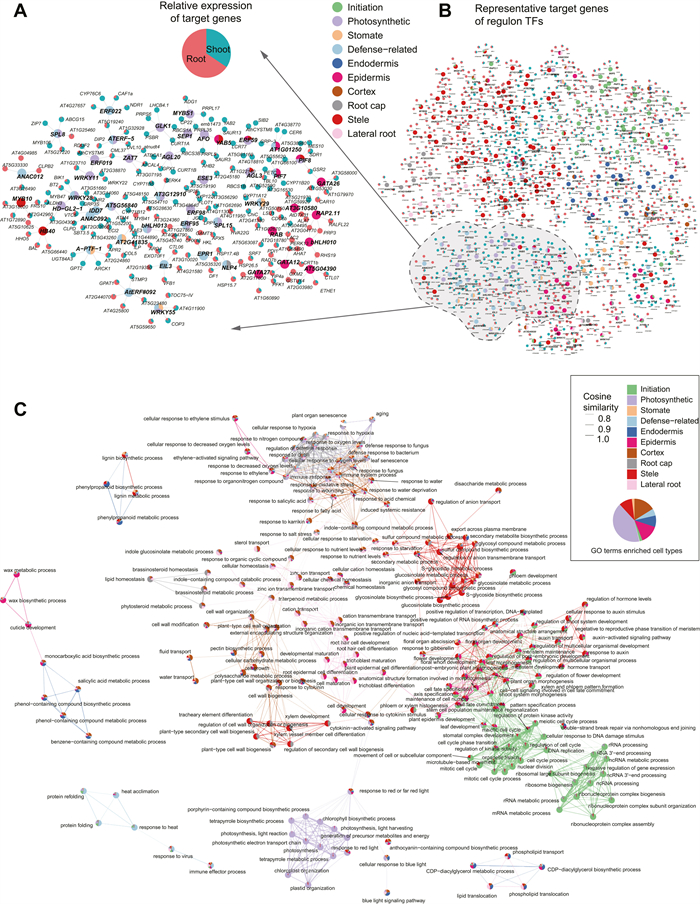
|
| Fig. 5 Expression and function specificity of cell type-related regulatory networks. (A) A network overview of representative target genes of cell-type specific regulons, revealing specific and common targets across regulons. Nodes with black circles represent TFs, colored according to the highly activated cell type; while nodes in pie charts indicate target genes. Note that only the top five target genes are shown for visualization purpose. (B) A detailed view of a subset of the network in (A). (C) A network analysis of GO terms (n = 174; nodes) based on similarity of enrichment scores across cell types (edges), laid out using the spring-based algorithm by Kamada and Kawaial(Kamada and Kawai, 1989). GO terms are colored by the contributing cell types (pie chart by the fraction of enrichment scores in terms of -log10P). |
Lastly, to demonstrate the concept of functional specificity within GRNs, we performed GO enrichment analysis using all target genes for regulons identified in each cell type. The enriched GO terms in different cell types were clustered according to the similarity of their enrichment scores. This co-enrichment analysis helps to uncover similarities and differences in biological processes across different cell types. The resulting co-enrichment was shown in a network which synoptically revealed several densely connected communities of biological processes with meaningful similarities (Fig. 5C). Unsurprisingly, chlorophyll biosynthetic process, light harvesting, light reaction and photosynthetic electron transport chain were specifically enriched in photosynthetic cells. Environmental stimulus related biological pathways such as response to bacterium, water and oxygen levels were specially enriched in defense-related cells, cortex, root cap and stomate cells, reflecting a complex and well-developed defence system of plants in response to environmental change and supporting a hypothesis that defense-related cells might be a previously uncharacterized cell type in response to stimulation of external environment. Similarly, biological processes related to cell cycle process, mRNA metabolic process, ribosomal biogenesis and nuclear division were highly enriched in initiation cells, consistent with their biological roles in cell fate reprogramming. Notably, some cell types shared a common set of GO terms with biologically meaningful similarities. For instance, nutrient level response related process was enriched in cortex, endodermis, epidermis and stele cells, consistent with the knowledge that absorption and transportation of nutrients are through concentric layers of those tissues (Barberon, 2017).
4. DiscussionRecently, empowered by the burst of single-cell sequencing techniques, references of single cell atlas across tissues or of the whole organisms have largely been generated in various model species especially in mammalian species (Han et al., 2018, 2020; Jones et al., 2022; Qu et al., 2022). To this end, the Human Cell Atlas (HCA) project aims to provide comprehensive reference maps of all human cells (Regev et al., 2017; Rozenblatt-Rosen et al., 2017). In the plant field, similar attempts such as the Plant Cell Atlas (PCA) framework has been proposed (Jha et al., 2021; Rhee et al., 2019). In fact, dozens of studies in plant sciences have fully embraced the single cell sequencing techniques, leading to generation of significant amounts of single-cell transcriptome datasets in specific tissues or organs over the past years (Seyfferth et al., 2021; Shaw et al., 2021). These advances have not only expanded our knowledge about plant cell compositions, but also laid the groundwork for PCA construction. One of the major challenges for PCA is to generate a reference cell map that includes all the cell types (or as much as possible) in various tissues (Cuperus, 2022). However, current plant cell atlases are usually constructed in specific tissues and a reference cell map of the entire plant is still missing. Therefore, collecting cells generated in representative tissues from different labs and development of data integration analysis pipelines would be the most effective way to achieve an “universal” annotation of cell types in plants. Consequently, here we have piloted an integration analysis of scRNA-seq data in Arabidopsis seedlings which represents a reference atlas at the early plant development stage. We attempt to provide valuable resources and experiences for PCA exploration, and also try to provide a guideline for re-evaluation of publicly plant single cell datasets.
The innate differences of snRNA-seq and scRNA-seq might result in protocol-related bias in integrative analysis, here we presented a nice fusion of snRNA-seq to the result of datasets, proved the effectiveness of the integrative strategy. However, it's worth noting that a cluster of putative suberin lamella cells (Cluster 78) is primarily originating from single-nucleus transcriptome data (Fig. 1E and Fig. S1E). This prevalence is likely due to suberin and cutin accumulation, which lead to the reduced digestibility of their cell wall, rather than being a result of methodological or study-related biases in clustering. This observation is consistent with similar findings reported by Farmer and its collaborators (Farmer et al., 2021) who noted a similar phenomenon when integrating root cells from isolated nuclei and protoplasts. This result indicates that snRNA-seq should be applied to hard-to-dissociate tissues to compensate for the limitations of scRNA-seq in order to provide a more comprehensive understanding.
We believe that successful data integration can yield new insights previously overlooked in the original studies. In our study, we identified a group of defense-related cells that response to external stimulus (Fig. 2A-B). Although experimental validations based on RNA-fluorescence in situ hybridization (FISH) and marker genes fused with reporters need to be conducted to further confirm the existence of those potentially novel cell types, we still believe that those cells have specific roles in plant defense system. The DEGs, key regulators and their target genes of those cell type are involved in plant defense responses process (Figs. 3D and 5A). In fact, two unknown clusters that enriched with genes “response to stress” were reported by Zhang et al. (2019), similar to the feature of our defense-related cell clusters. We hypothesise that transcriptional features of these cell types may have been diluted in bulk RNA-seq analysis. We were able to distinguish them in our large-scale data integration, perhaps because a sufficient number of rare cells from multiple studies are enriched into a specific population for cell type detection (Fig. S11). Furthermore, the putative novel cell type, suberin lamellae cells (cluster 11 and 78), displayed distinct regulon architectures compared to casparian strip cells (cluster 36 and 37) and the rest endodermis cells (Fig. 3A and D and Fig. S6). This suggests that these two cell types are governed by entirely different key regulators. These putative novel cell types demonstrate the significant potential of our high-resolution single cell atlas.
The development of plant organs and tissues relies on the accurate regulation of cellular differentiation. Cell fate specification is a progress of differential gene expression typically mediated by multiple TFs in a coordinated way (Drapek et al., 2017; Reiter et al., 2017). By presenting a comprehensive cell map of Arabidopsis, we delineate the architecture of GRNs in cell-type specific manner and explore the combination of key regulators (Figs. 4D and 5). That information could be useful to the ones who wish to explore the regulatory relationship of genes in a certain cell type.
Big dreams start small. Although references of plant cell atlases are far from complete, our integrative cell atlas provides a first reference cell map at the level of the entire plant. The generated cell-type specific GRNs in Arabidopsis will provides valuable resources and experiences to the public.
AcknowledgementsThis work is supported by the National Natural Science Foundation of China (No. 32070656), the Nanjing University Deng Feng Scholars Program and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions, China Postdoctoral Science Foundation funded project (No. 2022M711563) and Jiangsu Funding Program for Excellent Postdoctoral Talent (No. 2022ZB50).
Data availability
The processed and integrated single cell data in this study can be retrieved and viewed at https://biobigdata.nju.edu.cn/plantScGRN/.
Code availability
CellFunTopic is available at https://github.com/compbioNJU/CellFunTopic. R codes used to analyze data and generate figures are available upon reasonable request to the corresponding authors.
CRediT authorship contribution statement
Shanni Cao: Data curation, Formal analysis, Validation, Writing – original draft. Xue Zhao: Visualization, Writing – original draft, Writing – review & editing. Zhuojin Li: Formal analysis, Writing – review & editing. Ranran Yu: Writing – review & editing. Yuqi Li: Validation. Xinkai Zhou: Resources. Wenhao Yan: Conceptualization, Supervision. Dijun Chen: Conceptualization, Project administration. Chao He: Project administration, Validation, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.03.008.
Aibar, S., González-Blas, C.B., Moerman, T., et al., 2017. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods, 14: 1083-1086. DOI:10.1038/nmeth.4463 |
Armbruster, U., Labs, M., Pribil, M., et al., 2013. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell, 25: 2661-2678. DOI:10.1105/tpc.113.113118 |
Bakshi, M., Oelmüller, R., 2014. WRKY transcription factors: jack of many trades in plants. Plant Signal. Behav., 9: e27700. DOI:10.4161/psb.27700 |
Barberon, M., 2017. The endodermis as a checkpoint for nutrients. New Phytol., 213: 1604-1610. DOI:10.1111/nph.14140 |
Beisson, F., Li-Beisson, Y., Pollard, M., 2012. Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol., 15: 329-337. |
Bemer, M., van Dijk, A.D.J., Immink, R.G.H., et al., 2017. Cross-family transcription factor interactions: an additional layer of gene regulation. Trends Plant Sci., 22: 66-80. |
Blei, D.M., Ng, A.Y., Jordan, M.I., 2003. Latent dirichlet allocation. J. Mach. Learn. Res., 3: 993-1022. |
Brady, S.M., Zhang, L., Megraw, M., et al., 2011. A stele-enriched gene regulatory network in the Arabidopsis root. Mol. Syst. Biol., 7: 459. |
Burkart, R.C., Strotmann, V.I., Kirschner, G.K., et al., 2022. PLETHORA-WOX5 interaction and subnuclear localization control Arabidopsis root stem cell maintenance. EMBO Rep., 23: e54105. |
Butler, A., Hoffman, P., Smibert, P., et al., 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol., 36: 411-420. DOI:10.1038/nbt.4096 |
Büttner, M., Miao, Z., Wolf, F.A., et al., 2019. A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods, 16: 43-49. DOI:10.1038/s41592-018-0254-1 |
Chen, D., Yan, W., Fu, L.Y., et al., 2018. Architecture of gene regulatory networks controlling flower development in Arabidopsis thaliana. Nat. Commun., 9: 4534. |
Cuperus, J.T., 2022. Single-cell genomics in plants: current state, future directions, and hurdles to overcome. Plant Physiol., 188: 749-755. DOI:10.1093/plphys/kiab478 |
De Rybel, B., Mähönen, A.P., Helariutta, Y., et al., 2016. Plant vascular development: from early specification to differentiation. Nat. Rev. Mol. Cell Biol., 17: 30-40. DOI:10.1038/nrm.2015.6 |
Denyer, T., Ma, X., Klesen, S., et al., 2019. Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell, 48: 840-852.e845. |
Doblas, V.G., Geldner, N., Barberon, M., 2017. The endodermis, a tightly controlled barrier for nutrients. Curr. Opin. Plant Biol., 39: 136-143. |
Dorrity, M.W., Alexandre, C.M., Hamm, M.O., et al., 2021. The regulatory landscape of Arabidopsis thaliana roots at single-cell resolution. Nat. Commun., 12: 3334. |
Drapek, C., Sparks, E.E., Benfey, P.N., 2017. Uncovering gene regulatory networks controlling plant cell differentiation. Trends Genet., 33: 529-539. |
Dyson, B.C., Allwood, J.W., Feil, R., et al., 2015. Acclimation of metabolism to light in Arabidopsis thaliana: the glucose 6-phosphate/phosphate translocator GPT2 directs metabolic acclimation. Plant Cell Environ., 38: 1404-1417. DOI:10.1111/pce.12495 |
Farmer, A., Thibivilliers, S., Ryu, K.H., et al., 2021. Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol. Plant, 14: 372-383. |
Fornes, O., Castro-Mondragon, J.A., Khan, A., et al., 2020. Jaspar 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res., 48: D87-D92. |
Franke, R., Briesen, I., Wojciechowski, T., et al., 2005. Apoplastic polyesters in Arabidopsis surface tissues—a typical suberin and a particular cutin. Phytochemistry, 66: 2643-2658. |
Franke, R., Schreiber, L., 2007. Suberin—a biopolyester forming apoplastic plant interfaces. Curr. Opin. Plant Biol., 10: 252-259. |
Fujii, S., Kobayashi, K., Lin, Y.C., et al., 2022. Impacts of phosphatidylglycerol on plastid gene expression and light induction of nuclear photosynthetic genes. J. Exp. Bot., 73: 2952-2970. DOI:10.1093/jxb/erac034 |
Gala, H.P., Lanctot, A., Jean-Baptiste, K., et al., 2021. A single-cell view of the transcriptome during lateral root initiation in Arabidopsis thaliana. Plant Cell, 33: 2197-2220. DOI:10.1093/plcell/koab101 |
Granja, J.M., Corces, M.R., Pierce, S.E., et al., 2021. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet., 53: 403-411. DOI:10.1038/s41588-021-00790-6 |
Grant, C.E., Bailey, T.L., Noble, W.S., 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics, 27: 1017-1018. DOI:10.1093/bioinformatics/btr064 |
Gu, Z., 2022. Complex heatmap visualization. iMeta, 1: e43. |
Guiziou, S., Chu, J.C., Nemhauser, J.L., 2021. Decoding and recoding plant development. Plant Physiol., 187: 515-526. DOI:10.1093/plphys/kiab336 |
Guo, Y., Qin, G., Gu, H., et al., 2009. Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell, 21: 3518-3534. DOI:10.1105/tpc.108.064139 |
Hafemeister, C., Satija, R., 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol., 20: 296. |
Haga, N., Kobayashi, K., Suzuki, T., et al., 2011. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol., 157: 706-717. DOI:10.1104/pp.111.180836 |
Han, X., Wang, R., Zhou, Y., et al., 2018. Mapping the mouse cell atlas by microwell-seq. Cell, 172: 1091-1107.e1017. |
Han, X., Zhou, Z., Fei, L., et al., 2020. Construction of a human cell landscape at single-cell level. Nature, 581: 303-309. DOI:10.1038/s41586-020-2157-4 |
Hao, Y., Hao, S., Andersen-Nissen, E., et al., 2021. Integrated analysis of multimodal single-cell data. Cell, 184: 3573-3587.e3529. |
He, Z., Luo, Y., Zhou, X., et al., 2023. scPlantDB: a comprehensive database for exploring cell types and markers of plant cell atlases. Nucleic Acids Res., 52: D1629-D1638. |
Hernández-Reyes, C., Lichtenberg, E., Keller, J., et al., 2022. NIN-Like proteins: interesting players in rhizobia-induced nitrate signaling response during interaction with non-legume host Arabidopsis thaliana. Mol. Plant Microbe Interact., 35: 230-243. DOI:10.1094/mpmi-10-21-0261-r |
Hinckley, W.E., Keymanesh, K., Cordova, J.A., et al., 2019. The HAC1 histone acetyltransferase promotes leaf senescence and regulates the expression of ERF022. Plant Direct, 3: e00159. |
Holbein, J., Shen, D., Andersen, T.G., 2021. The endodermal passage cell - just another brick in the wall?. New Phytol., 230: 1321-1328. DOI:10.1111/nph.17182 |
Hosmani, P.S., Kamiya, T., Danku, J., et al., 2013. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based casparian strip in the root. Proc. Natl. Acad. Sci. U.S.A., 110: 14498-14503. DOI:10.1073/pnas.1308412110 |
Huysmans, M., Buono, R.A., Skorzinski, N., et al., 2018. NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. Plant Cell, 30: 2197-2213. DOI:10.1105/tpc.18.00293 |
Jean-Baptiste, K., McFaline-Figueroa, J.L., Alexandre, C.M., et al., 2019. Dynamics of gene expression in single root cells of Arabidopsis thaliana. Plant Cell, 31: 993-1011. DOI:10.1105/tpc.18.00785 |
Jha, S.G., Borowsky, A.T., Cole, B.J., et al., 2021. Vision, challenges and opportunities for a plant cell atlas. eLife, 10: e66877. |
Jolma, A., Yin, Y., Nitta, K.R., et al., 2015. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature, 527: 384-388. DOI:10.1038/nature15518 |
Jones, R.C., Karkanias, J., Krasnow, M.A., et al., 2022. The tabula sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science, 376: eabl4896. |
Kamada, T., Kawai, S., 1989. An algorithm for drawing general undirected graphs. Inf. Process. Lett., 31: 7-15. |
Kamiya, T., Borghi, M., Wang, P., et al., 2015. The MYB36 transcription factor orchestrates Casparian strip formation. Proc. Natl. Acad. Sci. U.S.A., 112: 10533-10538. DOI:10.1073/pnas.1507691112 |
Kareem, A., Durgaprasad, K., Sugimoto, K., et al., 2015. PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol., 25: 1017-1030. |
Kaufmann, K., Airoldi, C.A., 2018. Master regulatory transcription factors in plant development: a blooming perspective. Methods Mol. Biol., 1830: 3-22. DOI:10.1007/978-1-4939-8657-6_1 |
Kim, J.Y., Symeonidi, E., Pang, T.Y., et al., 2021. Distinct identities of leaf phloem cells revealed by single cell transcriptomics. Plant Cell, 33: 511-530. |
Liu, J., Sheng, L., Xu, Y., et al., 2014. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell, 26: 1081-1093. DOI:10.1105/tpc.114.122887 |
Liu, Y., Wang, T., Zhou, B., et al., 2021. Robust integration of multiple single-cell RNA sequencing datasets using a single reference space. Nat. Biotechnol., 39: 877-884. DOI:10.1038/s41587-021-00859-x |
Liu, Z., Zhou, Y., Guo, J., et al., 2020. Global dynamic molecular profiling of stomatal lineage cell development by single-cell RNA sequencing. Mol. Plant, 13: 1178-1193. |
Long, Y., Liu, Z., Jia, J., et al., 2021. FlsnRNA-seq: protoplasting-free full-length single-nucleus RNA profiling in plants. Genome Biol., 22: 66. |
Lopez-Anido, C.B., Vatén, A., Smoot, N.K., et al., 2021. Single-cell resolution of lineage trajectories in the Arabidopsis stomatal lineage and developing leaf. Dev. Cell, 56: 1043-1055.e1044. |
Lowe, K., Wu, E., Wang, N., et al., 2016. Morphogenic regulators baby boom and wuschel improve monocot transformation. Plant Cell, 28: 1998-2015. DOI:10.1105/tpc.16.00124 |
Luecken, M.D., Büttner, M., Chaichoompu, K., et al., 2022. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods, 19: 41-50. DOI:10.1038/s41592-021-01336-8 |
Mahmood, K., Zeisler-Diehl, V.V., Schreiber, L., et al., 2019. Overexpression of ANAC046 promotes suberin biosynthesis in roots of Arabidopsis thaliana. Int. J. Mol. Sci., 20: 6117. DOI:10.3390/ijms20246117 |
Moreno-Risueno, M.A., Sozzani, R., Yardımcı, G.G., et al., 2015. Transcriptional control of tissue formation throughout root development. Science, 350: 426-430. DOI:10.1126/science.aad1171 |
Nasios, N., Bors, A.G., 2006. Variational learning for Gaussian mixture models. IEEE Trans. Syst. Man Cybern., 36: 849-862. |
Newman, A.M., Steen, C.B., Liu, C.L., et al., 2019. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol., 37: 773-782. DOI:10.1038/s41587-019-0114-2 |
Ogasawara, H., Kaimi, R., Colasanti, J., et al., 2011. Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol. Biol., 77: 489-499. DOI:10.1007/s11103-011-9826-5 |
Ou, Y., Tao, B., Wu, Y., et al., 2022. Essential roles of SERKs in the ROOT MERISTEM GROWTH FACTOR-mediated signaling pathway. Plant Physiol., 189: 165-177. DOI:10.1093/plphys/kiac036 |
Procko, C., Lee, T., Borsuk, A., et al., 2022. Leaf cell-specific and single-cell transcriptional profiling reveals a role for the palisade layer in UV light protection. Plant Cell, 34: 3261-3279. DOI:10.1093/plcell/koac167 |
Qian, Y., Huang, S.-s.C., 2020. Improving plant gene regulatory network inference by integrative analysis of multi-omics and high resolution data sets. Curr. Opin. Struct. Biol., 22: 8-15. |
Qu, J., Yang, F., Zhu, T., et al., 2022. A reference single-cell regulomic and transcriptomic map of cynomolgus monkeys. Nat. Commun., 13: 4069. |
Ramirez-Parra, E., Desvoyes, B., Gutierrez, C., 2005. Balance between cell division and differentiation during plant development. Int. J. Dev. Biol., 49: 467-477. DOI:10.1387/ijdb.052001er |
Regev, A., Teichmann, S.A., Lander, E.S., et al., 2017. The human cell atlas. eLife, 6: e27041. |
Reiter, F., Wienerroither, S., Stark, A., 2017. Combinatorial function of transcription factors and cofactors. Curr. Opin. Genet. Dev., 43: 73-81. |
Reynoso, M.A., Borowsky, A.T., Pauluzzi, G.C., et al., 2022. Gene regulatory networks shape developmental plasticity of root cell types under water extremes in rice. Dev. Cell, 57: 1177-1192.e1176. |
Rhee, S.Y., Birnbaum, K.D., Ehrhardt, D.W., 2019. Towards building a plant cell atlas. Trends Plant Sci., 24: 303-310. |
Rich-Griffin, C., Eichmann, R., Reitz, M.U., et al., 2020. Regulation of cell type-specific immunity networks in Arabidopsis roots. Plant Cell, 32: 2742-2762. DOI:10.1105/tpc.20.00154 |
Roppolo, D., De Rybel, B., Dénervaud Tendon, V., et al., 2011. A novel protein family mediates Casparian strip formation in the endodermis. Nature, 473: 380-383. DOI:10.1038/nature10070 |
Roszak, P., Heo, J.O., Blob, B., et al., 2021. Cell-by-cell dissection of phloem development links a maturation gradient to cell specialization. Science, 374: eaba5531. |
Rozenblatt-Rosen, O., Stubbington, M.J.T., Regev, A., et al., 2017. The human cell atlas: from vision to reality. Nature, 550: 451-453. DOI:10.1038/550451a |
Ryu, K.H., Huang, L., Kang, H.M., et al., 2019. Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiol., 179: 1444-1456. DOI:10.1104/pp.18.01482 |
Ryu, Y., Han, G.H., Jung, E., et al., 2023. Integration of single-cell RNA-seq datasets: a review of computational methods. Mol. Cell., 46: 106-119. DOI:10.14348/molcells.2023.0009 |
Samaradivakara, S.P., Chen, H., Lu, Y.J., et al., 2022. Overexpression of NDR1 leads to pathogen resistance at elevated temperatures. New Phytol., 235: 1146-1162. DOI:10.1111/nph.18190 |
Santos-Mendoza, M., Dubreucq, B., Baud, S., et al., 2008. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J., 54: 608-620. |
Serrano-Ron, L., Perez-Garcia, P., Sanchez-Corrionero, A., et al., 2021. Reconstruction of lateral root formation through single-cell RNA sequencing reveals order of tissue initiation. Mol. Plant, 14: 1362-1378. |
Seyfferth, C., Renema, J., Wendrich, J.R., et al., 2021. Advances and opportunities in single-cell transcriptomics for plant research. Annu. Rev. Plant Biol., 72: 847-866. DOI:10.1146/annurev-arplant-081720-010120 |
Shahan, R., Hsu, C.W., Nolan, T.M., et al., 2022. A single-cell Arabidopsis root atlas reveals developmental trajectories in wild-type and cell identity mutants. Dev. Cell, 57: 543-560.e549. |
Shaw, R., Tian, X., Xu, J., 2021. Single-cell transcriptome analysis in plants: advances and challenges. Mol. Plant, 14: 115-126. |
Shulse, C.N., Cole, B.J., Ciobanu, D., et al., 2019. High-throughput single-cell transcriptome profiling of plant cell types. Cell Rep., 27: 2241-2247.e2244. |
Song, L.H., Hegie, A., Suzuki, N., et al., 2013. Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol. Plantarum, 148: 322-333. |
Stahl, Y., Simon, R., 2010. Plant primary meristems: shared functions and regulatory mechanisms. Curr. Opin. Plant Biol., 13: 53-58. |
Stuart, T., Butler, A., Hoffman, P., et al., 2019. Comprehensive integration of single-cell data. Cell, 177: 1888-1902.e1821. |
Subramanian, A., Tamayo, P., Mootha, V.K., et al., 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A., 102: 15545-15550. DOI:10.1073/pnas.0506580102 |
Suo, S., Zhu, Q., Saadatpour, A., et al., 2018. Revealing the critical regulators of cell identity in the mouse cell atlas. Cell Rep., 25: 1436-1445.e1433. |
Tanaka, H., Osakabe, Y., Katsura, S., et al., 2012. Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J., 70: 599-613. |
Taylor-Teeples, M., Lin, L., de Lucas, M., et al., 2015. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature, 517: 571-575. DOI:10.1038/nature14099 |
Toledo-Ortiz, G., Johansson, H., Lee, K.P., et al., 2014. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genetics, 10: e1004416. DOI:10.1371/journal.pgen.1004416 |
Tripathi, R.K., Wilkins, O., 2021. Single cell gene regulatory networks in plants: opportunities for enhancing climate change stress resilience. Plant Cell Environ., 44: 2006-2017. DOI:10.1111/pce.14012 |
Urano, K., Maruyama, K., Koyama, T., et al., 2022. CIN-like TCP13 is essential for plant growth regulation under dehydration stress. Plant Mol. Biol., 108: 257-275. DOI:10.1007/s11103-021-01238-5 |
Ursache, R., De Jesus Vieira Teixeira, C., Dénervaud Tendon, V., et al., 2021. GDSL-domain proteins have key roles in suberin polymerization and degradation. Nat. Plants, 7: 353-364. DOI:10.1038/s41477-021-00862-9 |
Vishwanath, S.J., Delude, C., Domergue, F., et al., 2015. Suberin: biosynthesis, regulation, and polymer assembly of a protective extracellular barrier. Plant Cell Rep., 34: 573-586. DOI:10.1007/s00299-014-1727-z |
Vlieghe, K., Boudolf, V., Beemster, G.T., et al., 2005. The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol., 15: 59-63. |
Walley, J.W., Kelley, D.R., Nestorova, G., et al., 2010. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol., 152: 866-875. DOI:10.1104/pp.109.149005 |
Weirauch, M.T., Yang, A., Albu, M., et al., 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell, 158: 1431-1443. |
Welch, D., Hassan, H., Blilou, I., et al., 2007. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev., 21: 2196-2204. DOI:10.1101/gad.440307 |
Wendrich, J.R., Yang, B., Vandamme, N., et al., 2020. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science, 370: eaay4970. |
Wu, S., Chen, D., Snyder, M.P., 2022. Network biology bridges the gaps between quantitative genetics and multi-omics to map complex diseases. Curr. Opin. Chem. Biol., 66: 102101. |
Wu, T., Hu, E., Xu, S., et al., 2021. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation, 2: 100141. |
Xiong, F., Zhang, B.K., Liu, H.H., et al., 2020. Transcriptional regulation of PLETHORA1 in the root meristem through an importin and its two antagonistic cargos. Plant Cell, 32: 3812-3824. DOI:10.1105/tpc.20.00108 |
Yadav, V., Molina, I., Ranathunge, K., et al., 2014. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell, 26: 3569-3588. DOI:10.1105/tpc.114.129049 |
Yan, W., Chen, D., Kaufmann, K., 2016. Molecular mechanisms of floral organ specification by MADS domain proteins. Curr. Opin. Plant Biol., 29: 154-162. |
Yang, B., Minne, M., Brunoni, F., et al., 2021. Non-cell autonomous and spatiotemporal signalling from a tissue organizer orchestrates root vascular development. Nat. Plants, 7: 1485-1494. DOI:10.1038/s41477-021-01017-6 |
Yu, Y., Zhang, H., Long, Y., et al., 2022. Plant Public RNA-seq Database: a comprehensive online database for expression analysis of ~45 000 plant public RNA-Seq libraries. Plant Biotechnol. J., 20: 806-808. DOI:10.1111/pbi.13798 |
Zhang, T.Q., Chen, Y., Wang, J.W., 2021. A single-cell analysis of the Arabidopsis vegetative shoot apex. Dev. Cell, 56: 1056-1074.e1058. |
Zhang, T.Q., Xu, Z.G., Shang, G.D., et al., 2019. A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Mol. Plant, 12: 648-660. |
Zheng, X., Lan, J., Yu, H., et al., 2022. Arabidopsis transcription factor TCP4 represses chlorophyll biosynthesis to prevent petal greening. Plant Commun., 3: 100309. |



