b. University of Chinese Academy of Sciences, Beijing 100049, China
The process of radiative evolution is often accompanied by complex biological events, such as hybridization, introgression, or incomplete lineage sorting (ILS) (Cai et al., 2021), which lead to incongruent phylogenetic relationships. As a result, the phylogenetic relationships between some species cannot be fully resolved as a bifurcating tree (Doolittle, 1999; Mallet et al., 2016; Yang et al., 2023), appearing the network-like pattern, which is called reticulate evolution (Linder and Rieseberg, 2004; Guo et al., 2023). Such a network-like pattern has been found throughout the evolutionary history of flowering plants (Rieseberg and Soltis, 1991; Soltis and Soltis, 2009; Pennisi, 2016), as a major impetus for species diversification and adaptation (Zhao et al., 2021). Therefore, disentangling the biological causes underlying the reticulate evolutionary history is crucial for our understanding of speciation and diversification.
As the major causes of reticulate evolution, hybridization and introgression are fundamental biological process, serving as major driving forces in plant evolution (Stewart et al., 2003; Hu et al., 2019; Zhang et al., 2021). Hybridization, introgression and ILS play a critical role in evolution: 1) to increase genetic diversity, such as spontaneous interspecific hybridization during soybean domestication showed a high level of genetic diversity (Wang et al., 2019); 2) to promote speciation, as interspecies hybridization may also lead to species fusion (Yu et al., 2023); and 3) to promote critical traits to enhance adaptability, for instance, a classic example was that hybrids Helianthus anomalus, H. deserticola and H. paradoxus showed the characteristics of being more tolerant to drought environment than their parents (Gross and Rieseberg, 2005). Often, closely related species hybridize with higher frequency than distant species (Mallet et al., 2016). Except for phylogenetic discordance including cytonuclear discordance and extensive gene tree-species tree conflict, complex reticulate evolution also makes profound impact on taxonomic treatment. In the genus Rosa, bright yellow-flowered species of Rosa only appear in two groups (R. subg. Hulthemia and R. sect. Pimpinellifoliae), but R. foetida and R. kokanica also have the character of bright yellow-flowered, were crossed by species of R. sect. Rosa and R. sect. Pimpinellifoliae (Debray et al., 2022). Therefore, for the study of reticulation, comprehensive evidence including genomic data from different genetic systems (plastid and nuclear), as well as morphological characters, and extensive sampling is critically important.
Most previous studies of reticulate evolution have focused on diploid lineages (Nitta et al., 2011; Parisod and Badaeva, 2020). However, it remains unknown whether these results are compatible with polyploid evolution. Woody bamboos, which have an allopolyploid origin (Guo et al., 2019a) and decades-long vegetative period (up to 120 years; Janzen, 1976), represent a unique system for understanding reticulate evolution. Following the divergence between the herbaceous bamboos and the diploid ancestors of woody bamboos about 22 million years ago (Ma), four extinct diploid ancestors of woody bamboos (named A, B, C, D) formed three clades of woody bamboos through three allopolyploid events, in which B/C and C/D hybridized to generate two allotetraploid lineages, namely the neotropical woody bamboo clade (BBCC) and the temperate woody bamboo clade (CCDD). The tetraploid ancestor of BBCC and the A progenitor again hybridized to form the allohexaploid palaeotropical woody bamboo clade (AABBCC) about 3 Ma later (Guo et al., 2019a). The temperate woody bamboos contain about 599 species in 36 genera (Soreng et al., 2022), all of which are tetraploid (CCDD), including Pseudosasa gracilis and its closely related species sampled in this study.
Previous studies have detected complex reticulate evolution at various taxonomic levels in the temperate woody bamboos (Guo et al., 2019a, 2019b; Ye et al., 2021). Phylogenetic analyses based on a few chloroplast and nuclear genes have indicated there are several cases of reticulate evolution involving intergeneric or interspecies hybridization and introgression of temperate woody bamboos (Triplett, 2008; Triplett et al., 2014; Triplett and Clark, 2010, 2021; Zhang et al., 2012; Yang et al., 2013). Recently, phylogenomic analyses based on ddRAD-seq and/or genome skimming data revealed extensive phylogenetic conflicts among different lineages of woody bamboos (Liu et al., 2020; Guo et al., 2021; Ye et al., 2021; Lv et al., 2023), indicating extensive reticulate evolution in the woody bamboo lineages. However, most of these speculations have yet to be validated using genome-scale integrative analyses, leaving the pattern of reticulate evolution for polyploidy bamboo lineages obscure.
Pseudosasa gracilis, like all extant temperate woody bamboo species, is a tetraploid with the CCDD genome (Guo et al., 2019a). Ps. gracilis is characterized morphologically by having slim culms, flat culm nodes, 2 or 3 branches per node, long bristles, and no ligules (Fig. 1). From a morphological point of view, compared with Sinosasa, Ps. gracilis is more similar to Ps. nanunica for possessing 3-branches and apically setulose on culms, whereas obviously distinguished from other species of Pseudosasa by unique morphological traits of the dwarf form of plants and 2 or 3 branches per node. In terms of phylogenetics, although Pseudosasa is not a monophyly, all species it contains were clustered together in one large subclade of the leptomorph lineage, except for Ps. gracilis. Ps. gracilis has isolated systematic position, was resolved to be closely related to Sinosasa longiligulata and S. guangxiensis, and stably clustered in an early-diverging subclade within the leptomorph lineage, which was distant from other species of Pseudosasa (Guo et al., 2021).

|
| Fig. 1 Comparison of morphological characters of Pseudosasa gracilis, and closely related species, Ps. nanunica, Sinosasa polytricha and S. longiligulata. |
Phylogenetic reconstruction of a large number of single or low-copy nuclear genes on genome-scale has solved many systematic problems that are difficult in traditional, morphology-based taxonomy (Berger et al., 2017; Vargas et al., 2019). One approach to solving problems unique to polyploid evolution is the use of "perfect-copy" syntenic genes in phylogenetic analysis. "Perfect-copy" syntenic genes represent those syntenic genes that match the ploidy level, such as syntenic genes with 1:2:3 copies in diploid, tetraploid, and hexaploid bamboos (Guo et al., 2019a). Compared with single-copy nuclear genes (SCNs), "perfect-copy" syntenic genes take conserved gene positions and ploidy-match gene copies into consideration. These "perfect-copy" syntenic genes have been used to successfully identify complex reticulate evolution of Bambusoideae at the tribal level (Guo et al., 2019a). Due to its low sequencing depth (usually less than 2–3×), genome skimming has been widely used to obtain maternally inherited (few are paternal inheritance) chloroplast genomes (Birky, 1995) and biparentally inherited nuclear ribosomal DNA (nrDNA) (Zhang et al., 2015; Guo et al., 2021; Wang et al., 2021a; Liu et al., 2023), although few high-quality nuclear genes. Deep genome skimming (DGS) is a newly developed approach to obtain a large number of nuclear genes at the genome level, and studies have shown that the best sequencing depth for obtaining high quality single-copy nuclear genes may be 10×or slightly higher (Liu et al., 2021). The greatest advantage of DGS is to obtain genome-level nuclear genes that can satisfy the phylogenetic requirements in both quantity and quality at relatively low cost. Its single sequencing can obtain large amount of molecular data of different genetic systems, and the informative sites are whole-genome wide. Therefore, DGS has gradually been applied in phylogenetically difficult taxa and reticulate evolution (Low et al., 2022; Guo et al., 2023). A large number of SCNs also frequently produce low resolution and intensely conflicting gene trees (Smith et al., 2015). Although this may complicate our understanding of phylogenetic relationships, it provides an opportunity to analyze the role of reticulate events such as hybridization and introgression through the cytonuclear conflict (Walker et al., 2019; Morales-Briones et al., 2021).
To decipher the causes of the unexpected phylogenetic position of Pseudosasa gracilis, we conducted a population-level sampling strategy for Ps. gracilis and its sympatric species Ps. nanunica and Sinosasa polytricha in this study. We first inferred the phylogenetic relationships among these species using parallel chloroplast genome sequences and genome-wide SNPs datasets generated from DGS data. Using gene trees inferred from 411 "perfect-copy" syntenic genes, we then quantified and visualized the phylogenetic incongruence among different gene trees, as well as between gene trees and the putative species tree. We further address the underlying biological process of the molecular-morphological conflict for Ps. gracilis based on analyses of 411 nuclear genes, plastid genomic data, and morphological characters. It is hoped that the comprehensive analyses performed in our study will not only validate the utility of DGS data for species-level phylogenetic study, but also provide insights into studies of plant speciation in the face of reticulate evolution.
2. Materials and methods 2.1. Taxon sampling and sequencingPseudosasa gracilis is a narrow-ranged endemic species distributed in Mangshan Mountain on the border of Hunan and Guangdong Provinces in South-central China. During our primary field investigation, we found that two other bamboo species, Ps. nanunica and Sinosasa polytricha, often grow in the same area (Fig. S1). We sampled four populations of Ps. gracilis, one population of Ps. nanunica, two populations of S. polytricha in the Mangshan Mountain area. In addition, we sampled one population of S. longiligulata (type species of Sinosasa) at Luofu Mountain, Guangdong Province to ensure that at least two species were sampled from each genus. All sampling information in this study is available in Table S1.
Fresh leaf materials were collected from two to five individuals from each population, and stored in silica gel for drying. A total of 22 individuals from eight populations of four species were sent to BENAGEN (Wuhan, China) for library preparation, and paired-end DGS data (150bp) were sequenced at 15×sequencing depths on the DNBSEQ-T7 sequencer.
2.2. Data processing, assembly and alignmentFor raw paired-end DGS data, we first filtered adapter sequences and low-quality sequences by Trimmomatic v.0.39 (Bolger et al., 2014). GetOrganelle v.1.9.77 (Jin et al., 2020) was used to assemble chloroplast genomes with the range of k-mers of 21, 45, 65, 85, and 105. The assembled contigs/circular plastome sequences were visualized by Bandage v.0.9.0 (Wick et al., 2015). Using plastome sequences of Pseudosasa japonica (NC_028328) and Sinosasa longiligulata (NC_036825) downloaded from NCBI as references, we checked and manually adjusted assembled plastome sequences using Genious v.11.1 (Kearse et al., 2012). Then, we used reference genomes to annotate the circular plastomes using PGA v.1.9.1 (Qu et al., 2019). Finally, plastome sequences of 22 individuals in the present study and 10 individuals (nine individuals from Pseudosasa and one individual from Indocalamus) sequenced in our previous study (Guo et al., 2021) were aligned in MAFFT v.7 (Katoh and Standley, 2013).
For genome-wide SNP assembly, we used the genome of Phyllostachys edulis (GCA_011038535.1) as a reference. The Genome Analysis Toolkit pipeline v.3.8 (GATK; McKenna et al., 2010) was used to detect and format the SNP matrix following these steps: read alignment of BWA v.0.7.13 (bwa men -R reference sequence paired-end sequencing files -o out file), data filtering (MarkDuplicates -I input file -O out file -CREATE_INDEX true -REMOVE_DUPLICATES true), detection of mutations (HaplotypeCaller -R reference sequence -I input file -ERC GVCF -O), combination of SNPs (ConbineGVCFs -R reference sequence -V sample list -O out file), extraction of SNPs (SelectVariants -R reference sequence -V SNPs file —select-type SNP -O out file) and filtering of SNPs (VariantFiltration -R reference sequence -V input file —filter-expression "QD < 2.0||MQ < 40.0||FS > 60.0||SOR > 3.0||MQRankSum < -12.5||ReadPosRankSum < -8.0" —filter-name 'SNP_filter' -O out file).
For nuclear gene assembly, 430 "perfect-copy" syntenic genes of Phyllostachys edulis extracted from a previous genomic study based on 11 chromosomal-level bamboo genomes were used as gene set of interest (Ma et al., 2024). Because temperate woody bamboos are tetraploid (Triplett et al., 2014; Guo et al., 2019a) with C and D subgenomes, each "perfect-copy" syntenic gene comprises two gene copies; thus, "perfect-copy" syntenic genes are low-copy nuclear genes with a conserved gene order. According to previous studies, the sequences of C and D subgenomes have differentiated to a certain extent, and the similarity of two gene copies of C and D subgenomes is generally lower than that of the different alleles between C and C or D and D (Ma et al., 2024), which allowed us to treat the two subgenomes as separate diploids in the following gene assembly. HybPiper v.2.0.2 (Johnson et al., 2016) was performed to assemble "perfect-copy" syntenic genes from 22 sequenced individuals. Paralogs were detected by the "paralog_retriever.py" script embedded in HybPiper. We assembled the C and D separately, and further compared and screened the assembly genes with paralog warnings in multiple contig. The selected "perfect-copy" syntenic genes were further aligned by MAFFT v.7.
2.3. Phylogenetic analysis of plastomesPlastome alignments of all 32 samples were trimmed using TrimAL v.1.4 (Capella-Gutierrez et al., 2009). We employed IQ-Tree v.2.0.6 (Nguyen et al., 2015) with the parameter of "-m MFP -bb 1000" to construct the plastid phylogenetic tree, Indocalamus sinicus was selected as an outgroup taxon.
2.4. Phylogenetic analyses of nuclear SNPs and "perfect-copy" syntenic gene data setsFor the nuclear SNP matrix, we reconstructed a Neighbor-Joining (NJ) tree using MEGA11 (https://www.megasoftware.net) with the following parameters. "Test of phylogeny: Bootstrap method; No. of bootstrap replications: 1000; Model/Method: Kimura 2-parameter model; Substitutions to include: d: Transitions + Transversions; Rates among sites: Gamma distributed (G); Gamma parameter: 1.00; Gaps/Missing data treatment: Partial deletion." Phyllostachys edulis was selected as outgroup taxon. For the "perfect-copy" syntenic gene data set, both concatenation and coalescent methods were used to reconstruct phylogenetic trees. For the concatenation approach, Geneious v.11.1 (Kearse et al., 2012) was first used to parallel concatenate "perfect-copy" syntenic genes alignments for C and D subgenome. The concatenated supermatrices were then visualized and manually adjusted to remove the inserted fragments. TrimAL v.1.4 was further used to trim the resulting alignments, and then IQ-Tree v.2.0.6 was adopted for phylogenetic tree reconstruction. For the coalescent approach, we selected RA×ML-HPC-MPI-AVX2 (Stamatakis, 2014) to reconstruct the gene trees of each "perfect-copy" syntenic gene under the GTRGAMMA + I model with 1000 bootstrap (BS). ASTRAL v.5.7.1 (Zhang et al., 2018) was used to reconstruct the species trees of the C and D subgenomes based on their inferred gene trees, respectively.
2.5. Quantification and visualizations of gene tree discordanceWe assessed the node-supporting information for putative species trees and both gene tree data sets using the Quartet Sampling (QS) method (Pease et al., 2018). The format of QS scores above the phylogenetic tree was Quartet Concordance (QC)/Quartet Differential (QD)/Quartet Informativeness (QI), in which QC measured the frequency of concordance over discordance, QD measured skew in the two discordant tree counts, QI measured the number of replicates that fail likelihood cutoff. Finally, we calculated the number of genes that support and conflict with the coalescent species tree for each node by PhyParts (Smith et al., 2015). We visualized the PhyParts output for both gene tree data sets in PhyParts PieCharts (https://github.com/mossmatters/MJPythonNotebooks/blob/master/PhyParts_PieCharts.ipynb).
2.6. Incomplete lineage sorting (ILS) and hybridization/introgression analysisTo determine the effect of ILS on conflicting observed phylogenetic patterns, we calculated a parameter called "theta", which reflect the level of ILS; high "theta" values indicate that ILS levels are high (Cai et al., 2021). The theta parameter was calculated by dividing the mutation units estimated from IQ-Tree v.2.0.6 and coalescent units inferred from ASTRAL v.5.7.1 (Ma et al., 2021). To detect the putative gene flow, we calculated the D-statistic (ABBA-BABA test) among Ps. gracilis, Ps. nanunica, S. longiligulata, and S. polytricha using D = (nABBA - nBABA)/(nABBA + nBABA). The condition for the existence of introgression is "D ≠ 0, Z > 3, p < 0.002" (Dong et al., 2022). In addition, we used PhyloNet v.0.9.1 (Solís-Lemus et al., 2017) to consider the effects of hybridization under ILS. The 411 gene trees of the C/D subgenome were used as inputs with the "InferNetwork_MPL" method. We assumed four parallel networks by allowing the existence of hybridization from 0 to 3 (max = 3) with 10 runs for each. Gene flow was further detected using Treemix v.1.12 (Pickrell and Pritchard, 2012).
3. Results 3.1. Plastid phylogenomic analysesAll plastid genomes of 22 individuals were completely circular. Assembly of Pseudosasa gracilis plastid genomes showed that 1) the length of plastome sequences was between 140, 070 and 143, 414 bp; 2) the number of genes was between 231 and 259; and 3) the final plastid sequence alignment was 141, 683 bp. The assembly and annotation results of HN4.1-Ps. gracilis are shown in Fig. S2. In our plastid phylogeny (Fig. 2a), 11 individuals of Ps. gracilis formed a highly supported clade (BS = 100%). This clade was sister to the Sinosasa clade, which includes three individuals of S. longiligulata and six individuals of S. polytricha, with strong support value (BS = 100%). In addition, three individuals of Ps. nanunica were clustered with other species of Pseudosasa. Most nodes within the S. longiligulata and S. polytricha clade were well-resolved (BS = 100%), whereas phylogenetic relationships among several individuals of the Ps. gracilis and Ps. nanunica clade were unresolved with low support (BS < 60%).
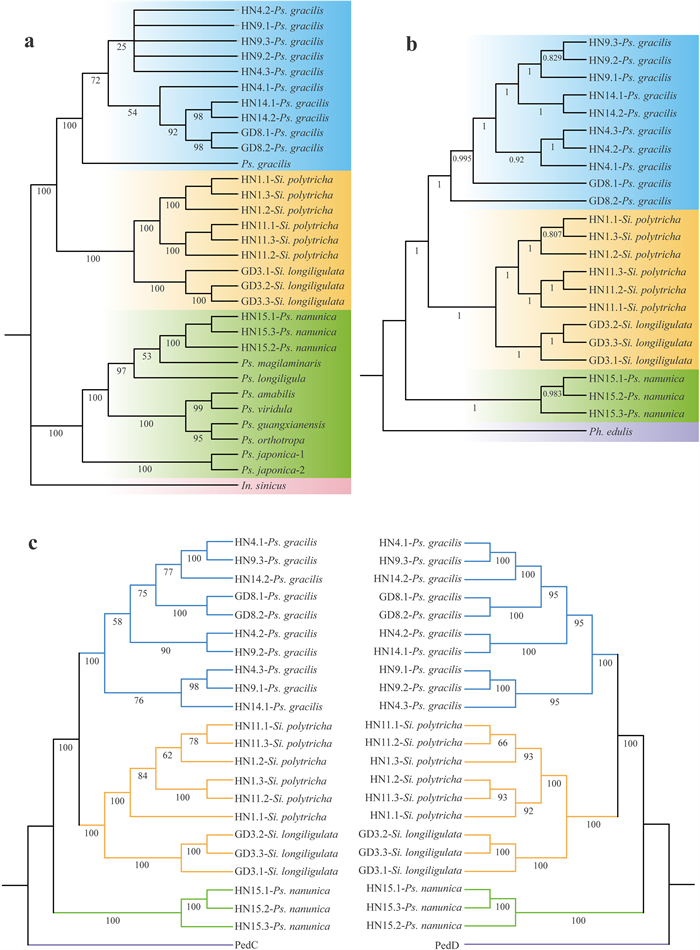
|
| Fig. 2 Phylogenomic analyses based on three data sets consistently recovered sister relationships between the Pseudosasa gracilis and the Sinosasa clades. a Plastid tree based on 32 chloroplast genome sequences. Maximum likelihood BS support values are shown below branches. b The NJ tree inferred from the SNP matrix. Support values are shown below branches. c Phylogenetic tree based on concatenated 411 "perfect-copy" syntenic gene data sets for the C (left) and D subgenome (right). Branches are colored by three clades: light blue (Ps. gracilis), light orange (Sinosasa), light green (Ps. nanunica), and the outgroup Ph. edulis is colored purple. |
A total of 6, 604, 607 genome-wide SNP sites were assembled and used to reconstruct a phylogenetic tree (Fig. 2b). In this phylogeny, all individuals of the four species were clustered into monophyletic clades with high statistical support, and the relationships among these clades were well-resolved (support value = 1). All individuals of Pseudosasa gracilis formed a group sister to Sinosasa (support value = 1) and were distinct from the other three individuals of Ps. nanunica, which is consistent with the plastid tree. Moreover, unlike the plastid tree, high resolution (support value > 0.8) for most relationships within each clade were resolved in this phylogeny.
3.3. Phylogenomic analyses based on 411 "perfect-copy" syntenic genesNuclear genes with parental genetic information were able to resolve complex phylogenetic relationships. We assembled ≥ 425 and ≥ 427 "perfect-copy" syntenic genes on the C/D subgenomes, respectively. The number of paralogs aligned to 22 individuals on the C subgenome ranged from 39 to 118, and on the D subgenome from 39 to 123 (Tables S2 and S3). Taking the highest similarity and uniqueness as our criteria, we obtained a total of 411 "perfect-copy" syntenic genes for further analysis.
The concatenated and coalescent analyses of 411 "perfect-copy" syntenic genes from both C and D subgenomes (Figs. 2c and 3a) consistently revealed the close relationship between the Ps. gracilis clade and Sinosasa (S. polytricha + S. longiligulata) clade (BS = 100% for both C and D subgenome tree), although BS support values between some individuals were relatively low (BS < 75%). The sister relationship of the Ps. gracilis and Sinosasa clade was also strongly supported by the QS score (0.7/0.75/1 for C, and 0.31/0.4/1 for D subgenome tree). The support value among different individuals within the branch was low (< 60%) when the QC < 0.2 or there was a serious conflict between the gene tree and the putative species tree.
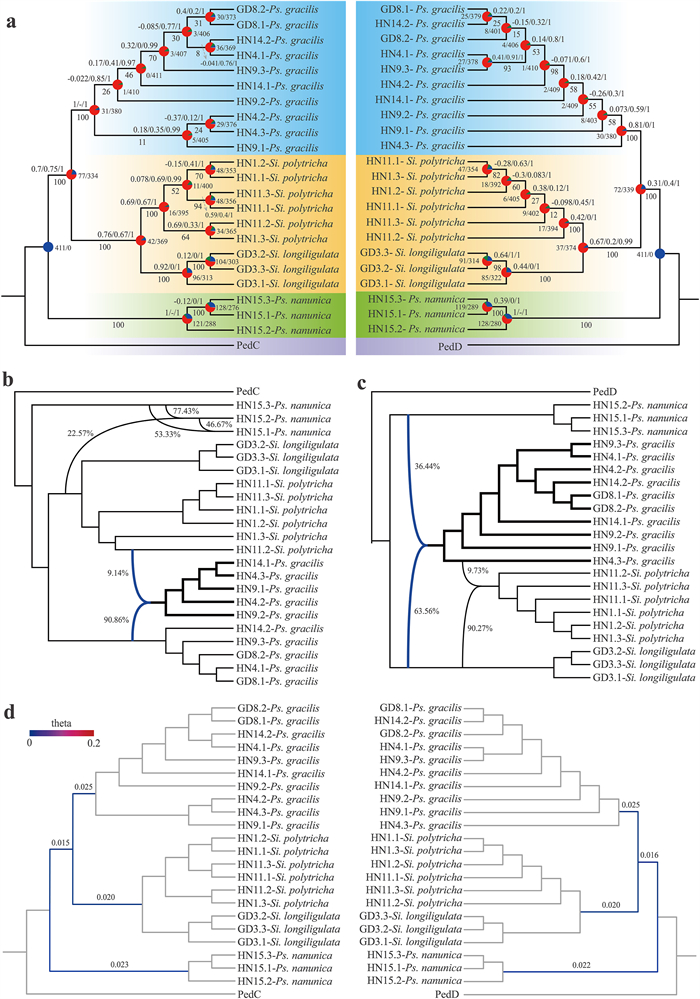
|
| Fig. 3 Extensive phylogenetic discordance and assessment of its underlying biological causes. a The ASTRAL tree inferred from 411 "perfect-copy" syntenic gene data sets of the C (left) and D subgenomes (right), and their topological comparison with the inferred gene trees. The QS scores are shown above branches, and the BS support values are shown below branches. Pie charts for major clades representing the proportion of gene trees supporting the ASTRAL species tree topology (blue), the main alternative bifurcation (green), the remaining alternatives (red), and BS values < 50% (gray). Numbers next to pie charts indicate the number of gene trees concordant/conflicting with the species tree for that node. b Three detected hybridization events in the C subgenome using PhyloNet. c Two detected hybridization events in the D subgenome using PhyloNet. d Estimated "theta" values for all internodes of three clades. Colder colors indicate lower "theta" values and thus lower ILS levels. Terminal branches are shown in grey due to the lack of specific branch lengths. |
PhyParts analyses also revealed extensive phylogenetic discordance between the gene trees and the ASTRAL tree. In this analysis, all 411 gene trees support the sister relationship between the Ps. nanunica clade and the Ps. gracilis + Sinosasa clade, whereas only 77/72 out of 411 genes recovered the congruent topology with the ASTRAL tree for the close relationship of the Ps. gracilis and Sinosasa clade. Moreover, most nodes within the clades revealed that most gene trees conflict with the putative species tree (Fig. 3a).
3.4. Detection of hybridization/introgression and ILSThe observed incongruence between the gene tree and the ASTRAL tree indicated the possibility of hybridization and/or ILS during the evolutionary history of Pseudosasa gracilis. D-statistic analysis showed that introgression has occurred between Ps. gracilis and S. longiligulata, Ps. nanunica, respectively (Fig. S4 and Table S4). We used 411 gene trees as input files to assess putative hybridization events in the C/D subgenomes by PhyloNet. When the number of hybridization events was assumed to be set to 2, a hybridization event was detected in the D subgenome (Fig. 3c), indicating Ps. gracilis was a hybrid descendant between S. longiligulata and Ps. nanunica, with 63.56% and 36.44% inheritance probabilities of each parent. Moreover, additional introgression events were detected in the C subgenome between Ps. gracilis (90.86%) and S. polytricha (9.14%) when the number of hybridization events was assumed to be set to 3 (Fig. 3b). In addition, gene flow events detected by Treemix analyses reveal a higher migration weight from S. longiligulata to Ps. gracilis when two gene flow events were set (Fig. S3), which suggests that more components of Ps. gracilis were derived from S. longiligulata in the hybridization event detected in the D subgenome.
To investigate the effect of ILS on the observed phylogenetic discordance, the values of "theta" parameter for both C and D subgenome trees were estimated. The calculated theta values for all internodes of three main clades ranged from 0.015 to 0.025 (Fig. 3d), indicating low ILS levels for the interspecies splits (as 0.01 and 0.1 reflect low and high ILS based on Cai et al., 2021). We found that the split of the Ps. gracilis and Sinosasa clade showed the minimum level of ILS, with "theta" values in the C/D subgenome estimated as 0.015 and 0.016, respectively.
4. Discussion 4.1. High resolution of inter- and intra-species relationships provided by deep genome skimming (DGS) dataFew studies have yet to adopt the DGS method for phylogenetic analyses of closely related species at the populational level (Low et al., 2022). In this study, we used the DGS strategy to obtain whole plastome sequences, genome-wide SNP sites, as well as 411 "perfect-copy" syntenic genes to examine the phylogenetic relationships among Ps. gracilis, S. polytricha, S. longiligulata, and Ps. nanunica at the population level. All three data sets provided full resolution of the phylogenetic relationships among the four species (Figs. 2 and 3a), indicating that the DGS data set is suitable and sufficient to resolve the phylogenetic relationships among closely related species. Similarly, the relationship between different populations within the species was also analyzed through two "perfect-copy" syntenic gene data sets. Individuals from different populations clustered together to form monophyletic groups, such as the population collected in Guangdong Province. However, although we assembled complete looped chloroplast genome sequences for all individuals from DGS data set, the chloroplast genome showed low support values between individuals within species, mainly due to its low variation rate and insufficient informative sites (Fig. 2a).
It is worth noting that, with a closely related reference genome or gene set of interest, DGS is a promising approach to generate a large set of lineage-specific nuclear genes for phylogenomic study. In our study, using 430 "perfect-copy" syntenic genes extracted from 11 bamboo genomes as reference (Ma et al., 2024), we were able to detect more than 97% of the genes of interest for tetraploid species. These resulting gene sequences enable us to perform coalescent analyses, as well as examine phylogenetic discordance between different gene trees. Moreover, DGS sequencing data can provide genome sequences from different genetic systems and sufficient evidence for hybridization analysis, in which chloroplast genome sequences supply clues for determining the maternal parent of hybrid species. In short, our study based on population sampling data from four closely related species, shows DGS sequencing data has great potential in resolving phylogenetic relationships among closely related species and individuals in different populations within a species.
4.2. Hybridization is the major driving force in the origin of Pseudosasa gracilisBased on phylogenetic analyses of data from eight combined plastid sequences (Zhang et al., 2012) and ddRAD data (Guo et al., 2021), an unexpected systematic position of Ps. gracilis clustered with Sinosasa was discovered. However, the conflict between morphological and molecular evidence existed of Ps. gracilis and Sinosasa (Fig. 1). Here, we analyzed the phylogenetic relationships between taxa in Pseudosasa using three different data sets as well as concatenated and coalescent methods congruently. Our results confirm those of previous studies that showed a close relationship between Ps. gracilis and Sinosasa (Figs. 2 and 3a).
Temperate woody bamboos belong to a tetraploid lineage with an allopolyploid origin (Guo et al., 2019a) and extremely long vegetative periods, representing a unique system for understanding ancient reticulate evolution. Previous studies have shown that hybridization played a significant role in bamboo evolution (Triplett, 2008; Triplett and Clark, 2010; Yang et al., 2013; Triplett et al., 2014). Interestingly, in our study, a hybridization event was detected based on 411 gene trees of the D subgenome, revealing that Ps. gracilis may have originated from the hybridization of S. longiligulata and Ps. nanunica (Fig. 3c). However, a different hybridization pattern was observed in the C subgenome. An introgression event of Ps. gracilis and S. polytricha in the same region was detected in the C subgenome. Previous genomic studies (Guo et al., 2019a; Ma et al., 2024) indicate that the C subgenome is shared by all extant woody bamboo lineages and has experienced complex chromosome rearrangements. This often obscures hybridization signals. The D subgenome is unique in the temperate woody bamboos, and underwent no large-scale chromosome rearrangement events, thus making the hybridization signal easier to detect. Therefore, we speculate that the hybrid signals in the D subgenome were more easily retained due to its relatively stable evolutionary history.
Combined with the nuclear and plastid trees, we speculate on possible parents. Based on the present geographical distribution of the two putative parents, it is speculated that Ps. gracilis may be originated from sympatric hybridization. Based on the dated results in our previous study (Guo et al., 2021), the differentiation of two major subclades within the leptomorph clade occurred circa 10.2 Ma (95%HPD: 6.75–13.96 Ma). A clade of other species of Pseudosasa diverged at about 9.33 Ma (95%HPD: 6.26–13.35 Ma), and the diversification of Ps. gracilis and the S. longiligulata + S. guangxiensis clade occurred at about 7.83 Ma (95%HPD: 4.33–10.84 Ma). Unlike the hybrids that emerged rapidly and possessed obscure morphological characters (Ramsey and Schemske, 2002), the evolutionary history of Ps. gracilis is on a million-year scale, which was long enough for speciation (Zou et al., 2022). Thus, we infer that Ps. gracilis is the result of hybrid origin rather than of hybrid swarms (Abbot, 2003; Wang et al., 2021b). Compared with the hybridization history of three species in Arundinaria from North America, we revealed similar origin of sympatric hybridization of Ps. gracilis, as a representative from the distribution center of the temperate woody bamboo in East Asia, suggesting that although the temperate woody bamboos are allotetraploid and have extremely long vegetative periods, many ancient hybridizations still existed in their evolutionary history (Stull et al., 2023).
One of the most important conditions for hybridization is overlapping geographical distribution. Ps. gracilis, Ps. nanunica, and S. polytricha are all distributed in Mangshan Mountain area (Fig. S1). Thus, the overlapping distribution of species increases their opportunity for gene exchange. However, our sampling may be insufficient to identify the concrete parent species of Ps. gracilis from Sinosasa, and species closely related to Ps. nanunica, necessitating further studies with more comprehensive species sampling be carried out, although we cannot rule out the possibility that the parents are already extinct.
By integrating analyses of both maternally inherited (i.e., chloroplast genome) and biparentally inherited (subgenome) molecular data, together with morphological and geographic information, for the first time we revealed the hybrid origin of a unique species in the temperate woody bamboos by employing datasets of syntenic genes and plastomes at the population level. The findings generated here will be significant for the studies of polyploid species with complicated reticulate evolutionary history.
AcknowledgementsWe thank Ms. Liang-Min Liu, Ms. Shuang-Xiu Xu (all from Kunming Institute of Botany, Chinese Academy of Sciences) for assistance in analytical processes. We are also grateful to Jun Chen, De-Sheng Chen (Mangshan National Nature Reserve Administration, Hunan Province), Zhang-Ping You (Nanling National Nature Reserve, Guangdong Province), and Hua-Ge Deng (Luofu Mountain Provincial Nature Reserve, Guangdong Province) for logistic support for sampling, and to the iFlora High Performance Computing Center of Germplasm Bank of Wild Species (iFlora HPC Center of GBOWS, KIB, CAS) for computation. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31000000) and the National Natural Science Foundation of China (32200193).
Author contributions
XZH, CG, and SYQ conducted the sampling. XZH conducted the chloroplast and SNPs phylogenetic analyses, detected the hybridization/introgression events. CG conducted the nuclear phylogenetic analyses and examined the conflicts between gene trees and species tree. SYQ analyzed the gene flow. XZH wrote the manuscript, CG, SYQ, DZL and ZHG revised the manuscript. All authors approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.06.001.
Abbott, R.J., 2003. Sex, sunflowers, and speciation. Science, 301: 1189-1190. |
Berger, B.A., Han, J., Sessa, E.B., et al., 2017. The unexpected depths of genome-skimming data: a case study examining Goodeniaceae floral symmetry genes. Appl. Plant Sci., 5: 1700042. |
Birky, C.W., 1995. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl. Acad. Sci. U.S.A., 92: 11331-11338. DOI:10.1073/pnas.92.25.11331 |
Bolger, A.M., Lohse, M., Usadel, B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30: 2114-2120. DOI:10.1093/bioinformatics/btu170 |
Cai, L.M., Xi, Z.X., Lemmon, E.M., et al., 2021. The perfect storm: gene tree estimation error, incomplete lineage sorting, and ancient gene flow explain the most recalcitrant ancient angiosperm clade. Malpighiales. Syst. Biol., 70: 491-507. DOI:10.1093/sysbio/syaa083 |
Capella-Gutierrez, S., Silla-Martinez, J.M., Gabaldon, T., 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 25: 1972-1973. DOI:10.1093/bioinformatics/btp348 |
Debray, K., Le Paslier, M.C., Bérard, A., et al., 2022. Unveiling the patterns of reticulated evolutionary processes with phylogenomics: hybridization and polyploidy in the genus Rosa. Syst. Biol., 71: 547-569. DOI:10.1093/sysbio/syab064 |
Dong, W.P., Li, E.Z., Liu, Y.L., et al., 2022. Phylogenomic approaches untangle early divergences and complex diversifications of the olive plant family. BMC Biology, 20: 92. |
Doolittle, W.F., 1999. Phylogenetic classification and the universal tree. Science, 284: 2124-2128. |
Gross, B.L., Rieseberg, L.H., 2005. The ecological genetics of homoploid hybrid speciation. J. Hered., 96: 241-252. DOI:10.1093/jhered/esi026 |
Guo, C., Guo, Z.H., Li, D.Z., 2019. Phylogenomic analyses reveal intractable evolutionary history of a temperate bamboo genus (Poaceae: Bambusoideae). Plant Divers., 41: 213-219. |
Guo, C., Ma, P.F., Yang, G.Q., et al., 2021. Parallel ddRAD and genome skimming analyses reveal a radiative and reticulate evolutionary history of the temperate bamboos. Syst. Biol., 70: 756-773. DOI:10.1093/sysbio/syaa076 |
Guo, C., Luo, Y., Gao, L.M., et al., 2023. Phylogenomics and the flowering plant tree of life. J. Integr. Plant Biol., 65: 299-323. DOI:10.1111/jipb.13415 |
Guo, Z.H., Ma, P.F., Yang, G.Q., et al., 2019. Genome sequences provide insights into the reticulate origin and unique traits of woody bamboos. Mol. Plant, 12: 1353-1365. |
Hu, Y.N., Zhao, L., Buggs, R.J.A., et al., 2019. Population structure of Betula albosinensis and Betula platyphylla: evidence for hybridization and a cryptic lineage. Ann. Bot., 123: 1179-1189. DOI:10.1093/aob/mcz024 |
Janzen, D.H., 1976. Why bamboos wait so long to flower. Annu. Rev. Ecol. Systemat., 7: 347-391. DOI:10.1146/annurev.es.07.110176.002023 |
Jin, J.J., Yu, W.B., Yang, J.B., et al., 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol., 21: 241. |
Johnson, M.G., Gardner, E.M., Liu, Y., et al., 2016. HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci., 4: 1600016. |
Katoh, K., Standley, D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Kearse, M., Moir, R., Wilson, A., et al., 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Linder, C.R., Rieseberg, L.H., 2004. Reconstructing patterns of reticulate evolution in plants. Am. J. Bot., 91: 1700-1708. DOI:10.3732/ajb.91.10.1700 |
Liu, B.B., Ma, Z.Y., Ren, C., et al., 2021. Capturing single-copy nuclear genes, organellar genomes, and nuclear ribosomal DNA from deep genome skimming data for plant phylogenetics: a case study in Vitaceae. J. Systemat. Evol., 5: 1124-1138. DOI:10.1111/jse.12806 |
Liu, J.X., Zhou, M.Y., Yang, G.Q., et al., 2020. ddRAD analyses reveal a credible phylogenetic relationship of the four main genera of Bambusa-Dendrocalamus-Gigantochloa complex (Poaceae: Bambusoideae). Mol. Phylogenet. Evol., 146: 106758. |
Liu, L.X., Deng, P., Chen, M.Z., et al., 2023. Systematics of Mukdenia and Oresitrophe (Saxifragaceae): insights from genome skimming data. J. Systemat. Evol., 61: 99-114. |
Low, L.W., Rajaraman, S., Tomlin, C.M., et al., 2022. Genomic insights into rapid speciation within the world's largest tree genus Syzygium. Nat. Commun., 13: 5031. |
Lv, S.Y., Ye, X.Y., Li, Z.H., et al., 2023. Testing complete plastomes and nuclear ribosomal DNA sequences for species identification in a taxonomically difficult bamboo genus Fargesia. Plant Divers., 45: 147-155. |
Ma, J.X., Sun, P.C., Wang, D.D., et al., 2021. The Chloranthus sessilifolius genome provides insight into early diversification of angiosperms. Nat. Commun., 12: 6929. |
Ma, P.F., Liu, Y.L., Guo, C., et al., 2024. Genome assemblies of 11 bamboo species highlight diversification induced by dynamic subgenome dominance. Nat. Genet., 56: 710-720. DOI:10.1038/s41588-024-01683-0 |
Mallet, J., Besansky, N., Hahn, M.W., 2016. How reticulated are species?. Bioessays, 38: 140-149. DOI:10.1002/bies.201500149 |
McKenna, A., Hanna, M., Banks, E., et al., 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20: 1297-1303. DOI:10.1101/gr.107524.110 |
Morales-Briones, D.F., Kadereit, G., Tefarikis, D.T., et al., 2021. Disentangling sources of gene tree discordance in phylogenomic data sets: testing ancient hybridization in Amaranthaceae s.l.. Syst. Biol., 70: 219-235. DOI:10.1093/sysbio/syaa066 |
Nguyen, L.T., Schmidt, H.A., Haeseler, A.V., et al., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol., 32: 268-274. DOI:10.1093/molbev/msu300 |
Nitta, J.H., Ebihara, A., Ito, M., 2011. Reticulate evolution in the Crepidomanes minutum species complex (Hymenophyllaceae). Am. J. Bot., 98: 1782-1800. DOI:10.3732/ajb.1000484 |
Parisod, C., Badaeva, E.D., 2020. Chromosome restructuring among hybridizing wild wheats. New Phytol., 226: 1263-1273. DOI:10.1111/nph.16415 |
Pease, J.B., Brown, J.W., Walker, J.F., et al., 2018. Quartet sampling distinguishes lack of support from conflicting support in the green plant tree of life. Am. J. Bot., 105: 385-403. DOI:10.1002/ajb2.1016 |
Pennisi, E., 2016. Shaking up the tree of life. Science, 354: 817-821. DOI:10.1126/science.354.6314.817 |
Pickrell, J.K., Pritchard, J.K., 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genetics, 8: e1002967. DOI:10.1371/journal.pgen.1002967 |
Qu, X.J., Moore, M.J., Li, D.Z., et al., 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 15: 50. |
Ramsey, J., Schemske, D.W., 2002. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Systemat., 33: 589-639. DOI:10.1146/annurev.ecolsys.33.010802.150437 |
Rieseberg, L.H., Soltis, D.E., 1991. Phylogenetic consequences of cytoplasmic gene flow in plants. Evol. Trends Plants, 5: 65-84. |
Smith, S.A., Moore, M.J., Brown, J.W., et al., 2015. Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evol. Biol., 15: 150. |
Solís-Lemus, C., Bastide, P., Ané, C., 2017. Phylonetworks: a package for phylogenetic networks. Mol. Biol. Evol., 34: 3292-3298. DOI:10.1093/molbev/msx235 |
Soltis, P.S., Soltis, D.E., 2009. The role of hybridization in plant speciation. Annu. Rev. Plant Biol., 60: 561-588. DOI:10.1146/annurev.arplant.043008.092039 |
Soreng, R.J., Peterson, P.M., Fernando, O.Z., et al., 2022. A worldwide phylogenetic classification of the Poaceae (Gramineae) Ⅲ: an update. J. Systemat. Evol., 60: 476-521. DOI:10.1111/jse.12847 |
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Stewart, N.C., Halfhill, M.D., Warwick, S.I., 2003. Transgene introgression from genetically modified crops to their wild relatives. Nat. Rev. Genet., 4: 806-817. |
Stull, G.W., Pham, K.K., Soltis, P.S., et al., 2023. Deep reticulation: the long legacy of hybridization in vascular plant evolution. Plant J., 114: 743-766. DOI:10.1111/tpj.16142 |
Triplett, J.K., 2008. Phyloenetic Relationships Among the Temperate Bamboos (Poaceae: Bambusoideae) with an Emphasis on Arundinaria and Allies. Lowa State University, Lowa, United States.
|
Triplett, J.K., Clark, L.G., 2010. Phylogeny of the temperate bamboos (Poaceae: Bambusoideae: Bambuseae) with an emphasis on Arundinaria and allies. Syst. Bot., 35: 102-120. DOI:10.1600/036364410790862678 |
Triplett, J.K., Clark, L.G., 2021. Hybridization in the temperate bamboos (Poaceae: Bambusoideae: Arundinarieae): a phylogenetic study using AFLPs and cpDNA sequence data. Syst. Bot., 46: 48-69. DOI:10.1600/036364421x16128061189503 |
Triplett, J.K., Clark, L.G., Fisher, L.G., et al., 2014. Independent allopolyploidization events preceded speciation in the temperate and tropical woody bamboos. New Phytol., 204: 66-73. DOI:10.1111/nph.12988 |
Vargas, O.M., Heuertz, M., Smith, S.A., et al., 2019. Target sequence capture in the Brazil nut family (Lecythidaceae): marker selection and in silico capture from genome skimming data. Mol. Phylogenet. Evol., 135: 98-104. |
Walker, J.F., Walker-Hale, N., Vargas, O.M., et al., 2019. Characterizing gene tree conflict in plastome-inferred phylogenies. PeerJ, 7: e7747. DOI:10.7717/peerj.7747 |
Wang, H.X., Morales-Briones, D.F., Moore, M.J., et al., 2021. A phylogenomic perspective on gene tree conflict and character evolution in Caprifoliaceae using target enrichment data, with Zabelioideae recognized as a new subfamily. J. Syst. Evol., 59: 897-914. DOI:10.1111/jse.12745 |
Wang, X.T., Chen, L.Y., Ma, J.X., 2019. Genomic introgression through interspecific hybridization counteracts genetic bottleneck during soybean domestication. Genome Biol., 20: 22-36. |
Wang, Z.F., Jiang, Y.Z., Bi, H., et al., 2021. Hybrid speciation via inheritance of alternate alleles of parental isolating genes. Mol. Plant, 14: 208-222. |
Wick, R.R., Schultz, M.B., Zobel, J., et al., 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics, 31: 3350-3352. DOI:10.1093/bioinformatics/btv383 |
Yang, F.M., Ge, J., Guo, Y.J., et al., 2023. Deciphering complex reticulate evolution of Asian Buddleja (Scrophulariaceae): insights into the taxonomy and speciation of polyploid taxa in the Sino-Himalayan region. Ann. Bot., 132: 15-28. DOI:10.1093/aob/mcad022 |
Yang, H.M., Zhang, Y.X., Yang, J.B., et al., 2013. The monophyly of Chimonocalamus and conflicting gene trees in Arundinarieae (Poaceae: Bambusoideae) inferred from four plastid and two nuclear markers. Mol. Phylogenet. Evol., 68: 340-356. |
Ye, X.Y., Ma, P.F., Guo, C., et al., 2021. Phylogenomics of Fargesia and Yushania reveals a history of reticulate evolution. J. Systemat. Evol., 59: 1183-1197. DOI:10.1111/jse.12719 |
Yu, J.R., Niu, Y.T., You, Y.C., et al., 2023. Integrated phylogenomic analyses unveil reticulate evolution in Parthenocissus (Vitaceae), highlighting speciation dynamics in the Himalayan-Hengduan Mountains. New Phytol., 238: 888-903. DOI:10.1111/nph.18580 |
Zhang, C., Rabiee, M., Sayyari, E., et al., 2018. ASTRAL-Ⅲ: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics, 19: 153. DOI:10.1007/978-3-319-93719-9_20 |
Zhang, N., Wen, J., Zimmer, E.A., 2015. Congruent deep relationships in the grape family (Vitaceae) based on sequences of chloroplast genomes and mitochondrial genes via genome skimming. PLoS One, 10: e0144701. DOI:10.1371/journal.pone.0144701 |
Zhang, X.T., Chen, S., Shi, L.Q., et al., 2021. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet., 53: 1250-1259. DOI:10.1038/s41588-021-00895-y |
Zhang, Y.X., Zeng, C.X., Li, D.Z., 2012. Complex evolution in Arundinarieae (Poaceae: Bambusoideae): incongruence between plastid and nuclear GBSSI gene phylogenies. Mol. Phylogenet. Evol., 63: 777-797. |
Zhao, X.B., Fu, X.D., Yin, C.B., et al., 2021. Wheat speciation and adaptation: perspectives from reticulate evolution. aBIOTECH, 2: 386-402. DOI:10.1007/s42994-021-00047-0 |
Zou, T.T., Kuang, W.M., Yin, T.T., et al., 2022. Uncovering the enigmatic evolution of bears in greater depth: the hybrid origin of the Asiatic black bear. Proc. Natl. Acad. Sci. U.S.A., 119: e2120307119. |



