b. Yunnan International Joint Laboratory for Biodiversity of Central Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, Yunnan, China;
c. Institute of Botany, Academy Sciences of Uzbekistan, Tashkent 100125, Uzbekistan;
d. University of Chinese Academy of Sciences, Beijing 100864, China
The Asphodelaceae Juss. (Asparagales) (APG IV, 2016) is divided into the subfamilies Hemerocallidoideae Lindl., Xanthorrhoeoideae M.W. Chase, Reveal & M.F. Fay, and Asphodeloideae Burnett. The last subfamily encompasses a total of 14 genera worldwide, including Eremurus M. Bieb (Chase et al., 2000). Eremurus was first described by Friedrich August Marschall von Bieberstein as having a distinct set of traits, including a leafless flowering stem with more than 50 flowers per inflorescence and a rhizomatous rootstock (Bieberstein, 1819). The first classification of Eremurus s.l. was by Baker (1877), who divided the genus into three subgenera based on filament length. The subsequent taxonomic classifications of Eremurus were based on the perianth structure, the number of nerves in the tepals, the surface of the capsule, and the filament length (Boissier, 1884; Vvedensky, 1941; Wendelbo, 1958, 1982; Khokhryakov, 1965). Using perianth structure, filament length and the number of nerves in the tepals as the main diagnostic characteristics for the taxonomy of the genus, Wendelbo (1982) produced the most recent revised classification and divided the genus into two subgenera, Eremurus Baker and Henningia (Kar. & Kir.) Baker, and three sections Eremurus (Baker) Wendelbo, Ammolirion (Kar. & Kir.) Boiss. and Henningia (Kar. & Kir.) Baker. Table 1 provides additional details on the historical background of the infrageneric classification of Eremurus. Based on Wendelbo's classification, Naderi et al. (2009) carried out the first morphological cladistic analysis of Eremurus and concluded that the genus, as well as subgenus Eremurus, was monophyletic, but subgenus Henningia was paraphyletic.
| Baker (1877) | Boissier (1884) | Vvedensky (1941) | Wendelbo (1958) | Khokhryakov (1965) | Wendelbo (1982) and current classification |
| subg. Eremurus | sect. Eremurus | sect. Eremurus | subg. Eremurus, sect. Eremurus | Eremurus | subg. Eremurus, sect. Eremurus (including genus Selonia) |
| subg. Ammolirion | sect. Ammolirion | sect. Ammolirion | subg. Eremurus, sect. Ammolirion | Ammolirion | subg. Eremurus, sect. Ammolirion |
| subg. Henningia | sect. Henningia | sect. Henningia (including sect. Trochantus) | subg. Henningia, sect. Henningia | Henningia | subg. Henningia, sect. Henningia |
| – | sect. Trochantus | – | subg. Henningia, sect. Trochantus | – | – |
| – | – | – | Selonia | – |
Molecular studies utilizing plastid (trnL-F) and nuclear DNA (ITS) data affirmed that Eremurus was monophyletic (Chase et al., 2000; Devey et al., 2006; Safar et al., 2014), but also that both subgenera were paraphyletic (Safar et al., 2014). Despite some progress in understanding the phylogeny of the genus (Naderi et al., 2009; Safar et al., 2014; Makhmudjanov et al., 2019), the limited sampling and small number of gene markers used in these studies precluded the phylogeny of the genus being fully resolved. Complete plastome sequencing, since becoming a relatively inexpensive procedure, has been widely used for phylogenetic reconstructions (Jansen et al., 2007; Moore et al., 2010; Malé et al., 2014; Yusupov et al., 2021; Lee et al., 2022; Liu et al., 2022; Su et al., 2023).
Eremurus comprises 59 species (Eker, 2020) distributed in Central Asia, the Caucasus region, Afghanistan, Iran, Pakistan, Iraq, Turkey, Lebanon, India and China (Wendelbo and Furse, 1969). Although southwest Asia harbors a large number of species (more than 24), Central Asia, where > 45 species occur, is considered to be the main center for Eremurus (Fedtschenko, 1935; Hedge and Wendelbo, 1963). In eastern Asia, however, the distribution of the endemic Eremurus chinensis O. Fedtsch. along the Hengduan Mountains exhibits an interesting biogeographical disjunction within this genus. This disjunction was attributed by Khokhryakov (1965) to the fact that Central Asia was once part of the ancient Mediterranean area with the dominance of a typical Mediterranean climate characterized by dry, hot summers and cool and humid winters (Popov, 1927, 1963). During that time, the xerophytic ancient Mediterranean flora extended much farther to the east than it does now, reaching even Japan. Popov (1941) attributed Eremurus to this flora. Khokhryakov (1965) hypothesized that subg. Ammolirion and Henningia originated from an ancient species of Aloe L. in what is present-day western and south central Asia in the late Paleogene to the early Neogene, most likely in the Oligocene. Those views have never been put to the test using molecular data or a formal biogeographical analysis. The first molecular estimates of divergence times carried out by McLay and Bayly (2016) for E. chinensis estimated its time of origin to be the early Oligocene. However, the divergence time of Eremurus lineages is not known yet, making it difficult to identify the most probable geological events responsible for the emergence and evolution of Eremurus.
In this study, we present the first analysis of Eremurus phylogeny and evolution based on both nuclear DNA (nrDNA) and complete chloroplast genomes (cp), covering ca. 60% of the described species. For these analyses, we sequenced, assemblad and analyzed the plastomes of 27 species (16 of which are new sequences), and nuclear data of 33 species (39 taxa) of Eremurus, and combined them with an additional 15 plastomes and 29 nuclear loci of the species from other genera of Asphodelaceae, Asparagaceae and Amaryllidaceae families. Specifically, we attempted to address the following issues: (1) resolve the infra-generic phylogeny and re-evaluate the taxonomic significance of the morphological characters that were used for systematic considerations in Eremurus; (2) estimate the divergence time and reconstruct the evolution of the genus. We hope that these findings will serve as a basis for future evolutionary reconstructions utilizing deeper and more comprehensive within-genus sampling.
2. Materials and methods 2.1. Taxon sampling and DNA extractionForty-two species representing three monocot families (Asphodelaceae, Asparagaceae and Amaryllidaceae), of which 27 species were from Eremurus, were studied using complete plastome data. Additionally, 61 species and 71 accessions (of which 33 species and 39 accessions belonged to Eremurus) were studied using nrDNA ITS. Information regarding sampling locations and voucher specimens of 27 (16 pt and 27 ITS) newly sequenced species of Eremurus is in Table S1. NCBI accessions of newly sequenced species and the species downloaded from NCBI are presented in Tables S2a and b. Leaf samples were dried in silica-gel upon collecting. Total genomic DNA was extracted from 1 g of silica-dried leaves using a modified CTAB protocol (Doyle and Doyle, 1987).
2.2. Sequence assembly and annotationDNA was sheared to construct a 350-bp (insert size) sequence library using the Genomic DNA Sample Prep Kit (Illumina) according to the manufacturer's protocol and then sequenced using 150 paired-end reads on the Illumina HiSeq 4000 at Beijing Novogene Bioinformatics Technology Co., Ltd, Beijing, China. For raw data processing, we used the Next Generation Sequencing (NGS) QC Tool Kit with default settings (Patel and Jain, 2012). The resulting clean reads were assembled in NovoPlasty v.3.8.3 (Dierckxsens et al., 2017) using Eremurus robustus NC_046772 (Makhmudjanov et al., 2019) as the reference genome. Contigs generated by NovoPlasty were sorted and joined into a single draft sequence with E. robustus as the reference in the software Geneious v.10.0.2. (Kearse et al., 2012). Gene annotation was performed in Geneious v.10.0.2. Start and stop codons and intron/exon boundaries for protein-coding genes were checked manually (Kearse et al., 2012). The nrDNA sequencing reads were assembled using GetOrganelle v.1.7.4.1 (Jin et al., 2020). Partial nrDNA sequences included the 18S ribosomal RNA gene, internal transcribed spacer 1, 5.8S ribosomal RNA gene and the internal transcribed spacer 2.
2.3. Phylogenetic analysisFor phylogenetic reconstruction, plastomes of three species representing two genera of Asphodeloideae (Aloe and Aloidendron (A. Berger) Klopper & Gideon F.Sm.), and one species of each of Hemerocallis L. (Hemerocallidoideae) and Xanthorrhoea Sm. (Xanthorrhoeoideae) and ITS sequences for 16 species (19 taxa) from nine genera of all three subfamilies (Aloe, Aloidendron, Bulbine, Bulbinella, Kniphofia, Asphodelus, Asphodeline, Xanthorrhoea and Hemerocallis) were chosen as outgroups. Their respective data were downloaded from NCBI (Tables S2a and b). In total, three data sets were produced: (1) 68 protein-coding sequences (CDS), (2) complete plastome sequences with the removal of inverted repeat A (IRA) and (3) nrDNA. The 68 plastid CDS genes were extracted from the annotated plastomes using the software Geneious v.10.0.2 (Kearse et al., 2012). All datasets were first aligned using MAFFT v.7.311 (Katoh and Standley, 2013) and then manually aligned with MEGA v.7.0.14 (Kumar et al., 2016). The 68 CDS were concatenated using Sequence Matrix software (Vaidya et al., 2011). Poorly aligned regions were removed. Maximum parsimony (MP) was performed using PAUP* v.4.10 (Swofford, 2002). All characters were equally weighted, gaps were treated as missing and character states were treated as unordered. A heuristic search was performed with TBR branch swapping and the Multrees option, and random stepwise addition with 1000 replications. All analyses used the best-fitting models of nucleotide substitutions selected in jModelTest v.2.1.4 (Darriba et al., 2012) under the Akaike information criterion (AIC). Maximum likelihood (ML) analyses were conducted using RAxML v.8.0 (Stamatakis, 2014) based on the best-fit (GTR + G for the CP data sets and GTR + I + G for the nrDNA data set) models and 1000 bootstrap replicates. Bayesian inference (BI) analyses were performed with MrBayes v.3.2 (Ronquist and Huelsenbeck, 2003). The Markov chain Monte Carlo (MCMC) algorithm was run for 10, 000, 000 generations with four incrementally heated chains, starting from random trees and sampling every 1000 generations. The first 25% of the trees were discarded as burn-in and the remaining trees were used to construct a 50% majority rule consensus tree. Internodes with posterior probabilities > 95% were considered statistically significant.
2.4. Ancestral state reconstruction of key morphological charactersWe traced the evolution of the following most informative taxonomic characters of Eremurus: 1, flower shape (campanulate, tubular or subrotate); 2, number of veins per tepal (3 or 5 veins, or 1 vein); and 3, stamen length (longer than perianth or shorter than perianth). The morphological data were obtained by examining fresh material and herbarium specimens at TASH, LE and MW and from the literature (Vvedensky, 1932, 1941, 1963; Kashenko, 1951; Goloskokov, 1958; Wendelbo, 1982; Xinqi and Turland, 2000; Naderi et al., 2009). Analyses were performed using an Mk1 evolutionary model in the software Mesquite v.2.0 (Maddison, 2007). The corresponding characteristics were mapped onto the species trees constructed from the CDS and nrDNA datasets (Fig. 1).
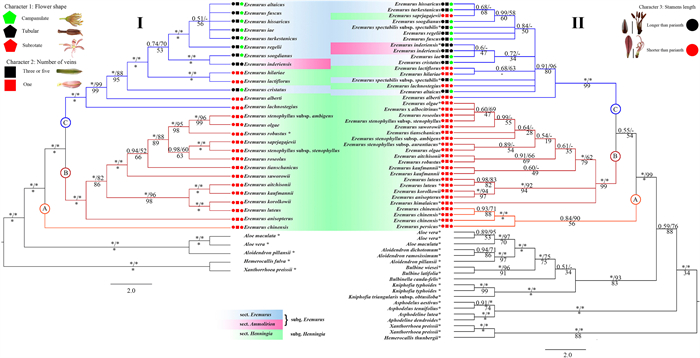
|
| Fig. 1 Majority rule consensus tree from the CDS (Ⅰ) and nrDNA (Ⅱ) data sets showing relationships among species of Eremurus and outgroups. Bayesian inference posterior probability (BI) and Maximum parsimony (MP) values are indicated above and maximum likelihood bootstrap (ML) values are indicated below the branches, respectively. Asterisks (*) indicate maximum support values (1.00 or 100%). The shapes refer to taxonomically important characters: 1-flower shape (pentagon), 2-number of veins per tepal (rectangle) and 3-stamen length (circle). |
For divergence time estimation based on the CDS dataset, we included seven species from the Asparagaceae (Hosta yingeri, Yucca filamentosa, Aphyllanthes monspeliensis, Bernardia japonica, Asparagus officinalis, Convallaria keiskei, Cordyline indivisa) and three species from the Amaryllidaceae (Allium galanthum, Lycoris aurea, Agapanthus coddii) (Fig. 2) as outgroups, in addition to five species used in the phylogenetic analysis (Fig. 1Ⅰ). For the nrDNA data set, in addition to the species of Asphodeloideae, Hemerocallidoideae and Xanthorrhoeoideae subfamilies used in the phylogenetic analysis (Fig. 1Ⅱ), we used the following outgroups: eight species from the Asparagaceae (Yucca mixtecana, Y. filifera, Y. lacandonica, Hosta plantaginea, Convallaria majalis, Cordyline cannifolia, C. indivisa, A. officinalis) and five species from the Amaryllidaceae (A. galanthum, Agapanthus campanulatus, A. caulescens, Lycoris squamigera, Lycoris radiata) (see Fig. 3 and Table S2a).
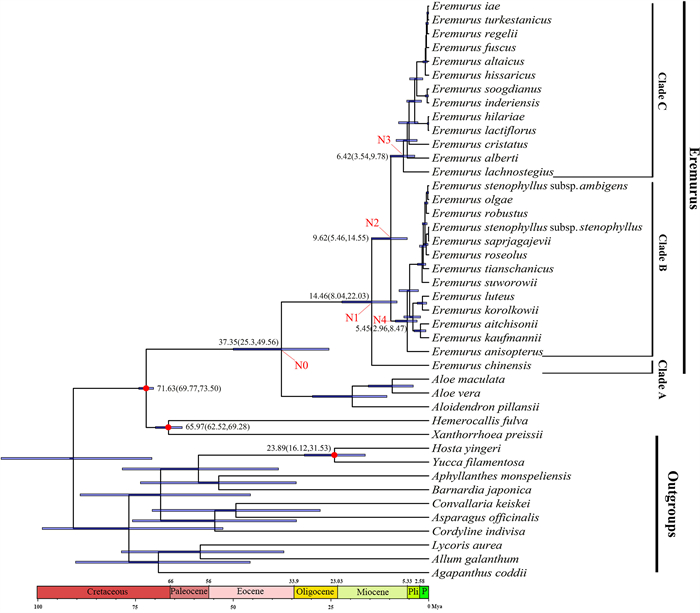
|
| Fig. 2 BEAST-derived chronogram of Eremurus based on the plastome data set. Red circles refer to calibration points. Bars on nodes indicate 95% posterior credibility intervals. On the timescale, Pli = Pliocene, P = Pleistocene. |
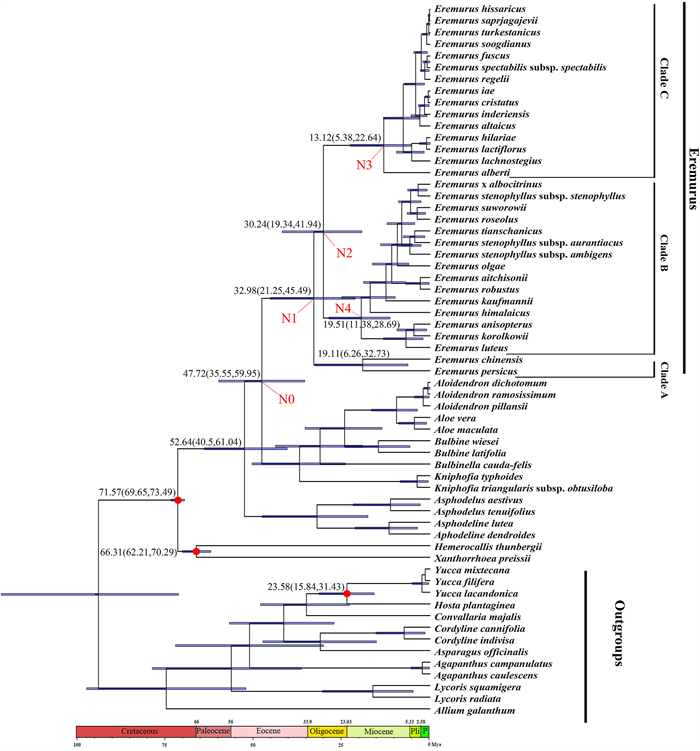
|
| Fig. 3 BEAST-derived chronogram of Eremurus based on nrDNA (ITS) data set. Red circles refer to calibration points. Bars on nodes indicate 95% posterior credibility intervals. On the timescale, Pli = Pliocene, P = Pleistocene. |
We estimated divergence times using BEAST v.1.8.4 (Drummond and Rambaut, 2007; Suchard and Rambaut, 2009) on XSEDE, with run parameters set using BEAUti v.1.8.4 (bundled with BEAST v.1.8.4). The best fit models GTR + G and GTR + I + G were selected for the CDS and the nrDNA sequence data respectively with a Yule speciation tree prior and an uncorrelated lognormal relaxed molecular clock model. Two independent analyses were run for 400, 000, 000 generations each using four Markov chains and sampling every 1000 generations. The first 10% generations were discarded as burn-in. The convergence of the MCMC log file was checked to ensure parameter effective sample size (ESS) values above 200 using Tracer 1.5 (Drummond and Rambaut, 2007). A maximum clade credibility (MCC) tree with mean and 95% HPD node ages was calculated using TreeAnnotator v.1.8.4 (Drummond and Rambaut, 2007) with 10% burn-in and a 0.5 posterior probability limit. The dating analysis utilized three age calibration points. Following McLay and Bayly (2016), a normal distribution with the mean of 71.36 Mya and a standard deviation of 1.0 was set to calibrate the crown group of Asphodelaceae, and a normal distribution with the mean of 67.37 Mya and a standard deviation of 2.3 was set to calibrate the crown group of Xanthorrhoeoidcae and Hemerocallidoideae. In addition, a normal distribution with the mean of 27.92 Mya and a standard deviation of 4.3 was set to calibrate the crown group of Yucca–Hosta according to McKain et al. (2016). The final tree was checked and edited in FigTree v.1.4.1 (Rambaut, 2018).
2.6. Ancestral area reconstructionAnalysis of potential ancestral distribution areas at internal nodes was conducted using RASP v.4.2 (Yu et al., 2020), which implements a dispersal-extinction-cladogenesis model (DEC) and a statistical dispersal–vicariance analysis (S-DIVA). Both analyses were performed using the 40, 000 post-burn-in trees of the BEAST analysis based on the nrDNA dataset. To represent the biogeographic range of the Eremurus alone, we removed all outgroups from the BEAST MCMC trees. Biogeographical data of all Eremurus species included in the study were compiled from natural distribution information described in regional floras (Vvedensky, 1932, 1941, 1963; Fedtschenko, 1935; Kashenko, 1951; Goloskokov, 1958; Vvedensky and Kovalevskaya, 1971; Wendelbo, 1982; Xinqi and Turland, 2000) and the GBIF database corresponding to these floras. We coded three biogeographical areas based on the natural distribution of the species of Eremurus: A–southwest Asia (SWA), B–central Asia (CA) and C–east Asia (SJ). We combined the outgroup-removed chronograms with the biogeographical data. The maximum number of areas occupied by each node was set to 3. The MCMC chains were run for 40, 000 generations with sampling every 1000 generations.
3. Results 3.1. Phylogenetic analyses and character evolutionComplete plastomes were recovered for all 27 species of Eremurus. The genome size ranged from 153, 763 (Eremurus hilariae) to 155, 759 bp (Eremurus tianschanicus), and the GC content varied from 37.3 to 37.5%. The alignment included 160, 171 characters, of which 1359 characters (0.85%) were parsimony informative. The length of the coding regions ranged from 67, 503 bp to 79, 377 bp, with the minimum and maximum lengths being 73, 955 bp and 76, 576 bp in the non-coding regions. Information on the LSC, SSC and IR regions is given in Table S3. The CDS data matrix comprised 56, 034 characters, of which 246 characters (0.44%) were parsimony informative. The nrDNA data matrix comprised 649 characters, of which 311 characters (47.9%) were parsimony informative. As mentioned above, two plastome and one nrDNA data sets were used for phylogenetic inference. The 50% majority-rule consensus tree obtained from Bayesian analysis based on the CDS (Ⅰ) and nrDNA (Ⅱ) data sets is presented in Fig. 1 and the IRA data set analysis can be found in Fig. S1B.
The Maximum Likelihood, Maximum Parsimony and Bayesian analyses based on the CDS and the IRA data sets produced highly similar topologies and all nodes were recovered as strongly supported (Figs. 1Ⅰ and S1A, B). Eremurus was found to be monophyletic based on all data sets (Figs. 2 and S1A, B). Within Eremurus, all phylogenetic reconstructions revealed the same three major clades (A, B, C) with maximum support (except Maximum Parsimony analysis based on nrDNA data sets) (Fig. 1Ⅰ), and clade C contained species representing all three sections. In the tree produced from the CDS data set, clade A was resolved as the earliest diverged clade of Eremurus. It contained only E. chinensis (subgenus Henningia). Clade B contained E. anisopterus, E. aitchisonii, E. kaufmannii, E. korolkowii, E. luteus, E. suworowii, E. roseolus, E. stenophyllus subsp. stenophyllus, E. saprjagajevii, E. robustus, E. olgae and E. stenophyllus subsp. ambigens (subgenus Henningia). Clade C contained species of both subgenera Henningia (E. lachnostegius, E. alberti, E. cristatus, E. hilariae and E. lactiflorus) and Eremurus (E. inderiensis, E. soogdianus, E. regelii, E. turkestanicus, E. iae, E. fuscus, E. altaicus and E. hissaricus) (Fig. 1Ⅰ). In the tree produced from the nrDNA data set, BI and ML analyses indicated that clade A (subgenus Henningia), with E. chinensis and E. persicus, was sister to clade B (the same subgenus Henningia) and clade C (both subgenera Henningia and Eremurus) (Fig. 1Ⅱ). MP analysis showed that all clades (A, B and C) diverged from a single node (not shown). The topology of the nrDNA-based tree differed from the CDS and IRA-based trees in the position of E. saprjagajevii, which was a part of clade B in the latter, but was a member of clade C in the former (Fig. 1).
Ancestral character state reconstructions (Fig. 1) indicated that the most probable ancestral state of Eremurus flower shape was subrotate, with an evolutionary trend within the genus towards the campanulate type. The tubular type emerged in Eremurus afterwards and only in one species (E. inderiensis). For number of veins per tepal (Fig. 1), a single vein was identified as the ancestral state, while 3 or 5 veins evolved in the genus only recently. Similarly, stamens exceeding the perianth in length evolved in several clades only recently (Fig. 1).
3.2. Divergence time estimatesIn the plastome dating analysis (Fig. 2), the most recent common ancestor (MRCA) of Aloe, Aloidendron, and Eremurus diverged in the Eocene (37 Mya, 95% highest posterior density (HPD) 25–50 Mya, Node 0 in Fig. 2). Clades B and C diverged from Clade A approximately 14.5 Mya in the Miocene (95% HPD = 8–22 Mya, Node 1 in Fig. 2); the separation of Clades B and C occurred approximately 10 Mya (95% HPD = 5.5–14.5 Mya, Node 2 in Fig. 2), again in the Miocene. The crown ages of the clades B and C were estimated to be 6.4 Mya (95% HPD = 3.5–9.8 Mya, Node 3 in Fig. 2) and 5.4 Mya (95% HPD = 3.0–8.5 Mya, Node 4), respectively.
In the nrDNA dating analysis (Fig. 3), the MRCA of Aloe, Aloidendron, Bulbine, Bulbinella, Kniphofia and Eremurus diverged also in the Eocene (47.72 Mya, 95% HPD = 35.55–59.95 Mya, Node 0 in Fig. 3). Clade B/C diverged from Clade A approximately 33 Mya in the Oligocene (95% HPD = 21.25–45.49 Mya, Node 1 in Fig. 3); the separation of Clades B and C occurred approximately 30.24 Mya (95% HPD = 19.34–42 Mya, Node 2 in Fig. 3) again in the Oligocene. The crown age of clades B and C was estimated to be 13.12 Mya (95% HPD = 5.4–22.6 Mya, Node 3 in Fig. 3) and 19.1 Mya (95% HPD = 6.3–32.7 Mya, Node 4 in Fig. 3), respectively.
3.3. Biogeographical analysesS-DIVA and DEC analyses produced different results regarding the MRCA of Eremurus and its major clades (Fig. 4). S-DIVA analysis indicated that the current representatives of the genus descended from a widespread ancestral species that was distributed over large parts of what today we know as southwest Asia, central Asia and eastern Asia (area ABC, Node 1 in Fig. 4). DEC analysis indicated the probability of the above scenario to be 26.41%, while other scenarios are also possible. Eremurus could have emerged in areas AB (29.53), BC (19.3), B (10.71), A (7.8) or AC (6.25). S-DIVA located the MRCA of Clades A and B in central Asia (100, Node 2 in Fig. 4), whereas DEC analysis identified the area to be either in central Asia (74.42) or in central Asia and adjacent southwest Asia (25.58). S-DIVA located the MRCA of Clade B also in central Asia (63) or in central Asia and adjacent southwest Asia (37, Node 4 in Fig. 4), whereas DEC analysis gave the following probabilistic scenarios: 55.05%, 38.3% and 6.65% for AB, B and A, respectively. Both analyses identified the location of MRCA of Clade C to be in central Asia (Node 3 in Fig. 4).
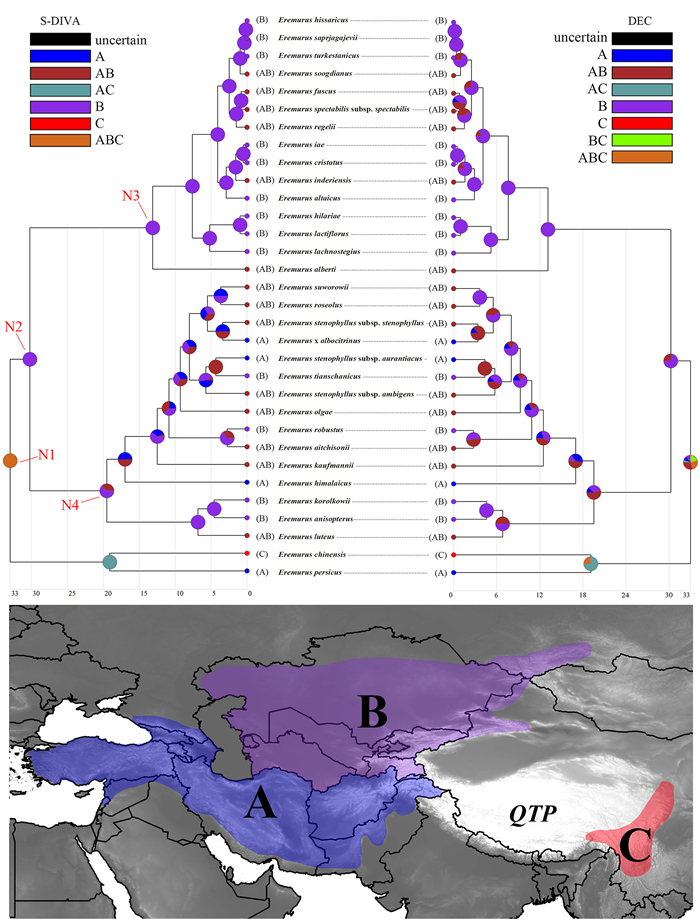
|
| Fig. 4 Ancestral geographical range estimation of Eremurus under the S-DIVA and DEC models in RASP v.4.2 and using the nrDNA data set-based phylogenetic tree from the BEAST analysis. Pie charts denote ancestral areas with probability values. Map shows the coding areas in different colors. |
Since the description of Eremurus in 1819, its circumscription has never been questioned. Our results based on plastome and nuclear datasets strongly support the monophyly of the genus, which has previously been supported by both morphological (Naderi et al., 2009) and molecular (nrITS and trnL-F) data (Safar et al., 2014). In our study, Eremurus was sister to Aloe and Aloidendron in the plastome analysis and to Aloe, Aloidendron, Bulbine, Bulbinella, Kniphofia, Asphodelus, Asphodeline, Xanthorrhoea and Hemerocallis in the nrDNA datasets. It must be noted, however, that our study did not include some genera close to Eremurus, such as Trachyandra, Kniphofia and Bulbinella, which were found to be closer to Eremurus than even Asphodelus and Asphodeline (Devey et al., 2006).
Traditional taxonomy (Wendelbo, 1982) recognized two subgenera and three sections of Eremurus. Subgenus Henningia differs from subgenus Eremurus in having (sub)rotate flowers with a single vein per tepal, mostly non-exserted filaments and long pedicels. Members of subgenus Eremurus, in turn, have tubular/campanulate incurved flowers with 3 or 5 veins and exserted filaments, long stamens and short pedicels. Within subgenus Eremurus, species with campanulate flowers comprise sect. Eremurus and those having tubular flowers belong to sect. Ammolirion. This taxonomy was not supported by either the plastome or nuclear data in our study. Both subgenera and the two sections were found to be paraphyletic, while sect. Ammolirion did not form a distinct clade but was embedded into a clade comprising species from the two other sections. Similar problems were detected earlier in one molecular (Safar et al., 2014) and two cladistic phylogenetic studies (Naderi et al., 2009; Makhmudjanov et al., 2022). These results cast doubt on the current subgeneric classification and validity of the subgenera and sections recognized. It is too early to propose a revised taxonomy of the genus based on molecular data because we do not have chloroplast sequence data for about 30% of the extant species of Eremurus. However, we can make some preliminary assertions. The length of the pedicels and stamens should not be used for infrageneric classifications beyond the species level. Instead, some previously overlooked morphological characters have taxonomic utility, e.g., seed shape and testa structure (Raza et al., 2022; Song et al., 2022; Yusupov et al., 2022).
The nuclear and plastome-based phylogenetic trees recovered three clades that are not reflected in current infra-generic classifications of Eremurus. The most basal line (clade A) is represented in the plastome-based tree by only the Chinese endemic, E. chinensis, which was separate from Clade B, which includes the currently recognized subg. Henningia species and in the ITS-based tree by E. chinensis and the southwest Asian endemic, E. persicus (also from subg. Henningia). These two species have racemes with lax flowers in which they resemble the closely related genera Trachyandra, Bulbinella, and Kniphofia. The second evolutionary line (clade B) also comprised species from subg. Henningia (13 taxa/species in the CDS data set and 18 taxa/15 species in the nrDNA data set) mainly distributed in southwestern and central Asia. The morphology of those species agrees well with the treatment by Wendelbo (1982) (i.e. (sub)rotate flowers with a single vein per tepal and mostly non-exserted filaments). The third evolutionary line (clade C) comprises mostly species from subg. Eremurus (9 out of 13 in the CDS data set; 10 out of 15 in the nrDNA data set) but includes also four and five species (plastome and nrDNA tree, respectively) from E. subg. Henningia.
The chloroplast and nuclear phylogenetic analyses produced conflicting results with respect to infrageneric relationships in clades B and C (Fig. 1). The difference between the two topologies is in the position of E. saprjagajevii (subg. Henningia). In the CDS data set tree, E. saprjagajevii was embedded within the strongly-supported Clade B and sister to E. stenophyllus subsp. stenophyllus with maximum possible support. In contrast, the nuclear topology of E. saprjagajevii located in the equally strongly supported clade C together with species from sect. Eremurus. Discordance between nuclear and cytoplasmic information is common in plants (Rieseberg and Soltis, 1991; Fehrer et al., 2007) due to the introgression of the cytoplasmic genome from one species into the nuclear background of another (or vice versa) through interspecific hybridization (Soltis and Kuzoff, 1995; Wendel and Doyle, 1998). As a result, such incongruent trees represent different histories of the cp- and nrDNA (Deng et al., 2015).
Our ancestral state reconstructions inferred that the most likely ancestral states of Eremurus were campanulate flower shape, one vein per tepal and stamens shorter than the perianth. Most of the members of clades A and B exhibit these character states. The third evolutionary lineage is more diverse (Clade C). Within this lineage, transitions from the conservative states to new ones occurred more often. Overall, the phylogenetic trees based on plastome and nuclear data suggest that evolution within Eremurus did not proceed as suggested by the current classification because transitions in the states of flower shape, number of veins per tepal and stamen length occurred more than once.
4.2. Evolutionary history and biogeographyTo understand the biogeographical history of Eremurus, we need to go back to as early as the Paleocene, or even Cretaceous. During those periods the climate was tropical or subtropical. The ancestor of all Asphodelaceae apparently was widely distributed in Africa and Eurasia. Since there are no extant species of Xanthorrhoeoideae in tropical regions, but numerous species in subtropical regions (e.g. Hemerocallis, Hemerocallidoideae), the ancestor was most probably a subtropical species. At the end of the Cretaceous and beginning of the Paleocene, the first lineages started to diverge from the common ancestor, representatives of which probably survive till today in eastern Asia (Hemerocallis) and which have cymes with only a few large flowers. One of the early diverging lineages somehow migrated to Australia in the Paleocene (apparently via long-distance seed dispersal), and gave rise to numerous species of Xanthorrhoea. Other lineages diverged later, apparently as a result of climate cooling. One of these events happened in the late Eocene when the African and Eurasian clades became separated. Unfortunately, we do not have cp genome data for such European genera as Asphodelus and Asphodeline, so we can only hypothesize that divergence of the Eremurus ancestor from the ancestor of those two genera happened at some time during the Oligocene–early Eocene. The separation of the European lineage occurred before separation of the African and Asian lineages as the latter two are closer to each other as suggested by evidence from this study as well as molecular (Devey et al., 2006) and karyological evidence. The chromosome number of 2n = 14 is shared by Eremurus (Cheng and Zhang, 1993; Ling et al., 2004), Aloe, Aloidendron, Trachyandra and Bulbinella (Zonneveld, 2002; Brandham, 1971; Thorsen et al., 2009; Naderi et al., 2009), while Asphodelus and Asphodeline have 2n = 28 (Lifante, 1996; Tuzlaci, 1986). The chromosome numbers of Asphodelus and Asphodeline suggest that their origin was associated with a polyploidization/hybridization event. So where did this happen? We will be better able to infer such origins when data on Asphodelus, Asphodeline, and Eremurus from the Crimea, Caucasus and the eastern Mediterranean become available.
In the context of interpreting the divergence time estimation and biogeography of Eremurus, for which we have two sets of results, we will use the nrDNA data set, as it provides enhanced taxon sampling compared to the CDS data set. Eremurus has been proposed to be an ancient Mediterranean element that originated in the southern part of present central Asia (Popov, 1941) at the end of the Paleogene or beginning of the Neogene (Khokhryakov, 1965). However, our results suggest that Eremurus originated earlier, in the Eocene (37 Mya according to the CDS data set and 47.7 Mya according to the ITS data set), while the distribution range of its ancestor was not limited to present-day central Asia, but included other regions too. Although we do not have molecular data for the species of Eremurus in the Caucasus, Crimea and eastern Mediterranean (with the exception of the wide-ranging E. spectabilis subsp. spectabilis), it is unlikely that areas west of Iran are the place of origin for Eremurus, because while large territories of the Irano-Turanian and Central Asian regions at that time were already terrestrial (Manafzadeh et al., 2014), the western part of Eurasia was covered by the Tethys Ocean (Zhang et al., 2012). In contrast, to the east the xerophytic proto-Mediterranean flora extended much farther than it does now, reaching even Japan (Popov, 1941). The first-diverging species in our phylogenetic trees, Clade A, E. chinensis, occurs in dry valleys of the Hengduan Mountains and apparently is a descendant or relict of the ancient Tethyan flora (Sun et al., 2017). Similar to some other genera (e.g., Kelloggia; Deng et al., 2017), the Tethyan origin of Eremurus was influenced by the uplift of the Qinghai-Tibet Plateau (QTP), which was a major cause of the aridification in Central Asia. The uplift blocked northward flow of moist, subtropical air and produced subsiding air over Central Asia, suppressing large-scale convective systems (Sato and Kimura, 2005; Molnar et al., 2010). The resulting climatic differences effectively separated Central Asia from other areas and caused disjunctions in the distribution of many phylogenetically related taxa.
As we have already acknowledged, our reconstruction of the early stages of the evolution of Eremurus is limited by the absence of some species from regions outside of central and southwest Asia that would be important for a more accurate reconstruction of the genus. Clade A likely diverged from the ancestral area (which we could not identify with certainty) in the early Miocene. One of its representatives, E. persicus is distributed in Iran, Afghanistan and Pakistan, while E. chinensis, already discussed above, is in the Hengduan Mountains of China. Apparently, the distribution range of their common ancestor included the territory of southwestern Asia and the ancient QTP during the Oligocene and early Miocene. The disjunct distribution of these two species can be explained by the formation and uplift of the QTP since the early Miocene (Harrison et al., 1992; Molnar et al., 1993; Copeland et al., 1995), which created a barrier for gene exchange. In general, the uplift of the QTP may have split Eremurus into two clades: one which evolved in central Asia and adjacent territories and the second, represented by the single species, that survived of the outskirts of the QTP in the Hengduan mountains under a milder climate.
In comparison with the reconstruction of the early stages of Eremurus evolution, the evolution of Eremurus in Central Asia can be inferred from our results with more certainty, although precise dating of the events is still problematic due to a difference between the plastome- and nrDNA-based estimates. Because nuclear genes generally have much faster substitution rates than plastid genes, nrDNA-based dating analyses often produce more ancient age estimates for nodes than do plastome-based analyses. According to the plastome-based dating, in central Asia the first split occurred in the mid Miocene; in contrast, according to the nrDNA-based dating, it happened much earlier, in the early Oligocene. From the early Oligocene to the late Miocene, serious topographic and climate changes occurred in central Asia. Aridification coincided with the retreat of the Paratethys (Ramstein et al., 1997), gradual global cooling (Lu et al., 2010) and the progressive uplift of the QTP (Molnar et al., 2010; Liu et al., 2015) and surrounding mountain belts (e.g., Altai, Pamir, Tian-Shan; Wang et al., 2019; Richter et al., 2022). Development of complex terrain including a great range of altitudes across many mountain chains, effectively isolated large areas from each other. Starting from the Miocene, speciation has proceeded rapidly in the transformed central Asia, which stands out for its geological heterogeneity and relatively stable continental climate. The primary evolutionary driver was apparently vicariance caused by numerous mountain chains preventing gene flow between populations, and localized specialization to a variety of climatic, topographic and soil conditions that exist in this region.
The majority of the members of the clade C (Eremurus lachnostegius, E. lactiflorus, E. hilareae, E. saprjagajevii, E. iae, E. hissaricus, E. cristatus, E. turkestanicus) are endemic to the Pamir-Alay, Tian Shan or both and are distributed only in mountainous central Asia. Therefore it is highly probable that the diversification of Clade C started in central Asia. In addition to the species diversification within this region, speciation occurred to the south (Iran, Afghanistan and Pakistan) (E. alberti, E. regelii, E. fuscus, E. soogdianus), to the west (Turkey) (E. spectabilis subsp. spectabilis) and to the north (Altai region) (E. altaicus). Among the species of Eremurus in our studies, the only species with a wide distribution covering most of central Asia, Afghanistan, Pakistan, Iran and the Xinjiang region of China is E. inderiensis from clade C. This is the only species that occupies sandy deserts, semi-deserts, and sandy hills and has specific adaptations to a desert climate and sandy soils (short vegetation period, narrow and long roots and few leaves).
In clade B, most of the species are endemic to (Eremurus robustus, E. tianschanicus and E. korolkowii), or have their major distribution in, central Asia (E. aitchisonii, E. kaufmannii, E. luteus, E. olgae, E. stenophyllus subsp. stenophyllus, E. stenophyllus subsp. ambigens, E. roseolus, E. suworowii), but the ranges of several species are outside of central Asia. E. himalaicus grows in northern Pakistan, and E.×albocitrinus and E. stenophyllus subsp. auranticus are found in Iran, Afghanistan and Pakistan. The most likely explanation for this pattern is an expansion of Eremurus in the Miocene from the southern part of central Asia (probably including the area north of Afghanistan) into adjacent areas after the uplift of the Iranian plateau and Himalayan orogeny (Clift et al., 2008; Mouthereau, 2011; McQuarrie and van Hinsbergen, 2013; Gupta et al., 2015; Ding et al., 2016; Ballato et al., 2017; Zhuang et al., 2017; Bialik et al., 2019).
Ecologically, the vast majority of the species of Eremurus grows in mountainous areas with a relatively humid climate, but with a few notable exceptions. Two species occur in steppes (Eremurus anisopterus and E. korolkowii) and one species occupies deserts and semi-deserts (E. inderiensis). Those species are specifically adapted to a xeric climate and sandy soils (a short growing period, long and narrow roots and few, thin leaves). The wider distribution of these species, compared to other montane Eremurus species, can be attributed to their specific habitat. Eremurus anisopterus occupies not only on the sandy steppes of central Asia (Kazakhstan, Uzbekistan, Turkmenistan) but also the steppes of western China (Xinjiang). Eremurus inderiensis grows in sandy deserts and semi-deserts, and on sandy hills throughout most of central Asia, in Afghanistan, Pakistan, Iran and the Xinjiang region of China. As for E. korolkowii, which is limited to bedrock outcrops of the Kizilkum desert, it is considered by a majority of taxonomist to be a subspecies or a form of E. anisopterus.
5. ConclusionsOur study shows that Central Asia played a key role in the origin and evolution of Eremurus. Most species of Eremurus originated in the Eocene in situ in this area, which served as the source area for migration and colonization of adjacent areas. Future studies will allow reconstruction of the routes and timing of migrations from central Asia toward the southwest (Iran, Afghanistan, Pakistan), and west (Turkey, the Caucasus, the Crimea and the eastern Mediterranean).
AcknowledgmentsThe authors are thankful to Dr. David Boufford from Harvard University Herbaria (USA) for editing the English. We would like to thank Davlatali Abdullaev (Institute of Botany, Academy of Sciences of Uzbekistan) and Isakul Turakulov (Khujand State University, Tajikistan) for their help in identifying the species and for collecting materials used in this study. We are also grateful to Davron Dekhkonov (Namangan State University, Uzbekistan) and Xianhan Huang (Kunming Institute of Botany, China) for their useful comments and suggestions. This study was supported by grants from the Key Projects of the Joint Fund of the National Natural Science Foundation of China (U23A20149), the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA20050203), International Partnership Program of the Chinese Academy of Sciences (151853KYSB20180009), the state research project 'Taxonomic revision of polymorphic plant families of the flora of Uzbekistan' (FZ-20200929321) and the State Programs for the years 2021–2025 'Grid mapping of the flora of Uzbekistan' and the 'Tree of life: monocots of Uzbekistan' of the Institute of Botany of the Academy of Sciences of the Republic of Uzbekistan.
Authors contributions
Dilmurod Makhmudjanov: Investigation, Analyses, Visualization, Writing, Original draft preparation. Sergei Volis: Writing, Original draft preparation, Editing. Ziyoviddin Yusupov, Inom Juramurodov: Analyses, Visualization. Tao Deng, Komiljon Tojibaev, Hang Sun: Conceptualization, Supervision, Reviewing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that might appear to influence the work presented in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.08.004.
APG IV, 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc., 181: 1-20. |
Baker, J.G., 1877. Revision of the genera and species of Anthericeae and Eriospermeae. J. Linn. Soc., 15: 253-363. |
Ballato, P., Cifelli, F., Heidarzadeh, G., et al., 2017. Tectono-sedimentary evolution of the northern Iranian Plateau: insights from middle–late Miocene foreland-basin deposits. Basin Res., 29: 417-446. DOI:10.1111/bre.12180 |
Bialik, O.M., Frank, M., Betzler, C., et al., 2019. Two-step closure of the Miocene Indian Ocean Gateway to the Mediterranean. Sci. Rep., 9: 8842. |
Bieberstein, F.A.M. v., 1819. Flora Taurico-Caucasica. Typis Academicis, Charkouiae (Kharkov), pp. 269-270.
|
Boissier, E., 1884. Eremurus. In: Boissier, E. (Ed.), Flora Orientalis. Georg, H., Geneve, pp. 321-328.
|
Brandham, P., 1971. The chromosomes of the Liliaceae: Ⅱ: Polyploidy and karyotype variation in the Aloineae. Kew Bull., 25: 381-399. DOI:10.2307/4103181 |
Chase, M.W., De Bruijn, A.Y., Cox, A.V., et al., 2000. Phylogenetics of Asphodelaceae (Asparagales): an analysis of plastid rbcL and trnL-F DNA sequences. Ann. Bot., 86: 935-951. |
Cheng, L., Zhang, Y.J., 1993. Chromosome number and karyotype of Eremurus chinensis Fedtsch. Wuhan. Bot. Res., 11: 281-282. |
Clift, P.D., Hodges, K.V., Heslop, D., et al., 2008. Correlation of Himalayan exhumation rates and Asian monsoon intensity. Nat. Geosci., 1: 875-880. DOI:10.1038/ngeo351 |
Copeland, P., Harrison, T.M., Pan, Y., et al., 1995. Thermal evolution of the Gangdese batholith, southern Tibet: a history of episodic unroofing. Tectonics, 14: 223-236. |
Darriba, D., Taboada, G.L., Doallo, R., et al., 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods, 9: 772. DOI:10.1038/nmeth.2109 |
Deng, T., Nie, Z.-L., Drew, B.T., et al., 2015. Does the Arcto-Tertiary biogeographic hypothesis explain the disjunct distribution of Northern Hemisphere herbaceous plants? The case of Meehania (Lamiaceae). PLoS One, 10: e0117171. DOI:10.1371/journal.pone.0117171 |
Deng, T., Zhang, J.-W., Meng, Y., et al., 2017. Role of the Qinghai-Tibetan Plateau uplift in the Northern Hemisphere disjunction: evidence from two herbaceous genera of Rubiaceae. Sci. Rep., 7: 13411. |
Devey, D.S., Leitch, I., Pires, J.C., et al., 2006. Systematics of Xanthorrhoeaceae sensu lato, with an emphasis on Bulbine. Aliso, 22: 345-351. DOI:10.5642/aliso.20062201.28 |
Dierckxsens, N., Mardulyn, P., Smits, G., 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res., 45: e18. |
Ding, H., Zhang, Z., Dong, X., et al., 2016. Early Eocene (c. 50 Ma) collision of the Indian and Asian continents: Constraints from the North Himalayan metamorphic rocks, southeastern Tibet. Earth Planet Sci. Lett., 435: 64-73. |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull., 19: 11-15. |
Drummond, A.J., Rambaut, A., 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol., 7: 1-8. |
Eker, I., Bieb, Eremurus M., 2020. In: Guner, A., Kandemir, A., Menemen, Y., et al. (Eds.), The Illustrated Flora of Turkey web version. ANG Foundation Nezahat Gökyiǧit Botanik Bahçesi Publications, Istanbul, pp. 1-9.
|
Fedtschenko, B., 1935. Eremurus M. Bieb. In: Komarov, V. (Ed.), Flora of the USSR. USSR Acad. Sci. Leningrad, pp. 37-52.
|
Fehrer, J., Gemeinholzer, B., , et al., 2007. Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Mol. Phylogenet. Evol., 42: 347-361. |
Goloskokov, V., 1958. Eremurus. In: Pavlov, N. (Ed.), Flora of Kazakhstan. Acad. Sci. KazSSR, Almaty, pp. 109-117.
|
Gupta, A.K., Yuvaraja, A., Prakasam, M., et al., 2015. Evolution of the South Asian monsoon wind system since the late Middle Miocene. Palaeogeogr. Palaeoclimatol. Palaeoecol., 438: 160-167. |
Harrison, T.M., Copeland, P., Kidd, W., et al., 1992. Raising Tibet. Science, 255: 1663-1670. DOI:10.1126/science.255.5052.1663 |
Hedge, I., Wendelbo, P., 1963. Notes on the giant Asphodels of Afghanistan. J. R. Hortic., 88: 402-406. |
Jansen, R.K., Cai, Z., Raubeson, L.A., et al., 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A., 104: 19369-19374. DOI:10.1073/pnas.0709121104 |
Jin, J.J., Yu, W.B., Yang, J.B., et al., 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol., 21: 1-31. DOI:10.1117/1.oe.59.1.016110 |
Kashenko, L., 1951. Eremurus. In: Vvedensky, A. (Ed.), Flora of Kirgizii. Kirgiztan USSR, Frunze, pp. 29-36.
|
Katoh, K., Standley, D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Kearse, M., Moir, R., Wilson, A., et al., 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Khokhryakov, А., 1965. Eremuruses and its Culture. Nauka. Moskow.
|
Kumar, S., Stecher, G., Tamura, K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol., 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Lee, S.Y., Xu, K.W., Huang, C.Y., et al., 2022. Molecular phylogenetic analyses based on the complete plastid genomes and nuclear sequences reveal Daphne (Thymelaeaceae) to be non-monophyletic as current circumscription. Plant Divers., 44: 279-289. |
Lifante, Z.D., 1996. A karyological study of Asphodelus L. (Asphodelaceae) from the Western Mediterranean. Bot. J. Linn. Soc., 121: 285-344. |
Ling, W., Lingji, W., Miao, M., et al., 2004. Karyotype analysis of Eremurus anisopterus. J. Shihezi Univ., 22: 417-418. DOI:10.1007/s11766-004-0008-y |
Liu, C., Chen, H.H., Tang, L.Z., et al., 2022. Plastid genome evolution of a monophyletic group in the subtribe Lauriineae (Laureae, Lauraceae). Plant Divers., 44: 377-388. |
Liu, X., Sun, H., Miao, Y., et al., 2015. Impacts of uplift of northern Tibetan Plateau and formation of Asian inland deserts on regional climate and environment. Quat. Sci. Rev., 116: 1-14. DOI:10.1155/2015/846308 |
Lu, H., Wang, X., Li, L., 2010. Aeolian sediment evidence that global cooling has driven late Cenozoic stepwise aridification in central Asia. Geol. Soc. Spec. Publ., 342: 29-44. |
Maddison, W.P., 2007. Mesquite: a modular system for evolutionary analysis. Version 2.0. http://mesquiteproject.org.
|
Makhmudjanov, D., Juramurodov, I., Kurbonalieva, M., et al., 2022. Genus Eremurus (Asphodelaceae) in the flora of Uzbekistan. Plant Divers. Cen. As., 2: 82-127. |
Makhmudjanov, D., Yusupov, Z., Abdullaev, D., et al., 2019. The complete chloroplast genome of Eremurus robustus (Asphodelaceae). Mitochondrial DNA B Resour., 4: 3366-3367. DOI:10.1080/23802359.2019.1674198 |
Malé, P.J.G., Bardon, L., Besnard, G., et al., 2014. Genome skimming by shotgun sequencing helps resolve the phylogeny of a pantropical tree family. Mol. Ecol. Resour., 14: 966-975. DOI:10.1111/1755-0998.12246 |
Manafzadeh, S., Salvo, G., Conti, E., 2014. A tale of migrations from east to west: the Irano-Turanian floristic region as a source of Mediterranean xerophytes. J. Biogeogr., 41: 366-379. DOI:10.1111/jbi.12185 |
McKain, M.R., McNeal, J.R., Kellar, P.R., et al., 2016. Timing of rapid diversification and convergent origins of active pollination within Agavoideae (Asparagaceae). Am. J. Bot., 103: 1717-1729. DOI:10.3732/ajb.1600198 |
McLay, T.G., Bayly, M., 2016. A new family placement for Australian blue squill, Chamaescilla: Xanthorrhoeaceae (Hemerocallidoideae), not Asparagaceae. Phytotaxa, 275: 97-111. DOI:10.11646/phytotaxa.275.2.2 |
McQuarrie, N., van Hinsbergen, D.J., 2013. Retrodeforming the Arabia-Eurasia collision zone: age of collision versus magnitude of continental subduction. Geology, 41: 315-318. |
Molnar, P., Boos, W.R., Battisti, D.S., et al., 2010. Orographic controls on climate and paleoclimate of Asia: thermal and mechanical roles for the Tibetan Plateau. Annu. Rev. Earth Planet Sci., 38: 77-102. DOI:10.1146/annurev-earth-040809-152456 |
Molnar, P., England, P., Martinod, J., 1993. Mantle dynamics, uplift of the Tibetan Plateau, and the Indian monsoon. Rev. Geophys., 31: 357-396. |
Moore, M.J., Soltis, P.S., Bell, C.D., et al., 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. U.S.A., 107: 4623-4628. DOI:10.1073/pnas.0907801107 |
Mouthereau, F., 2011. Timing of uplift in the Zagros belt/Iranian plateau and accommodation of late Cenozoic Arabia–Eurasia convergence. Geol. Mag., 148: 726-738. |
Naderi, S.K., Kazempour, O.S., Zareei, M., 2009. Phylogeny of the genus Eremurus (Asphodelaceae) based on morphological characters in the Flora Iranica area. Iran. J. Bot., 15: 7-35. |
Patel, R.K., Jain, M., 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One, 7: e30619. DOI:10.1371/journal.pone.0030619 |
Popov, M.G., 1927. Geographical and morphological methods in systematics and processes of hybridization in nature. Tr. Prik. Bot. Sel., 17: 221-229. |
Popov, M.G., 1941. Geographic and genetic elements of the flora of the Alma-Ata Reserve. In: Vegetation of Kazakhstan. Publishing House of the Academy of Sciences of the USSR, Moscow.
|
Popov, M.G., 1963. Basic Florogenetics. Academy of Sciences of the USSR, MoscowLeningrad.
|
Rambaut, A., 2018. FigTreeeTree Figure Drawing Tool. Version v. 1.4.4. http://tree.bio.ed.ac.uk/software/figtree/.
|
Ramstein, G., Fluteau, F., Besse, J., et al., 1997. Effect of orogeny, plate motion and land–sea distribution on Eurasian climate change over the past 30 million years. Nature, 386: 788-795. |
Raza, J., Ahmad, M., Zafar, M., et al., 2022. Systematic significance of seed morphology and foliar anatomy among Acanthaceous taxa. Biologia, 77: 3125-3142. DOI:10.1007/s11756-022-01137-0 |
Richter, F., Pearson, J., Vilkas, M., et al., 2022. Growth of the southern Tian Shan-Pamir and its impact on central Asian climate. GSA Bull., 135: 1859-1878. |
Rieseberg, L.H., Soltis, D., 1991. Phylogenetic consequences of cytoplasmic gene flow in plants. Evol. Trends Plants, 5: 65-84. |
Ronquist, F., Huelsenbeck, J.P., 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572-1574. |
Safar, K.N., Osaloo, S.K., Assadi, M., et al., 2014. Phylogenetic analysis of Eremurus, Asphodelus, and Asphodeline (Xanthorrhoeaceae-Asphodeloideae) inferred from plastid trnL-F and nrDNA ITS sequences. Biochem. Syst. Ecol., 56: 32-39. |
Sato, T., Kimura, F., 2005. Impact of diabatic heating over the Tibetan Plateau on subsidence over northeast Asian arid region. Geophys. Res. Lett., 32: L05809. |
Soltis, D.E., Kuzoff, R.K., 1995. Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae). Evolution, 49: 727-742. |
Song, Y.-X., Peng, S., Mutie, F.M., et al., 2022. Evolution and taxonomic significance of seed micromorphology in Impatiens (Balsaminaceae). Front. Plant Sci., 13: 215. |
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Su, N., Hodel, R.G., Wang, X., et al., 2023. Molecular phylogeny and inflorescence evolution of Prunus (Rosaceae) based on RAD-seq and genome skimming analyses. Plant Divers., 45: 397-408. |
Suchard, M.A., Rambaut, A., 2009. Many-core algorithms for statistical phylogenetics. Bioinformatics, 25: 1370-1376. DOI:10.1093/bioinformatics/btp244 |
Sun, H., Zhang, J., Deng, T., et al., 2017. Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Divers., 39: 161. DOI:10.1201/b14970-8 |
Swofford, D.L., 2002. PAUP*, Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4.10. Sinauer Associates. Sunderland.
|
Thorsen, M.J., Dickinson, K.J., Seddon, P.J., et al., 2009. Seed dispersal systems in the New Zealand flora. Perspect. Plant Ecol. Evol. Syst., 11: 285-309. |
Tuzlaci, E., 1986. Chromosome numbers of some Asphodeline species. MARMARA Pharm. J., 2: 113-117. |
Vaidya, G., Lohman, D.J., Meier, R., 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics, 27: 171-180. DOI:10.1111/j.1096-0031.2010.00329.x |
Vvedensky, A., 1932. Eremurus. In: Fedtschenko, B.A., Popov, M.G. (Eds.), Flora of Turkmenii. USSR Acad. Sci. and Bot. Ins. Turkm. USSR, Leningrad, pp. 250-257.
|
Vvedensky, A., 1941. Eremurus. In: Kudryashev, S. (Ed.), Flora of Uzbekistan. Uzb. Sec. USSR Acad. Sci. Tashkent, pp. 398-410.
|
Vvedensky, A., 1963. Eremurus. In: Ovchinnikov, P. (Ed.), Flora of Tadjikistan. USSR Acad. Sci. Moscow-Leningrad, pp. 186-212.
|
Vvedensky, A., Kovalevskaya, S., 1971. Eremurus. In: Kovalevskaya, S. (Ed.), Conspectus Florae Asiae Mediae. Fan UzSSR Tashkent, pp. 14-27.
|
Wang, X., Carrapa, B., Chapman, J.B., et al., 2019. Parathethys last gasp in central Asia and late Oligocene accelerated uplift of the Pamirs. Geophys. Res. Lett., 46: 11773-11781. DOI:10.1029/2019gl084838 |
Wendel, J.F., Doyle, J.J., 1998. Phylogenetic Incongruence: Window into Genome History and Molecular Evolution. Molecular Systematics of Plants Ⅱ: DNA Sequencing. Springer, Boston, pp. 265-296.
|
Wendelbo, P., 1958. Eremurus. In: Køie, M., Rechinger, K.H. (Eds.), Symbolae Afghanicae, pp. 150-191.
|
Wendelbo, P., 1982. Asphodeloideae: Asphodelus, Asphodeline & Eremerus. In: Rechinger, K. (Ed.), Flora Iranica, pp. 3-31.
|
Wendelbo, P., Furse, P., 1969. Eremurus of South West Asia. Lily Year Book, pp. 56-69.
|
Xinqi, C., Turland, N., 2000. Eremurus. In: Wu, Z., Raven, P. (Eds.), Flora of China. Science Press and Missouri Botanical Garden Press, Beijing and St. Louis (MO), pp. 159-160.
|
Yu, Y., Blair, C., He, X., 2020. RASP 4: ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol., 37: 604-606. DOI:10.1093/molbev/msz257 |
Yusupov, Z., Deng, T., Volis, S., et al., 2021. Phylogenomics of Allium section Cepa (Amaryllidaceae) provides new insights on domestication of onion. Plant Divers., 43: 102-110. |
Yusupov, Z., Ergashov, I., Volis, S., et al., 2022. Seed macro- and micromorphology in Allium (Amaryllidaceae) and its phylogenetic significance. Ann. Bot., 129: 869-911. DOI:10.1093/aob/mcac067 |
Zhang, Z., Flatøy, F., Wang, H., et al., 2012. Early Eocene Asian climate dominated by desert and steppe with limited monsoons. J. Asian Earth Sci., 44: 24-35. |
Zhuang, G., Pagani, M., Zhang, Y.G., 2017. Monsoonal upwelling in the western Arabian Sea since the middle Miocene. Geology, 45: 655-658. |
Zonneveld, B.J., 2002. Genome size analysis of selected species of Aloe (Aloaceae) reveals the most primitive species and results in some new combinations. Bradleya, 2002: 5-12. DOI:10.25223/brad.n20.2002.a3 |



