b. Key Laboratory of Biodiversity Conservation and Sustainable Utilization in Mongolian Plateau for College and University of Inner Mongolia Autonomous Region, Hohhot 010022, China;
c. College of Landscape and Art, Henan Agricultural University, Zhengzhou 450002, China;
d. State Key Laboratory of Tree Genetics and Breeding, Chinese Academy of Forestry, Beijing 100091, China
Chinese wingnut (Pterocarya stenoptera C. DC.) is of great importance in traditional Chinese medicine. Its leaves and bark of these trees have been found to possess antipyretic and insecticidal effects, as well as the ability to dispel pathogenic wind and dampness. Its leaves have also shown significant therapeutic effects in treating skin allergies, toothaches, and bacterial infections (Nanjing University of Traditional Chinese Medicine, 1997). Recent study revealed that Chinese wingnut contain a high concentration of volatile oils, such as terpenes, steroids, flavonoids, and other chemical compounds, which contribute to their antimicrobial, antioxidant, and anti-tumor activities (Li et al., 2021a). As a result, the extracts from its leaves are frequently used as natural additives in food and cosmetics industries for their antibacterial and antioxidant properties (Yin et al., 2020).
Chinese wingnut is a deciduous tree belonging to Juglandaceae. Juglandaceae consists of a total of 8 genera and 63 species, with discontinuous distribution in Eurasia and the Americas. Regions from southern China to the Indochinese Peninsula, as well as southern United States to Central America, are known to harbor the highest species diversity within the Juglandaceae (Ding et al., 2023). The fruits of Juglandaceae can be categorized into two groups based on the presence or absence of wings in their fruits. The winged group includes genera such as Pterocarya, Cyclocarya, Platycarya, and Oreomunnea, while the wingless group comprises of Juglans, Annamocarya, Carya, and Engelhardia.
A total of eight species are in the genus of Pterocarya. P. fraxinifolia (Poir.) Spach is found in the Caucasus region, P. rhoifolia Siebold & Zucc is distributed in Shandong Peninsula and Japanese archipelago, and P. tonkinensis (Franch.) Dode can be found in northern Vietnam and southeastern Yunnan Province, China. The remaining five species are endemic to China and include P. stenoptera, P. delavayi Franch., P. insignis Rehd. et Wils, P. macroptera Batalin, and P. hupehensis Skan (Kuang and Li, 1979). As an important dominant tree in deciduous broad-leaved forests, Chinese wingnut is widely distributed in southern and southwestern of China (Xu et al., 2002). The winged fruits of Chinese wingnut have evolved to aid in their dispersal by wind and water currents. Consequently, this species is commonly found along the banks of streams and rivers in its natural habitat (Li et al., 2018).
The trunk of Chinese wingnut is tall and straight, with a thick and sturdy body. Its crown is full and spread out, and the fruits are winged and arranged in clusters, hanging on the tree for up to six months. Thus, it has a high ornamental value (Zhang et al., 2023). Therefore, it is commonly used as a greening tree species in courtyards, parks, and urban streets. As a fast-growing afforestation tree species, Chinese wingnut has a wide and deep root system with abundant lateral roots. It can be used as an excellent tree species for waterbank protection, water conservation, and soil preservation. As an extremely water-resistant tree species for waterbank protection, it also has a strong ability to remove total nitrogen (TN), total phosphorus, and chemical oxygen demand from water bodies (Gao, 2009).
Due to its high medicinal value, ornamental value, and ecological value, studies on Chinese wingnut have been increasing in recent years. Molecular mechanisms of stress response (Ye et al., 2020; Li et al., 2021b, Li et al., 2021c; Zhang et al., 2023), genetic diversity and structure (Qian et al., 2019), adaptive evolution (Li et al., 2018, 2022), and medicinal effective components (Yin et al., 2020; Bo and Yu, 2021) have been gradually reported. However, the lack of a high-quality genome has hindered research on this species. Therefore, assembling a high-quality genome for Chinese wingnut is essential.
The assembly of the high-quality genome of Chinese wingnut was successfully achieved in this study. By conducting comparative genomic analysis, we elucidated the evolutionary history of Chinese wingnut. Furthermore, through integrated analysis of transcriptome and metabolome, the candidate genes involved in the synthesis pathway of terpenoid volatiles were identified. Our study provides important genomic resources for unraveling the evolutionary history and synthesis pathways of medicinal components in Chinese wingnut.
2. Materials and methods 2.1. Plant materialsFruits, stems, leaves, and roots of Chinese wingnut were collected in May from Henan Agricultural University in Zhengzhou, China. All the samples utilized in this study were in compliance with the regulations of the Chinese government. To maintain their freshness, the aforementioned samples were rapidly treated with liquid nitrogen before being stored in an ultra-low temperature refrigerator set at −80 ℃.
2.2. Genome sequencingThe extraction of total genomic DNA from Chinese wingnut leaves was conducted using the Cetyl Trimethyl Ammonium Bromide method (Doyle and Doyle, 1987). To determine the concentration of DNA, NanoDrop One (Thermo Fisher, Wilmington, USA) was utilized. Genome size of Chinese wingnut was determined using the K-mer approach (Liu et al., 2013), relying on the short sequencing data generated from the Illumina platform (Illumina, CA, USA). DNA library for genome sequencing was generated in accordance with the PacBio SMRT construction protocol. The concentration of the DNA library was determined using the Qubit 3.0 fluorometer from Thermo Fisher (Wilmington, USA).
2.3. Genome assemblyHigh-throughput chromatin conformation capture (Hi-C) method, along with HiFi sequencing approach, were utilized to generate chromosome-level genomes for Chinese wingnut. A single SMRT cell from the PacBio Sequel Ⅱ platform generated a substantial amount of 19.44 Gb Hi-Fi subreads for the Chinese wingnut. For the assembly of highly accurate HiFi data, Hifiasm software (Cheng et al., 2021) was employed. We utilized four approaches to evaluate the genome assembly results of Chinese wingnut. The first assessment method is known as the Core Eukaryotic Genes Mapping Approach (CEGMA; Parra et al., 2007). The second approach is Benchmarking Universal Single-Copy Orthologs (BUSCO; Simao et al., 2015) assessment. The third and fourth approaches were the comparing results of the second and third generations sequencing reads to assemble the genome sequence. The comparison is conducted with BWA (Li and Durbin, 2009) and Minimap2 (Li, 2021), respectively. Library construction and sequencing procedures for Hi-C were executed in accordance with the standardized protocol described in Li et al. (2023). Utilizing LACHESIS (Burton et al., 2013), the genome sequences are systematically partitioned, arranged, and oriented into clusters, followed by manual verification and inspection, ultimately resulting in a chromosome-level genome assembly.
2.4. Genome annotationFirstly, RepeatModeler2 (Flynn et al., 2020) was employed for de novo prediction, which primarily utilized two de novo prediction software, RECON (Bao and Eddy, 2002) and RepeatScout (Price et al., 2005). The prediction results were then classified using RepeatClassifier with the assistance of Dfam (v.3.5) database. Secondly, LTR_retriever (Ou and Jiang, 2018) was specifically utilized for de novo prediction of LTRs, primarily relying on the prediction results from LTRharvest (Ellinghaus et al., 2008) and LTR_FINDER (Xu and Wang, 2007). Next, the aforementioned de novo prediction results were merged with the known database and removed redundancies to obtain a species-specific repeat sequence database. Finally, RepeatMasker (Tarailo-Graovac and Chen, 2009) was used to predict transposable element (TE) sequences.
The prediction of genes was accomplished through three methods: homology-based prediction, ab initio prediction, and transcriptome-based prediction. When conducting homology-based predictions using GeMoMa (Keilwagen et al., 2016), we selected four species, namely Arabidopsis thaliana, Carya illinoinensis, Juglans mandshurica, and J. regia. Augustus (Stanke et al., 2008) and SNAP (Korf, 2004) were utilized for de novo prediction. Based on transcriptome data, we performed gene prediction using GeneMarkS-T (Tang et al., 2015a) and PASA (Haas et al., 2003). Ultimately, the prediction results were obtained by integrating the aforementioned three methods using EVM (Haas et al., 2008). Then, annotation analysis was performed using various databases such as Non-Redundant (nr; Deng et al., 2006), eggNOG (Huerta-Cepas et al., 2019), Gene Ontology (GO; Ashburner et al., 2000), Kyoto Encyclopedia of Genes and Genomes (KEGG) (KEGG; Kanehisa et al., 2016), TrEMBL (Boeckmann et al., 2003), EuKaryotic Orthologous Groups (KOG, Tatusov et al., 2003), SWISS-PROT (Boeckmann et al., 2003), and Pfam (Finn et al., 2006).
In the process of pseudogene prediction, GenBlastA (She et al., 2009) was utilized for comparative analysis to identify homologous gene sequences in the genome after filtering out the true gene loci. Then, GeneWise (Birney et al., 2004) was employed to detect immature stop codons and frameshift mutations within the identified gene sequences, ultimately leading to the prediction of pseudogenes.
To identify tRNA, tRNAscan-SE (Lowe and Eddy, 1997) was utilized. For predicting rRNA, barrnap (Loman, 2017) was performed. miRNA, snoRNA, and snRNA were predicted based on the Rfam (Griffiths-Jones et al., 2005) database by utilizing Infenal (Nawrocki and Eddy, 2013).
2.5. Evolutionary analysisOrthoFinder v.2.4 (Emms and Kelly, 2019) was employed to identify the single-copy genes in Arabidopsis thaliana, Forsythia suspensa, Glycine max, Oryza sativa, Sesamum indicum, Vitis vinifera, Cyclocarya paliurus, Juglans mandshurica, J. regia, Carya illinoinensis, and Chinese wingnut. Sequences of each single-copy gene were aligned using MAFFT (Katoh et al., 2009). Then, Gblocks (Talavera and Castresana, 2007) was applied to remove poorly aligned or highly divergent regions from the alignments. Afterwards, the aligned sequences of each gene from all species were concatenated to generate a super gene. The ModelFinder tool (Kalyaanamoorthy et al., 2017) integrated in IQ-TREE was utilized to determine the best-fit model, which was found to be JTT + F + I + G4. Subsequently, the maximum likelihood (ML) method was employed to construct the phylogenetic tree with a bootstrap value set at 1000. Finally, we employed MCMCTREE in PAML (Yang, 1997) to calculate divergence time. The time correction points for P. stenoptera - O. sativa (142.1–163.5 million years ago; Mya), G. max - V. vinifera (109.8–124.4 Mya), G. max - O. sativa (142.1–163.5 Mya), P. stenoptera - C. paliurus (0.0–21.8 Mya) were from TimeTree website (http://www.timetree.org; Kumar et al., 2017).
With the assistance of Orthofinder v.2.4 (Emms and Kelly, 2019), the protein sequences of species were classified into gene families using the diamond alignment method. The annotated gene families were obtained using the PANTHER database (Mi et al., 2019). Lastly, the unique gene families of this species were subjected to GO and KEGG enrichment analysis by using clusterProfile (Yu et al., 2012).
Utilizing the CAFE (Han et al., 2013), the results obtained after analyzing the evolutionary tree and gene family clustering with divergence times can be used to estimate the number of gene family members for each ancestor branch. This information can then be used to predict the expansion or contraction of gene families relative to their ancestors in a given species. The criteria for determining significant expansion or contraction are defined as having family-wide and viterbi P values both less than 0.05.
Positive selection analysis was performed using the CodeML module in PAML (Yang, 1997). Specifically, single-copy gene families were obtained among Carya illinoinensis, Cyclocarya paliurus, Juglans mandshurica, J. regia and Chinese wingnut. The protein sequences were aligned using MAFFT (Katoh et al., 2009). Then, the aligned protein sequences were converted into codon alignment using PAL2NAL (Suyama et al., 2006). Finally, the CodeML program was applied with the F3x4 model of codon frequencies and the Branch-site model in PAML (Yang, 1997). The likelihood ratio test was performed by comparing Model A (assuming positive selection on the foreground branch, i.e., ω > 1) to the null model (which does not allow any site to have ω > 1). Significantly different results (P < 0.01) were obtained, and the Bayes empirical Bayes method was used to calculate the posterior probabilities of sites being under positive selection (> 0.95 was considered significant).
By employing diamond (Buchfink et al., 2015), the gene sequences were aligned to identify similar gene pairs. Subsequently, with the assistance of MCScanX (Wang et al., 2012), the adjacency of the identified similar gene pairs on the chromosomes was determined based on the gff3 file. This analysis program ultimately provided all the genes within collinear blocks. The figure illustrates the linear pattern of collinearity among the different species was generated using JCVI (Tang et al., 2015b).
Ks distribution was employed to identify potential whole-genome duplication (WGD) events in Carya illinoinensis, Cyclocarya paliurus, Juglans mandshurica, J. regia, and Chinese wingnut. Ks value of gene pairs within these blocks was calculated using WGD (Zwaenepoel and Van de Peer, 2019). The density distribution of Ks was visualized by employing ggplot (Wickham, 2009). The WGD event date was obtained based on the Ks values calculation with a mutation rate of 7.5 × 10−9 per nucleotide per year (Sollars et al., 2017).
Pairwise Sequentially Markovian Coalescent (PSMC) method (Li and Durbin, 2011) was employed to examine changes in population demographics within Chinese wingnut. The simplified genomic sequencing data from nine genetic groups (GZFJ1: SRR10014614; HNHM1: SRX6752859; HNTM1: SRX6752881; JSLM1: SRX6752801; JXSQ1: SRX6752621; SCWD1: SRX6752904; SDTM1: SRX6752754; ZJTM1: SRX6752582; YNYB2: SRX6752710) of Chinese wingnut (Li et al., 2022) was adopted. The analysis parameters were defined as follows: an initial h = q ratio of 5, conducting 100 iterations, resampling chunks consisting of 500,000 base pairs each time, and performing 100 bootstrap replicates. Additionally, a generation time of 8 years (Pan, 2021) and a mutation rate of 7.5 × 10−9 per nucleotide per year (Sollars et al., 2017) were considered.
2.6. Transcriptome sequencing analysisTranscriptome sequencing was performed on fruits, stems, leaves, and roots of Chinese wingnut. RNA extraction was carried out using the plant RNA Isolation kit DP432 (Tiangen, Beijing, China). Purity and quality of RNA were assessed using NanoDrop One (Thermo Fisher, DE, USA) and Agilent 2100 (Agilent, CA, USA). RNA samples with an OD260/OD280 ratio higher than 2.0 were retained for sequencing experiments. To construct the RNA sequencing libraries, the RNA library prep kit (New England BioLabs, MA, USA) was employed, utilizing 10 μg of RNA from each sample. Quality of the libraries was further evaluated using Agilent 2100 (Agilent, CA, USA), and the qualified libraries were sequenced using the Illumina HiSeq X-ten (Illumina, CA, USA).
The initial step in data processing involved the removal of adapter and low-quality sequences from the raw data. Afterward, the remaining high-quality reads were aligned to the genome of Chinese wingnut using HISAT2 (Kim et al., 2015). The assembled transcripts were then generated using StringTie (Pertea et al., 2015). The assembled reads were annotated based on the existing Chinese wingnut genome. Furthermore, for the unannotated genes discovered within the Chinese wingnut genome, additional annotation was performed utilizing various databases as mentioned in the section of Genome Annotation.
Expression levels of all genes of Chinese wingnut were evaluated using StringTie (Pertea et al., 2015). The calculations involved measuring fragments per kilobase of transcript per million fragments mapped (FPKM). To elucidate the overall trends of variation among different tissues, we conducted Principal Component Analysis (PCA) using the plotPCA function in DESeq2 (Love et al., 2014). During the identification of differentially expressed genes (DEGs), a filtering criterion was employed where Fold Change ≥ 2 and FDR < 0.01.
2.7. Detection of the metabolites in different tissues of Chinese wingnutTo analyze the samples of Chinese wingnut, a specific internal standard known as 2-octanol (10 mg/L) dissolved in H2O was employed. Measure out approximately 500 mg of each sample and transfer them into 20 mL headspace bottle. In the prep and load solution rail system's solid-phase microextraction cycle, the temperature during incubation was maintained at 60 ℃. The preheating period lasted for 15 min, followed by a 30-min incubation time. Finally, the desorption process occurred over a 4-min duration. GC-MS analysis was conducted utilizing Agilent 7890 in combination with a 5977B mass spectrometer. An DB-Wax (30 m × 0.25 mm × 0.25 μm, Agilent Technologies, USA) was used to separate and detect the volatile compounds. The sample was introduced in Splitless Mode. Helium served as the carrier gas, with a front inlet purge flow rate of 3 mL min−1, and the gas flow rate through the column was maintained at 1 mL min−1. Initially, the temperature was set at 40 ℃ for a duration of 4 min, followed by an increase to 245 ℃ at a rate of 5 ℃ min−1, which was then held steady for 5 min. The injection, transfer line, ion source, and quad temperatures were maintained at 250, 250, 230, and 150 ℃, respectively. In electron impact mode, the energy used was −70 eV. Mass spectrometry data were obtained using scan mode, with a m/z range of 20–400 and a solvent delay of 2.13 min. The raw peaks extraction, data baseline filtering, baseline calibration, peak alignment, deconvolution analysis, peak identification, integration, and spectrum matching of the peak area were performed using the Chroma TOF 4.3X software provided by LECO Corporation and the NIST database (Kind et al., 2009).
2.8. Correlation analyses between metabolites and gene expressionWeighted co-expression network analysis (WGCNA) was employed to identify co-expression gene modules and discover potential regulators associated with biosynthesis by examining correlations between gene expression values. To construct a weighted gene co-expression network, R package WGCNA (Langfelder and Horvath, 2008) was utilized. According to Tommasini and Fogel (2023), a minimum sample size of 12 can be used for WGCNA analysis. In this study, we utilized transcriptomic and metabolomic data from 12 samples each to perform WGCNA analysis. The construction of the gene co-expression network was achieved through the automatic network construction function known as "blockwiseModules". To visualize the co-expression network, Cytoscape (Shannon et al., 2003) was utilized.
R package was utilized to calculate the Pearson correlation coefficients (r) between the expression levels of genes and the concentrations of volatile compounds. To identify candidate genes associated with the volatile compounds synthesis, the genes were considered if their r exceeded 0.7, with a P value less than 0.01.
3. Results 3.1. Genome information of Chinese wingnutIn this study, Chinese wingnut genome was successfully assembled. Through k-mer analyses, it was determined that Chinese wingnut genome had a heterozygosity rate of 1.37%, a repetitive sequence content of approximately 36.42%, and an estimated genome size of 521.12 Mb. Subsequently, Chinese wingnut genome was assembled, resulting in a contig N50 value of 25.23 Mb and genome sizes of 550.86 M. Hi-C sequencing was then employed to anchor the sequences to 16 pseudochromosomes (Fig. 1). The anchoring success rate for the sequences of Chinese wingnut to the pseudochromosomes reached 99.99%. The pseudochromosomes consisted of 1–9 assembled contigs, with lengths ranging from 22, 735, 625 bp to 50, 287, 276 bp.
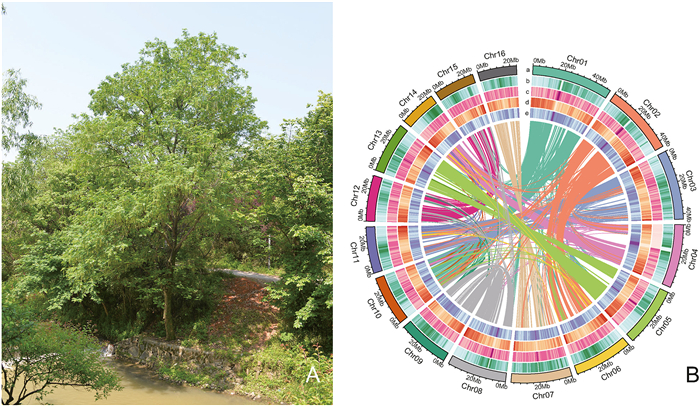
|
| Fig. 1 The assembly genome of Chinese wingnut. (A) Chinese wingnut is scattered along the banks of the stream. (B) The collinearity within Chinese wingnut genome by paralogous pair genes. The chr1-16 with different colors represent the different linkage groups. Each line represents a duplicated gene pair. The circles from the outside to the inside indicate the TE density, SSR density, gene density and GC content with the window of 4 Mb. |
Evaluation using BUSCO and CEGMA showed integrality scores of 98.4% and 98.5% respectively. The results of the second generation reads exhibited a return ratio of 98.76%, a coverage of 99.99%, and an average sequencing depth of 119×. The results of the third generation reads displayed a return ratio of 99.78%, a coverage of 99.99%, and an average sequencing depth of 33×.
In the genome of Chinese wingnut, a significant proportion of TE sequences, accounting for 214.2 Mb, were discovered. Among these TEs, retroelements were identified as the most abundant (Table S1). Tandem repeat sequences, totaling 23.9 Mb, were also detected in the genome (Table S2), constituting approximately 4.35% of its genome. A total of 39,648 protein-coding genes (36,824 of them were annotated; Table S3) and 232 pseudogenes were identified in Chinese wingnut genome. Additionally, various types of non-coding RNAs were identified, including 99 microRNAs, 526 transfer RNAs, 1794 ribosomal RNAs, 101 small nucleolar RNAs, and 97 small nuclear RNAs.
3.2. Evolutionary history of the Chinese wingnutA total of 635 single-copy protein-coding genes were identified across all 11 species, and these genes were concatenated and used to create a super matrix. This upper matrix was utilized to construct the phylogenetic tree (Fig. 2A) wherein it was revealed that Chinese wingnut and Cyclocarya paliurus diverged approximately 11.69 Mya, whilst the differentiation between the winged group (the Chinese wingnut and C. paliurus) and the wingless group (Carya illinoinensis, J. mandshurica, J. regia) in Juglandaceae was estimated to have occurred approximately 13.66 Mya. The divergence of Carya from other genera in Juglandaceae occurred approximately 19.88 Mya.
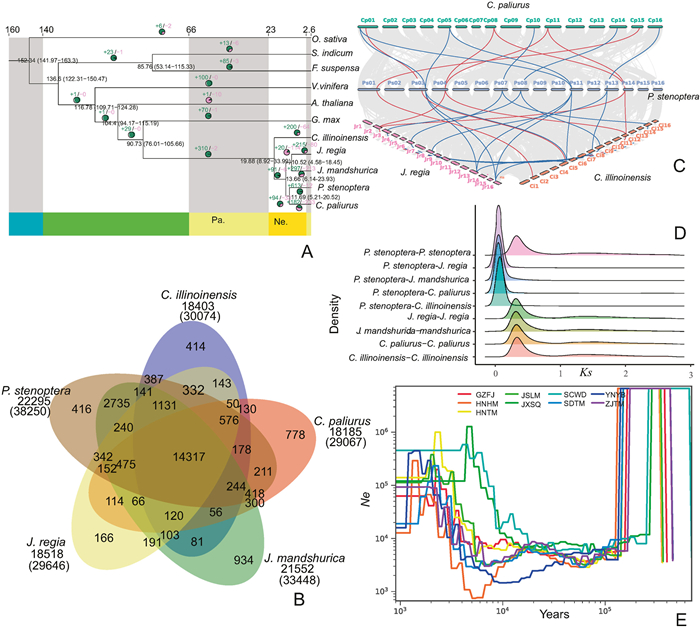
|
| Fig. 2 Phylogenetic relationship, gene family comparison, colinearity, WGD of Chinese wingnut and its related species and the effective population size change of all genetic groups of Chinese wingnut. (A) Phylogenetic tree of the Chinese wingnut, one monocot species, and nine dicot species. Green number indicates the expansion number of gene family, purple number indicates the contraction number of gene family, black number indicates the mean divergence time (out of brackets) and the 95% confidence interval divergence time (in brackets). (B) Venn diagram of all shared and unique gene families among Chinese wingnut, Cyclocarya paliurus, Carya illinoinensis, Juglans mandshurica, and J. regia. (C) Linear collinearity diagram of chromosomes among the Chinese wingnut, C. paliurus, J. regia, and C. illinoinensis. Lines in blue, green, purple, and brick red represent chromosomes of Chinese wingnut, C. paliurus, J. regia, and C. illinoinensis. Gray line shows each syntenic gene pairs, red line shows syntenic gene pairs of terpene synthase 2 gene family, and blue line shows syntenic gene pairs of terpene synthase 10 gene family. (D) Ks distribution density plots. The x- and y-axes indicate the Ks values and the density of paralog or ortholog genes. (E) Historical change of effective population size of all genetic groups of Chinese wingnut. The x-axis represents the scale of the time. The y-axis represents the effective population size. |
To explore the unique and shared gene families of Chinese wingnut and other Juglandaceae species, we conducted a comparative analysis of their gene families (Fig. 2B). Five species of the Juglandaceae family share a total of 14,317 gene families, Juglans mandshurica possesses the highest number of unique gene families with 934, while J. regia has the fewest with 166. Chinese wingnut have 416 unique gene families. The enrichment analysis of unique genes evolved in Chinese wingnut revealed that these genes were significantly enriched in 5 pathways and 14 ontologies (Table S4). Among them, a significant evolution of unique genes related to the synthesis of arginine (ko00220) was found.
To predict which traits and metabolic functions have been enhanced during the evolution of winged group and Chinese wingnut after their divergence from their ancestors, we conducted an analysis of gene family contraction and expansion. In our evolutionary tree branches (Fig. 2A), we have observed the most distinct differentiation between Chinese wingnut and Cyclocarya paliurus. Following their divergence, Chinese wingnut showed a significant expansion of 613 gene families, whereas C. paliurus experienced a contraction in 233 gene families. The expanded gene family in Chinese wingnut was notably enriched in 16 pathways and 77 ontologies (Table S5). Based on the enrichment analysis of pathways, there was a significant expansion of gene families associated with terpene biosynthesis (ko00904, ko00909, and ko00902) and other benzene compound metabolism (ko00402, ko00240, ko00360, ko00380). Additionally, there has been an expansion of gene families associated with the stress response, such as gene family related to ascorbate and aldarate metabolism (ko00053), galactose metabolism (ko00052), and linoleic acid metabolism (ko00591). Due to the presence of various terpene volatile compounds in Chinese wingnut leaves, we conducted a comprehensive investigation on the specific terpene synthase gene families of Chinese wingnut. Surprisingly, we discovered that two terpene synthase gene families significantly expanded in comparison to other species in the Juglandaceae family. Terpene synthase 2 family consists of 16 members in Carya illinoinensis, 12 members in C. paliurus, 8 members in Juglans mandshurica, 11 members in J. regia, and an increased count of 18 members in Chinese wingnut. Terpene synthase 10 family consists of 15 members in C. illinoinensis, 15 members in C. paliurus, 6 members in J. mandshurica, 21 members in J. regia, and a significant increase to 30 members in Chinese wingnut. The co-linearity relationship of two gene families among four species, namely C. illinoinensis, C. paliurus, J. regia, and Chinese wingnut, was demonstrated (Fig. 2C).
In the winged group, we have discovered that a total of 94 gene families have undergone expansion since its divergence from the wingless group within Juglandaceae. These gene families have been enriched in 7 specific pathways and 55 ontologies in Chinese wingnut and 7 specific pathways and 57 ontologies in C. paliurus (Table S6). We did not find any expansion of gene families related to fruit development in the winged group, but we did find a significant expansion of gene families related to root development, including lateral root formation (GO:0010311), lateral root morphogenesis (GO:0010102), lateral root development (GO:0048527), post-embryonic root morphogenesis (GO:0010101), post-embryonic root development, root morphogenesis (GO:0010015), root system development (GO:0022622), root development (GO:0048364), and adventitious root development (GO:0048830).
When using Chinese wingnut as the foreground branch and the other species of Juglandaceae as background branch, we detected a total of 239 positively selected genes. These genes have been enriched in 38 ontologies (Table S7), with 10 ontologies (GO:0070469, GO:0044429, GO:0005747, GO:0045271, GO:0098803, GO:0005746, GO:0005759, GO:0098798, GO:0098800, and GO:0044455) specifically related to mitochondrial aerobic respiration. When the winged group is regarded as the foreground branch and the wingless group is as the background branch, 65 gene families have been subject to positive selection, including 66 genes in both Chinese wingnut and C. paliurus. These positively selected genes have been found to be enriched in 1 ontology of Chinese wingnut and 2 ontologies of Cyclocarya paliurus (Table S8). The CCR4-NOT complex (GO:0030014) is an ontology that is shared by two species and is closely associated with the regulation of gene expression.
The distribution diagram of Ks values (Fig. 2d) demonstrated that the five Juglandaceae species we investigated experienced one round of WGD event before their divergence. The peak of Ks at 0.31 suggested the WGD event occurred at approximately 20.7 Mya. The results of PSMC analysis indicated that the effective population size of nine genetic groups of Chinese wingnut experienced a significant decrease around 0.14 Mya and a rapid recovery around 0.01 Mya (Fig. 2e). The reduction and recovery time of the effective population of Chinese wingnut closely aligns with the coming and ending of the last glacial (LG) period.
3.3. Transcription expression in different tissues of Chinese wingnutTranscriptome sequencing was conducted on twelve samples consisting of four tissues - leaves, stems, roots, and fruits - from Chinese wingnut (Fig. 3A). A total of 251.42 million reads were obtained, resulting in 75.23 Gb of clean data. Each sample yielded a clean data amount of 5.79 Gb, with a Q30 base percentage of 93.43% or higher. In this study, we conducted a PCA clustering analysis to evaluate the differences in overall gene expression patterns among the four tissues (Fig. 3B). The overall gene expression patterns of the four tissues show significant differences. The gene expression in leaves is noticeably separated from roots and stems along PC1 axis, while it exhibits significant distinction from fruits along PC2 axis. The gene expression between stems and fruits exhibits minimal differences, yet they can still be clearly distinguished. After comparing the gene expression between leaves and roots, stems, and fruits, it was found that the expression of genes related to volatile biosynthesis and metabolism pathways of 'terpenoid backbone biosynthesis' and 'limonene and pinene degradation' and 'ubiquinone and other terpenoid-quinone biosynthesis' in leaves and fruits was significantly higher than in roots, while the gene expression of 'monoterpenoid biosynthesis' was significantly higher in fruits compared to stems (Table S9).
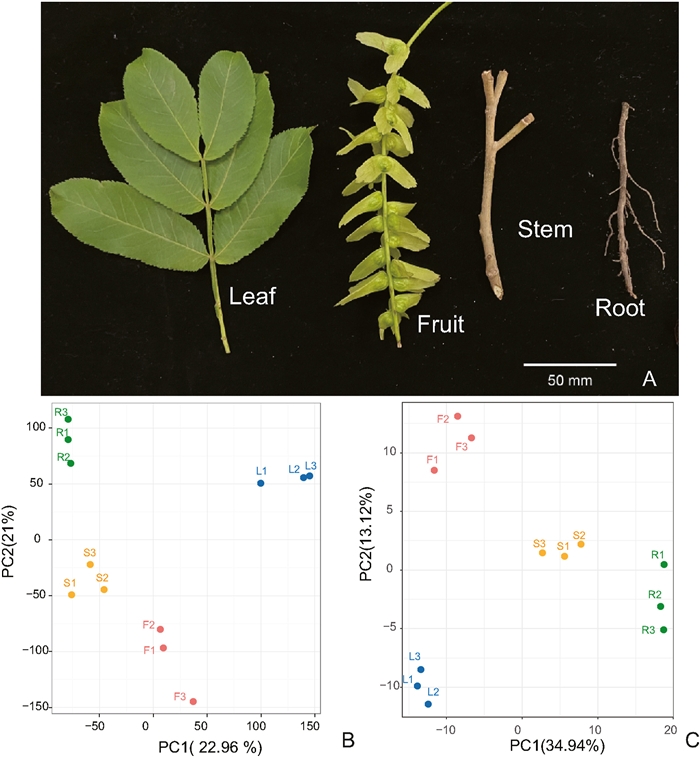
|
| Fig. 3 PCA clustering of different tissue samples of Chinese wingnut based on transcriptome and metabolome data. (A) Different tissue samples of the Chinese wingnut. (B) PCA of gene expression for different tissue samples of the Chinese wingnut. (C) PCA of metabolites for different tissue samples of the Chinese wingnut. |
GC-MS was utilized to analyze the metabolites present in 12 samples of leaves, stems, roots, and fruits from Chinese wingnut. A total of 423 volatile compounds were detected across the four tissues (Table S10). There are significant differences in the volatile components present among the four tissue samples in Chinese wingnut (Fig. 3C). After conducting a thorough analysis, we have compared the concentrations of the top 10 volatile compounds found in the leaves and fruits of Chinese wingnut. It has been revealed that terpenes, specifically d-limonene, α-pinene and 3-carene, exhibited the highest levels of concentration. On the other hand, when examining the roots, we observed that low molecular weight alcohols and aldehydes, such as 1-hexanol and ethanol, had the highest content. Among the top 10 volatiles in stems, the contents of aldehydes, alcohols and acids were highest, such as hexanal, 1-hexanol and hexanoic acid, and we also found that terpenes, such as α-pinene and 3-carene also account for a certain amount.
3.5. Candidate modules and genes for the synthesis of volatile compoundsClusters of co-expressed genes resulted a total of 17 gene modules (Fig. 4A). Correlation analysis between pairs of modules revealed that the MElightslateblue module correlated with the highest number of gene modules (Fig. 4B). The MElightslateblue module exhibited significant positive correlations with d-limonene, α-pinene, 3-carene, and caryophyllene, while the MEblue4 module showed significant correlations with d-limonene, α-pinene, and 3-carene. Similarly, the MEcyan module displayed significant positive correlations with d-limonene and caryophyllene, while the MEthistle module was significantly correlated with α-pinene and 3-carene. Additionally, the MEfirebrick4 module exhibited a significant positive correlation with 3-carene (Fig. 4C). The MElightslateblue module contained a total of 433 genes, which were found to be significantly enriched in the pathways related to photosynthesis and carotenoid synthesis. The MEblue4 module consisted of 74 genes that were significantly enriched in the phenylpropanoid biosynthesis pathway. Within the MEcyan module, 866 genes were significantly enriched in 7 pathways, with carotenoid biosynthesis pathway still showing significant enrichment. The MEthistle module included 1388 genes that were enriched in 12 pathways, while the MEfirebrick4 module contained 964 genes enriched in 2 pathways (Table S11). Overall, the enrichment analysis from WGCNA suggests a close relationship between genes involved in carotenoid and phenylpropanoid biosynthesis and the volatile terpenes d-limonene, α-pinene, 3-carene, and caryophyllene.
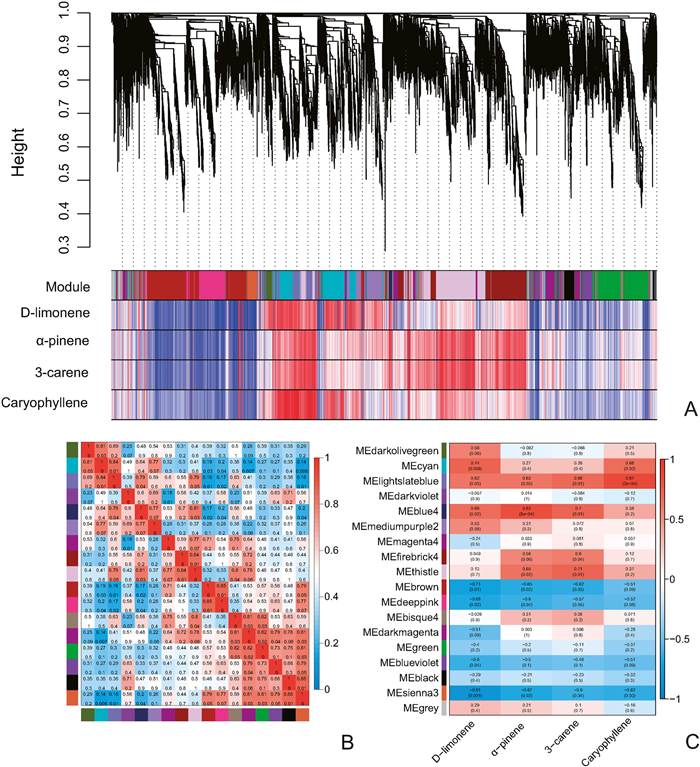
|
| Fig. 4 WGCNA analysis of gene expression and volatile metabolism in different tissues of Chinese wingnut. (A) Cluster tree of the expressed genes and the Correlation heat map between the expressed genes and the terpene volatiles. (B) The correlation heatmap of gene expression modules. The upper numbers in the color blocks represent the correlation coefficients, while the lower numbers represent the P-values. (C) Correlation heatmap between gene expression modules and four terpenoid volatiles. Numbers in the colored boxes have the same significance as mentioned above. |
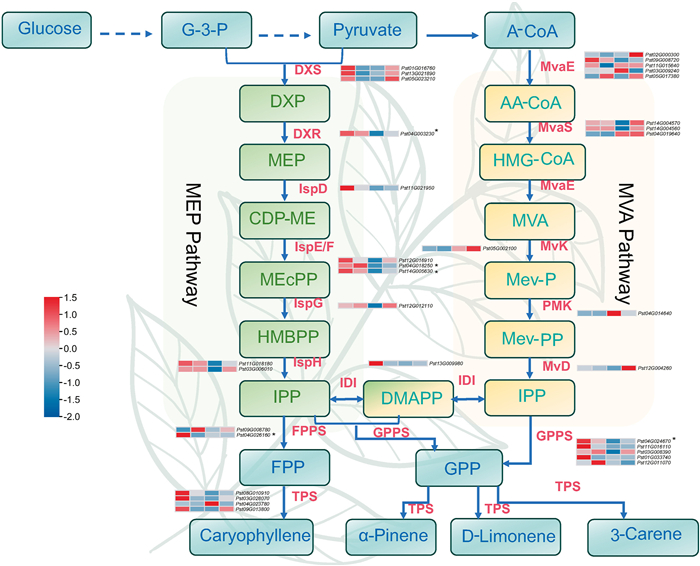
Synthesis of d-limonene, α-pinene, 3-carene, and caryophyllene, four volatile terpene compounds, can be achieved through the methylerythritol phosphate (MEP) and the mevalonic acid (MVA) pathways (Fig. 5). Based on the synthesis pathways of these four volatile compounds in other species, we screened for candidate genes involved, resulting in 119 genes identified (Supporting Information Table S12). Among these genes, 34 showed effective expression (log2FPKM > 1) in both fruits and leaves, but only five genes, i.e., 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR, Pst04G003230), 2-C-methyl-D-erythritol 2, 4-cyclopyrophosphate synthase (IspF, Pst04G018250 and Pst14G005630), geranylgeranyl pyrophosphate synthase (GPPS, Pst04G024670), and farnesyl pyrophosphate synthase (FPPS, Pst04G026160) were found to be significantly associated with at least one of the four volatile compounds (Fig. 5). FPPS (Pst04G026160) belongs to the MEfirebrick4 module, while the other four candidate genes are not part of these 17 co-expressed gene modules.
|
| Fig. 5 Identification the candidates for the biosynthesis of four terpenoid volatiles. The color blocks in the gene expression heat map, from left to right, represent fruits, leaves, roots, and stems. Enzymes involved in the biosynthesis of four terpenoid volatiles: DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; IspD, 4-pyrophosphocytidyl-2-C-methyl-D-erythritol synthase; IspE, 4-pyrophosphocytidyl-2-C-methylerythritol kinase; IspF, 2-C-methyl-D-erythritol 2, 4-cyclopyrophosphate synthase; IspG, 4-hydroxy-3-methylbut-2-enyl pyrophosphate synthase; IspH, 1-hydroxy-2-methyl-butenyl 4-pyrophosphate reductase; IDI, IPP isomerase; MvaE, acetyl-CoA acetyltransferase/HMG-CoA reductase; MvaS, HMG-CoA synthase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; MVD, mevalonate pyrophosphate decarboxylase; GPPS, geranylgeranyl pyrophosphate synthase; FPPS, farnesyl pyrophosphate synthase; TPS, terpene synthase. G-3-P, glyceraldehyde 3-phosphate; DXP, 1-deoxy-d-xylulose 5-phosphate; MEP, 2C-methyl-D-erythritol 4-phosphate; CDP-ME, 4-diphosphocytidyl-2C-methyl-D-erythritol; MEcPP, 2C-methyl-D-erythritol 2, 4-cyclodiphosphate; HMBPP, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate; IPP, isopentenyl pyrophosphate; A-CoA acetyl-CoA, AA-CoA acetoacetyl-CoA, HMG-CoA 3-hydroxy-3-methylglutaryl-CoA; Mev-P, mevalonate 5-phosphate; Mev-PP, mevalonate 5-diphosphate; DMAPP, dimethylallyl pyrophosphate; GPP, geranyl diphosphate; FPP, farnesyl pyrophosphate. * means five candidate key genes (Pst04G003230, Pst04G018250, Pst14G005630, Pst04G024670, and Pst04G026160) that gene expression is significantly related to metabolite content. Colour scale from blue to red represents expression levels (log10FPKM) from low to high. |
Due to its high medicinal value, ornamental value, and ecological value, significant attention has been given to the study of Chinese wingnut in recent years (Zhang et al., 2023). However, due to the lack of a high-quality genome, we do not have a clear understanding of its evolutionary history and the synthesis mechanism of the main medicinal compounds. In this study, we assembled a high-quality chromosome-level genome for Chinese wingnut and elucidated its evolutionary history through comparative genomics. Furthermore, we identified candidate genes in the synthesis pathway of the main medicinal compounds in its leaves through a combined analysis of transcriptomics and metabolomics.
4.1. Evolutionary history of Chinese wingnutPhylogenetic tree indicated that the wingless group was the ancestral type in Juglandaceae, while the winged group originated from the ancestral type around 13.66 Mya. Among the five Juglandaceae species we examined, their ancestors experienced one WGD event before their divergence. The WGD events during this period were a common occurrence found in multiple species (Wang et al., 2022; Liu et al., 2023). In Juglandaceae, the ancestral type of wingless fruit is large, nut-like drupes, while the newly differentiated the winged species have smaller fruits with wings developed from bracts. Fossil records indicated that a significant distribution of these species already existed in the Bangmai region of Yunnan during the late Miocene (11.63–5.33 Mya; Shao, 2016). Moreover, the vegetation in this region during that time was predominantly subtropical and tropical, suggesting a moist subtropical climate in the late Miocene (Xie et al., 2013). Therefore, it is speculated that the differentiation of the winged group may be linked to historic climate fluctuations.
The large amount of precipitation stimulated the divergence of Juglandaceae into winged species that rely on both wind and streams for propagation. Moreover, the reduced fruit size facilitates better dispersal through streams. Through the analysis of gene family expansion in the winged group, significant gene expansion associated with root, lateral root, and adventitious root morphology and development was detected. In the detected genes related to root growth expansion (Pst01G013290.1, Pst02G002390.1, Pst06G023960.1, Pst09G006290.1, Pst13G009870.1, Pst14G011580.1), they all belong to the Lateral Organ Boundaries Domain (LBD) gene family, which has been found in Arabidopsis that mutations in LBD genes lead to a significant reduction in lateral roots (Berckmans et al., 2011). The winged groups are mainly distributed on both sides of streams or in habitat areas with high humidity, where they are likely to encounter frequent flooding events in historical environmental changes. This ecological environment forces them to evolve strong root systems to withstand being washed away by floods. The expansion of gene families related to the development and morphogenesis of roots, lateral roots, and adventitious roots has also endowed winged group with the ability to adapt to waterlogging stress. For example, under prolonged stress, Chinese wingnut generate adventitious roots to enhance oxygen acquisition (Li et al., 2010). The divergence of Pterocarya and Cyclocarya is estimated to have occurred approximately 11.69 Mya, as indicated by the phylogenetic tree. Fossil records of the Pterocarya suggest that it mainly appeared during the Paleogene to Neogene periods (66–1.75 Mya), with a peak occurrence in the Neogene (23.5–1.75 Mya; Shao, 2016). Our molecular evolutionary data further support the separation of Pterocarya from Cyclocarya during the Neogene period. The current distribution patterns of Pterocarya and Cyclocarya indicate ecological niche differentiation following their divergence. Species of Pterocarya predominantly inhabit streamside environments, whereas species of Cyclocarya are found in moist forests. The long-term occupancy of the streamside habitat by Pterocarya suggests that it may frequently face waterlogging stress. In this study, the mitochondrial aerobic respiration-related genes that underwent positive selection were identified as NADH dehydrogenase (Pst02G001400.1, Pst12G001640.1). Previous research has demonstrated that the expression of NADH dehydrogenase can enhance the survival rate of plants under oxidative stress conditions (Jethva et al., 2023). Our results reveal significant positive selection on mitochondrial aerobic respiration-related genes in Chinese wingnut, which may confer stronger tolerance to waterlogging stress.
After the divergence of Pterocarya and Cyclocarya, Chinese wingnut has not only evolved stronger tolerance to waterlogging but may also possess higher capacity to remove TN. Comparative analysis of walnut species genomes revealed that Chinese wingnut have evolved unique genes related to arginine synthesis. Among the 21 protein amino acids, arginine has the highest N/C ratio (Winter et al., 2015). This make it have higher nitrogen storage capacity and can effectively absorb nitrogen in water. The increase in TN removal capacity may also be related to their ecological niche, as the decomposition of excessive plant litter in streams promotes the increase of the gene number related to arginine synthesis. Our results showed that all genetic groups of Chinese wingnut experienced significant population decline during the LG period in the Pleistocene, but rapidly rebounded after the glacial period. Previous phylogeographic study on Chinese wingnut also suggested that Pleistocene climate changes led to significant fluctuations in their effective populations, which subsequently experienced a rapid recovery after LG (Qian et al., 2019).
4.2. Candidate modules and genes involved in the synthesis of terpene volatilesChinese wingnut leaves contain a significant amount of volatile oils, with terpenes being the predominant component (Yin et al., 2020). Our findings indicate that terpene volatiles are also present in high concentrations in its fruits. According to a recent analysis of volatile compounds in Chinese wingnut leaves, it was found that caryophyllene and caryophyllene oxide accounted for the highest content (Bo and Yu, 2021). However, our study reveals that although caryophyllene and caryophyllene oxide have relatively high content in volatiles, compounds such as d-limonene, α-pinene, and 3-carene may have higher content in the leaves and fruits of Chinese wingnut. During pest control in tea gardens, it has been discovered that the crude extract of Chinese wingnut leaves has insecticidal effects on various insects such as diamondback moth, leaf roller, tea caterpillar, tea looper, and tussock moth (Li et al., 2007). In the crude extract, the volatile oils including d-Limonene, α-pinene, caryophyllene, and 3-carene have been found to exhibit significant insecticidal properties against herbivorous insects (Tripathi et al., 2003; Kumbasli and Bauce, 2013; Chohan et al., 2023). Therefore, we speculate that the abundance of terpenoid volatile compounds in the leaves and fruits of Chinese wingnut may have evolved as a means to defend against herbivorous insects when distributed along the streamsides.
Our analysis of gene families shows that a significant expansion occurs in the terpene synthase 2 and 10 families in Chinese wingnut and Cyclocarya paliurus following their divergence. In fact, among the Juglandaceae species we examined, Chinese wingnut has the highest number of members in these two gene families. We speculate that it is due to this significant expansion of terpene synthase gene family members in Chinese wingnut that its leaves and fruits are able to produce more terpenoid volatiles, which help to deter large numbers of herbivorous insects near streams. In addition to their role in defending against herbivorous insects, terpenoid volatiles also possess antimicrobial, antioxidant, and anti-tumor activities (Li et al., 2021c). As a result, Chinese wingnut leaves are widely used in traditional Chinese medicine.
The synthesis of terpenoid volatiles primarily occurs through the MEP and MVA pathways (Gao et al., 2018; Cheng et al., 2022). In this study, we conducted WGCNA analysis and Pearson correlation analysis to investigate the candidate modules and genes associated with the synthesis of major terpenoid volatiles, including d-limonene, α-pinene, 3-carene, and caryophyllene. The WGCNA analysis results suggest a significant correlation between carotenoid and phenylpropanoid biosynthesis and the synthesis of the four investigated terpene volatiles. It has been found that carotenoid and phenylpropanoid biosynthesis share common pathways with d-limonene, α-pinene, 3-carene, and caryophyllene (Dang et al., 2022; Zhu et al., 2022). Therefore, the genes related to carotenoid synthesis and phenylpropanoid biosynthesis may impact the synthesis of these terpene volatiles. A total of 34 candidate genes for effective expression in fruits and leaves were screened based on the synthesis pathways of other species such as d-limonene, α-pinene, 3-carene, and caryophyllene (Adal et al., 2017; Gao et al., 2018; Cheng et al., 2022). However, only 5 genes (Pst04G003230, Pst04G018250, Pst14G005630, Pst04G024670, and Pst04G026160) were found to be significantly correlated with at least one of the four volatile compounds. Five key candidate genes that we have identified have been confirmed to be involved in the synthesis of these volatile oils in previous studies. For instance, DXR has been shown to be significantly correlated with the content of Caryophyllene (Rai et al., 2024). The IspF participates in the synthesis of isoprenoids in the MEP pathway (Chen et al., 2018). GPPS has been found to significantly increase the levels of α-pinene and d-limonene in plants (Niu et al., 2018; Xie et al., 2023), while FPPS has been shown to enhance the content of caryophyllene in maize (Richter et al., 2015). Apart from these five genes, other potential genes may also be involved in the formation of these volatile oils, but they have not been effectively identified by Pearson correlation analysis. This phenomenon may be attributed to the existence of two pathways in the synthesis pathway of these volatile compounds, and there might be multiple possible synthetic enzymes involved in each synthesis step. Therefore, the expression levels of these genes may not necessarily show a strict positive correlation with the volatile compound content. In the process of synthesizing metabolites, where multiple pathways and lengthy routes exist, Pearson correlation analysis often only identifies a small number of candidate genes (Li et al., 2023). Therefore, in this study, we consider genes that are effectively expressed in tissues with high content as candidate genes. Integrated analysis of the metabolome and transcriptome data demonstrates that 34 genes effectively expressed in the fruits and leaves of Chinese wingnut can be considered as candidate genes for the synthesis of d-limonene, α-pinene, 3-carene, and caryophyllene.
5. ConclusionsIn this study, we have successfully assembled a high-quality chromosomal-level genome of Chinese wingnut. The divergence between the winged group and the wingless group within the Juglandaceae family may have arisen from shifts in ancient climatic conditions, characterized by increased precipitation and humidity. The reduction in fruit size and the change in dispersal method after wing development in the winged group of Juglandaceae led to a significant expansion of gene families associated with root growth, thus enabling the development of a well-established root system. Furthermore, the ecological niche differentiation between Chinese wingnut and Cyclocarya paliurus may have led to significant positive selection on mitochondrial genes related to aerobic respiration in Chinese wingnut occupying the ecological niche on both sides of the stream, enhancing its ability to withstand waterlogging stress. Comparative genomic analysis also revealed that Chinese wingnut has evolved unique genes associated with arginine synthesis, potentially equipping them with a higher capacity for purifying nutrient-rich water bodies. Metabolite analysis of different tissues in Chinese wingnut reveals higher levels of terpenoid volatiles, such as d-limonene, α-pinene, 3-carene, and caryophyllene, in its leaves and fruits. This may be attributed to the possibility that Chinese wingnut, being distributed along streamsides, are more exposed to herbivorous insects. Furthermore, analysis of gene family contraction and expansion suggests significant expansion of the terpene synthase 2 and 10 families in Chinese wingnut after its divergence from C. paliurus. Additionally, through combined transcriptome and metabolome analysis, we have identified 34 candidate genes involved in the synthesis of d-limonene, α-pinene, 3-carene, and caryophyllene. Our study provides genomic resources for Chinese wingnut, elucidating its evolutionary history and identifying candidate genes involved in the synthesis of terpenoid volatiles.
AcknowledgmentsWe are very grateful to Professor Jin-Ling Huang of East Carolina University for his advice on our manuscript. This work was supported by National Natural Science Foundation of China (32360307), the Natural Science Foundation of Inner Mongolia (2023MS03031), Inner Mongolia Grassland Talents Project (3211002406), and the Open Fund of State Key Laboratory of Tree Genetics and Breeding (Chinese Academy of Forestry) (Grant No. TGB2021004).
CRediT authorship contribution statement
Zi-Yan Zhang: Methodology, Investigation, Formal analysis, Data curation. He-Xiao Xia: Methodology, Investigation, Formal analysis, Data curation. Meng-Jie Yuan: Methodology, Investigation, Formal analysis, Data curation. Feng Gao: Methodology, Investigation, Formal analysis, Data curation. Wen-Hua Bao: Methodology, Investigation, Formal analysis, Data curation. Lan Jin: Methodology, Investigation, Formal analysis, Data curation. Min Li: Methodology, Investigation, Formal analysis, Data curation. Yong Li: Writing – review & editing, Supervision, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
All raw data for the Chinese wingnut genome and transcriptome sequencing were deposited in NCBI with accession numbers of PRJNA1047036. Genome assembly and annotation data were deposited in the Figshare database: https://doi.org/10.6084/m9.figshare.25237942. Raw data of GC-MS were deposited in the Figshare database: https://doi.org/10.6084/m9.figshare.24717660. The other data that support the findings of this study in the supplementary material of this article.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.03.010.
Adal, A.M., Sarker, L.S., Lemke, A.D., et al., 2017. Isolation and functional characterization of a methyl jasmonate-responsive 3-carene synthase from Lavandula x intermedia. Plant Mol. Biol., 93: 641-657. DOI:10.1007/s11103-017-0588-6 |
Ashburner, M., Ball, C.A., Blake, J.A., et al., 2000. Gene ontology: tool for the unification of biology. Nat. Genet., 25: 25-29. DOI:10.1038/75556 |
Bao, Z., Eddy, S.R., 2002. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res., 12: 1269-1276. DOI:10.1101/gr.88502 |
Berckmans, B., Vassileva, V., Schmid, S.P., et al., 2011. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell, 23: 3671-3683. DOI:10.1105/tpc.111.088377 |
Birney, E., Clamp, M., Durbin, R., 2004. GeneWise and genomewise. Genome Res., 14: 988-995. DOI:10.1101/gr.1865504 |
Bo, X., Yu, K., 2021. Study on extraction and chemical constituents of volatile oil from the leaves of Pterocarya stenoptera C. DC. Hubei Agr. Sci., 60: 119-123. |
Boeckmann, B., Bairoch, A., Apweiler, R., et al., 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31: 365-370. DOI:10.1093/nar/gkg095 |
Buchfink, B., Xie, C., Huson, D.H., 2015. Fast and sensitive protein alignment using DIAMOND. Nat. Methods, 12: 59-60. DOI:10.1038/nmeth.3176 |
Burton, J.N., Adey, A., Patwardhan, R.P., et al., 2013. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol., 31: 1119-1125. DOI:10.1038/nbt.2727 |
Chen, N.G., Wang, P.R., Li, C.M., et al., 2018. A Single nucleotide mutation of the gene participating in the MEP pathway forisoprenoid biosynthesis causes a green-revertible yellow leaf phenotype in rice. Plant Cell Physiol., 59: 1905-1917. DOI:10.1093/pcp/pcy108 |
Cheng, H., Concepcion, G.T., Feng, X., et al., 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods, 18: 170-175. DOI:10.1038/s41592-020-01056-5 |
Cheng, T., Zhang, K., Guo, J., et al., 2022. Highly efficient biosynthesis of β-caryophyllene with a new sesquiterpene synthase from tobacco. Biotechnol. Biof. Biop., 15: 39. DOI:10.4324/9781003316688-3 |
Chohan, T.A., Chohan, T.A., Mumtaz, M.Z., et al., 2023. Insecticidal potential of α-pinene and β-caryophyllene against Myzus persicae and their impacts on gene expression. Phyton-Int. J. Exp. Bot., 92: 1943-1954. |
Dang, J.J., Lin, G.Y., Liu, L.C., et al., 2022. Comparison of pulegone and estragole chemotypes provides new insight into volatile oil biosynthesis of Agastache rugosa. Front. Plant Sci., 13: 850130. DOI:10.3389/fpls.2022.850130 |
Deng, Y.Y., Li, J.Q., Wu, S.F., et al., 2006. Integrated NR database in protein annotation system and its localization. Comp. Eng., 32: 71-74. |
Ding, Y.M., Pang, X.X., Cao, Y., et al., 2023. Genome structure-based Juglandaceae phylogenies contradict alignment-based phylogenies and substitution rates vary with DNA repair genes. Nat. Commun., 14: 617. DOI:10.1038/s41467-023-36247-z |
Doyle, J.J., Doyle, J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. bull, 19: 11-15. |
Ellinghaus, D., Kurtz, S., Willhoeft, U., 2008. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics, 9: 18. DOI:10.1186/1471-2105-9-18 |
Emms, D.M., Kelly, S., 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol., 20: 238. DOI:10.1186/s13059-019-1832-y |
Finn, R.D., Mistry, J., Schuster-Bockler, B., et al., 2006. Pfam: clans, web tools and services. Nucleic Acids Res., 34: D247-D251. DOI:10.1093/nar/gkj149 |
Flynn, J.M., Hubley, R., Goubert, C., et al., 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. U.S.A., 117: 9451-9457. DOI:10.1073/pnas.1921046117 |
Gao, F.Z., Liu, B.F., Li, M., et al., 2018. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia x hybrida. J. Exp. Bot., 69: 4249-4265. DOI:10.1093/jxb/ery224 |
Gao, N., 2009. Study on Pllutant in the Water Removal Efficiency of Several Trees Commonly Used in Urban. Beijing Forestry University, Beijing (Master thesis).
|
Griffiths-Jones, S., Moxon, S., Marshall, M., et al., 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res., 33: D121-D124. |
Haas, B.J., Delcher, A.L., Mount, S.M., et al., 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res., 31: 5654-5666. DOI:10.1093/nar/gkg770 |
Haas, B.J., Salzberg, S.L., Zhu, W., et al., 2008. Automated eukaryotic gene structure annotation using EVidence Modeler and the Program to Assemble Spliced Alignments. Genome Biol., 9: R7. DOI:10.1186/gb-2008-9-1-r7 |
Han, M.V., Thomas, G.W.C., Lugo-Martinez, J., et al., 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol., 30: 1987-1997. DOI:10.1093/molbev/mst100 |
Huerta-Cepas, J., Szklarczyk, D., Heller, D., et al., 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res., 47: D309-D314. DOI:10.1093/nar/gky1085 |
Jethva, J., Lichtenauer, S., Schmidt-Schippers, R., et al., 2023. Mitochondrial alternative NADH dehydrogenases NDA1 and NDA2 promote survival of reoxygenation stress in Arabidopsis by safeguarding photosynthesis and limiting ROS generation. New Phytol., 238: 96-112. DOI:10.1111/nph.18657 |
Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., et al., 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods, 14: 587-589. DOI:10.1038/nmeth.4285 |
Kanehisa, M., Sato, Y., Kawashima, M., et al., 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res., 44: D457-D462. DOI:10.1093/nar/gkv1070 |
Katoh, K., Asimenos, G., Toh, H., 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol., 537: 39-64. DOI:10.1007/978-1-59745-251-9_3 |
Keilwagen, J., Wenk, M., Erickson, J.L., et al., 2016. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res., 44: e89. DOI:10.1093/nar/gkw092 |
Kim, D., Landmead, B., Salzberg, S.L., 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods, 12: 357-360. DOI:10.1038/nmeth.3317 |
Kind, T., Wohlgemuth, G., Lee, D.Y., et al., 2009. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem., 81: 10038-10048. DOI:10.1021/ac9019522 |
Korf, I., 2004. Gene finding in novel genomes. BMC Bioinf., 5: 59. DOI:10.1186/1471-2105-5-59 |
Kuang, K.R., Li, P.Q., 1979. Flora of China (Volume 21). Science Press, Beijing, pp. 21–30.
|
Kumar, S., Stecher, G., Suleski, M., et al., 2017. Timetree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol., 34: 1812-1819. DOI:10.1093/molbev/msx116 |
Kumbasli, M., Bauce, É., 2013. Spruce budworm biological and nutritional performance responses to varying levels of monoterpenes. iForest-Biogeosci. Fores., 6: 117. |
Langfelder, P., Horvath, S., 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf., 9: 559. DOI:10.1186/1471-2105-9-559 |
Li, C.X., Wei, H., Lv, Q., Zhang, Y., 2010. Effects of water stresses on growth and contents of oxalate and tartarate in the roots of Chinese wingnut (Pterocarya stenoptera) seedlings. Sci. Silvae Sin., 46: 81-88. DOI:10.3901/JME.2010.12.081 |
Li, D.X., Cui, C.B., Cai, B., et al., 2007. Research progress of Pterocarya. Pharm. J. Chinese P. L. A., 23: 365-369. |
Li, H., 2021. New strategies to improve minimap2 alignment accuracy. Bioinformatics, 37: 4572-4574. DOI:10.1093/bioinformatics/btab705 |
Li, H., Durbin, R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25: 1754-1760. DOI:10.1093/bioinformatics/btp324 |
Li, H., Durbin, R., 2011. Inference of human population history from individual whole-genome sequences. Nature, 475: 493-496. DOI:10.1038/nature10231 |
Li, X.R., Zhang, X.X., Xing, M.Y., et al., 2021a. Antioxidant and antibacterial activities of Pterocarya stenoptera bark extract and its mechanism on Staphylococcus aureus through cell membrane damage. Bioresources, 16: 3771-3782. DOI:10.15376/biores.16.2.3771-3782 |
Li, J.X., Zhu, X.H., Li, Y., et al., 2018. Adaptive genetic differentiation in Pterocarya stenoptera (Juglandaceae) driven by multiple environmental variables were revealed by landscape genomics. BMC Plant Biol., 18: 306. DOI:10.1007/978-3-319-91250-9_24 |
Li, L.F., Cushman, S.A., He, Y.X., et al., 2022. Landscape genomics reveals genetic evidence of local adaptation in a widespread tree, the Chinese wingnut (Pterocarya stenoptera). J. Systemat. Evol., 60: 386-397. DOI:10.1111/jse.12699 |
Li, Y., Shi, L.C., Yang, J., et al., 2021b. Physiological and transcriptional changes provide insights into the effect of root waterlogging on the aboveground part of Pterocarya stenoptera. Genomics, 113: 2583-2590. DOI:10.1016/j.ygeno.2021.06.005 |
Li, Y., Si, Y.T., He, Y.X., et al., 2021c. Comparative analysis of drought-responsive and -adaptive genes in Chinese wingnut (Pterocarya stenoptera C. DC.). BMC Genomics, 22: 155. DOI:10.1186/s12864-021-07470-z |
Li, Y., Wang, F., Pei, N.C., et al., 2023. The updated weeping forsythia genome reveals the genomic basis for the evolution and the forsythin and forsythoside A biosynthesis. Hortic.Plant J., 9: 1149-1161. DOI:10.3390/ma16031149 |
Liu, Y., Schröder, J., Schmidt, B., 2013. Musket: a multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics, 29: 308-315. DOI:10.1093/bioinformatics/bts690 |
Liu, Z.K., Fu, Y.H., Wang, H., et al., 2023. The high-quality sequencing of the Brassica rapa 'XiangQingCai' genome and exploration of genome evolution and genes related to volatile aroma. Hortic. Res., 10: uhad187. DOI:10.1093/hr/uhad187 |
Loman, T., 2017. A novel method for predicting ribosomal RNA genes in prokaryotic genomes. Degree Projects in Bioinformatics. http://lup.lub.lu.se/studentpapers/record/8914064.
|
Love, M.I., Huber, W., Anders, S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15: 550. DOI:10.1186/s13059-014-0550-8 |
Lowe, T.M., Eddy, S.R., 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25: 955-964. DOI:10.1093/nar/25.5.955 |
Mi, H., Muruganujan, A., Ebert, D., et al., 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res., 47: D419-D426. DOI:10.1093/nar/gky1038 |
Nanjing University of Traditional Chinese Medicine, 1997. Dictionary of Traditional Chinese Medicine. Shanghai: Shanghai Science & Technology Press.
|
Nawrocki, E.P., Eddy, S.R., 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics, 29: 2933-2935. DOI:10.1093/bioinformatics/btt509 |
Niu, F.X., He, X., Wu, Y.Q., et al., 2018. Enhancing Production of Pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering. Front. Microbiol., 9: 1623. DOI:10.3389/fmicb.2018.01623 |
Ou, S., Jiang, N., 2018. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol., 176: 1410-1422. DOI:10.1104/pp.17.01310 |
Pan, Y., 2021. Propagation and cultivation techniques for ginkgo and Pterocarya stenoptera trees in Changji prefecture. Forest. Xinjiang, 1: 22-24. |
Parra, G., Bradnam, K., Korf, I., 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics, 23: 1061-1067. DOI:10.1093/bioinformatics/btm071 |
Pertea, M., Pertea, G.M., Antonescu, C.M., et al., 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol., 33: 290-295. DOI:10.1038/nbt.3122 |
Price, A.L., Jones, N.C., Pevzner, P.A., 2005. De novo identification of repeat families in large genomes. Bioinformatics, 21: i351-i358. DOI:10.1093/bioinformatics/bti1018 |
Qian, Z.H., Li, Y., Li, M.W., et al., 2019. Molecular phylogeography analysis reveals population dynamics and genetic divergence of a widespread tree Pterocarya stenoptera in China. Front. Genet., 10: 1089. DOI:10.3389/fgene.2019.01089 |
Rai, N., Kumari, S., Singh, S., et al., 2024. Modulation of morpho-physiological attributes and in situ analysis of secondary metabolites using Raman spectroscopy in response to red and blue light exposure in Artemisia annua. Environ. Exp. Bot., 217: 105563. DOI:10.1016/j.envexpbot.2023.105563 |
Richter, A., Seidl-Adams, I., Köllner, T.G., et al., 2015. A small, differentially regulated family of farnesyl diphosphate synthases in maize (Zea mays) provides farnesyl diphosphate for the biosynthesis of herbivore-induced sesquiterpenes. Planta, 241: 1351-1361. DOI:10.1007/s00425-015-2254-z |
Shannon, P., Markiel, A., Ozier, O., et al., 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13: 2498-2504. DOI:10.1101/gr.1239303 |
Shao, Y., 2016. Study on Juglandaceae Fossils from the Late Miocene of Lincang, Yunnan Province, China. Lanzhou University, Lanzhou (Master's thesis).
|
She, R., Chu, J.S., Wang, K., et al., 2009. GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res., 19: 143-149. DOI:10.1101/gr.082081.108 |
Simao, F.A., Waterhouse, R.M., Ioannidis, P., et al., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31: 3210-3212. DOI:10.1093/bioinformatics/btv351 |
Sollars, E.S., Harper, A.L., Kelly, L.J., et al., 2017. Genome sequence and genetic diversity of European ash trees. Nature, 541: 212-216. DOI:10.1038/nature20786 |
Stanke, M., Diekhans, M., Baertsch, R., et al., 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics, 24: 637-644. DOI:10.1093/bioinformatics/btn013 |
Suyama, M., Torrents, D., Bork, P., 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res., 34: W609-W612. DOI:10.1093/nar/gkl315 |
Talavera, G., Castresana, J., 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol., 56: 564-577. DOI:10.1080/10635150701472164 |
Tang, H., Krishnakumar, V., Li, J., et al., 2015a. Jcvi: JCVI Utility Libraries. Zenodo.
|
Tang, S., Lomsadze, A., Borodovsky, M., 2015b. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res., 43: e78. DOI:10.1093/nar/gkv227 |
Tarailo-Graovac, M., Chen, N., 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform., 25,(4.10.1–4.10.14). |
Tatusov, R.L., Fedorova, N.D., Jackson, J.D., et al., 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics, 4: 41. DOI:10.1186/1471-2105-4-41 |
Tommasini, D., Fogel, B.L., 2023. multiWGCNA: an R package for deep mining gene co-expression networks in multi-trait expression data. BMC Bioinformatics, 24: 115. DOI:10.1186/s12859-023-05233-z |
Tripathi, A.K., Prajapati, V., Khanuja, S.P.S., et al., 2003. Effect of d-limonene on three stored-product beetles. J. Econ. Entomol., 96: 990-995. DOI:10.1603/0022-0493-96.3.990 |
Wang, W., Shao, A., Xu, X., et al., 2022. Comparative genomics reveals the molecular mechanism of salt adaptation for zoysiagrasses. BMC Plant Biol., 22: 355. DOI:10.1186/s12870-022-03752-0 |
Wang, Y., Tang, H., Debarry, J.D., et al., 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res., 40: e49. DOI:10.1093/nar/gkr1293 |
Winter, G., Todd, C.D., Trovato, M., et al., 2015. Physiological implications of arginine metabolism in plants. Front. Plant Sci., 6: 534. |
Xie, S., Wu, G., Ren, R.H., et al., 2023. Transcriptomic and metabolic analyses reveal differences in monoterpene profiles and the underlying molecular mechanisms in six grape varieties with different flavors. LWT–Food Sci. Technol., 174: 114442. DOI:10.1016/j.lwt.2023.114442 |
Xie, S.P., Manchester, S.R., Liu, K.N., et al., 2013. Sp N., A leaf fossil of Rutaceae from the late Miocene of Yunnan, China. Int. J. Plant Sci., 174: 1201-1207. DOI:10.1086/671796 |
Xu, Y.M., Zhou, M.H., Shi, Y.H., et al., 2002. Advance on the biological properties and resources utilization of Pterocarya stenoptera. J. Northeast For. Univ., 30: 42-48. DOI:10.3901/JME.2002.10.042 |
Xu, Z., Wang, H., 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res., 35: W265-W268. DOI:10.1093/nar/gkm286 |
Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci., 13: 555-556. DOI:10.1093/bioinformatics/13.5.555 |
Ye, X.F., Li, Y., Liu, H.L., et al., 2020. Physiological analysis and transcriptome sequencing reveal the effects of drier air humidity stress on Pterocarya stenoptera. Genomics, 112: 5005-5011. DOI:10.1016/j.ygeno.2020.09.027 |
Yin, C., Sun, F., Rao, Q., et al., 2020. Chemical compositions and antimicrobial activities of the essential oil from Pterocarya stenoptera C. DC. Nat. Prod. Res., 34: 2828-2831. DOI:10.1080/14786419.2019.1587426 |
Yu, G.C., Wang, L.G., Han, Y.Y., et al., 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 16: 284-287. DOI:10.1089/omi.2011.0118 |
Zhang, W., Wang, S.C., Li, Y., 2023. Molecular mechanism of thiamine in mitigating drought stress in Chinese wingnut (Pterocarya stenoptera): insights from transcriptomics. Ecotoxicol. Environ. Saf., 263: 115307. DOI:10.1016/j.ecoenv.2023.115307 |
Zhu, C.Y., Peng, C., Qiu, D.Y., et al., 2022. Metabolic profiling and transcriptional analysis of carotenoid accumulation in a red-fleshed mutant of pummelo (Citrus grandis). Molecules, 27: 4595. DOI:10.3390/molecules27144595 |
Zwaenepoel, A., Van de Peer, Y., 2019. WGD-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics, 35: 2153-2155. DOI:10.1093/bioinformatics/bty915 |



