b. National Engineering Research Center of Eco-Environment Protection for Yangtze River Economic Belt, China Three Gorges Corporation, Beijing 100083, China;
c. Rare Plants Research Institute of Yangtze River, China Three Gorges Corporation, Yichang 443133, China;
d. Research Institute of Economic Forest Cultivation and Processing, Anhui Academy of Forestry, Hefei 230031, China
Endangered species are often characterized by severe population decline or even near-extinction over their demographic histories, due to historical climate change and/or contemporary human activities (Pimm et al., 2014). For small isolated populations, genetic drift can lead to inbreeding depression, erosion of genetic diversity, and accumulation of deleterious mutations (Abascal et al., 2016; Benazzo et al., 2017; Chen et al., 2020), which further decrease the adaptive potential and increase extinction risk when facing volatile natural habitats or climate. Elucidating the demographic history of threatened species and the impact of genomic erosion on endangerment is fundamental to implementing effective conservation efforts (Yang et al., 2018; Ma et al., 2021a, 2022; Dedato et al., 2022).
The subtropical forests of China are among the most threatened forest types in East Asia (Wei et al., 2009). Many of the threatened plant species in these forests are Plant Species with Extremely Small Populations (PSESP), which have restricted geographical ranges and are subject to serious interference (Ma et al., 2013; Sun et al., 2019). One approach researchers have used to determine the genetic diversity and demographic history of these PSESPs is conservation genomics, e.g., Magnolia fistulosa (Yang et al., 2022a), Acer yangbiense (Ma et al., 2022), Populus ilicifolia (Chen et al., 2020), and Ostrya rehderiana (Yang et al., 2018). Compared to PSESPs, few studies have used conservation genomics approaches to understand the maintenance and genetic effects of endangered plants with relatively wide distribution. Such species have historically occupied larger geographic ranges than they do now (Werth and Shear, 2014; Lidgard and Love, 2018), but their populations may have severely declined due to past climate fluctuations and recent habitat fragmentation. Nonetheless, certain species have exhibited remarkable resilience and adaptability across various temporal and spatial dimensions. For instance, Ostrya chinensis underwent multiple population declines during the Last Glacial Maximum (LGM), but subsequently rebounded to pre-LGM levels. Additionally, this species purged many severely deleterious mutations, potentially mitigating inbreeding depression and contributing to its future survival (Yang et al., 2018). Hence, these taxa represent exceptional models for investigating diverse demographic histories.
Genomic approaches are becoming increasingly useful in answering important evolutionary issues and guiding conservation (Funk et al., 2012; Beichman et al., 2018). Unlike traditional conservation genetics, which relies on a limited number of near-neutral markers, conservation genomic strategies have enabled the development of genome-wide conservation frameworks. This is particularly true for genome resequencing, which allows for the identification of nucleotide variations throughout the entire genome (Cammen et al., 2016; Hohenlohe et al., 2021). Major achievements facilitated by genome-wide data include, but are not limited to: (ⅰ) delineating species/population subdivisions more accurately; (ⅱ) depicting changes in effective population size (Ne), using temporal patterns to infer whether changes are related to historical natural processes or recent anthropogenic activities; (ⅲ) quantifying genomic-scale inbreeding and evaluating the risk of species extinction based on mutation accumulation, especially deleterious ones in the homozygous state; and (ⅳ) tracing the distribution patterns of deleterious mutations in populations, which will aid conservation and further translocation actions.
The Corylus genus contains a wide diversity of deciduous tree and shrub species that are prominent elements of northern temperate forests, with the diversity center in Southeast Asia (Zhao et al., 2020). Members of the genus are typically not threatened or vulnerable to extinction, and almost all species demonstrate high degrees of diversity and adaptability. However, there is one exception, C. chinensis Franch., the sole macrophanerophyte species within Corylus, which naturally occurs in subtropical China. Fossil records indicate that C. chinensis has an ancient origin and was extensively distributed in Asia during the Tertiary. C. chinensis has successfully endured intense geological movements and climatic fluctuations since then (Whitcher and Wen, 2001), and the extant populations are only disjunctively distributed in the mid-high mountains of subtropical forests. Climate change and human activities have severely devastated its habitats, leading to a dramatic decline in population size. This plant's ability to renew seedlings is weak, further hindering its natural recovery. C. chinensis is currently included on the red list of endangered species by the International Union for Conservation of Nature (Beech, 2018). Its sympatric relatives, C. kweichowensis and C. yunnanensis, have considerably larger natural populations. The two are genetically difficult to separate from each other, thus constituting the C. kweichowensis–C. yunnanensis species complex (hereafter CKY). Both C. chinensis and CKY are wind-pollinated, monoecious, and primarily outcrossing. However, despite similar regional conditions, noticeable disparities in population dynamics have been observed between them (He et al., 2022, 2023). Moreover, there is phylogenetic evidence that introgressive hybridization and genetic admixture has frequently occurred within Corylus, particularly between sympatric species, such as C. chinensis and CKY (Zhao et al., 2020). Thus, the population dynamics of C. chinensis and CKY have likely been affected by a combination of natural processes, anthropogenic disturbances, and differences in life history.
In this study, we generated the first chromosome-level genome of Corylus Chinenis and re-sequenced 47 individuals (12 populations) for conservation genomic research. We also re-sequenced 44 individuals (10 populations) of CKY for selection and introgression tests. Utilizing these genomic data, we aimed (ⅰ) to estimate the population structure and genomic diversity of C. chinensis; (ⅱ) to compare demographic histories of C. chinensis and CKY, as well as whether and how these histories were connected with climatic fluctuations; (ⅲ) to evaluate inbreeding and endangerment of C. chinensis in terms of accumulation of deleterious mutations; (ⅳ) to examine evidence for adaptation within C. chinensis and interspecific introgression between C. chinensis and CKY. This research will elucidate the interplay between demographic history and other factors in determining population dynamics, while simultaneously laying a foundation for the conservation of an endangered species.
2. Materials and methods 2.1. Plant materials, library construction, and sequencingThe Corylus chinensis accession used for genome sequencing was collected from the Huoditang Forestry District, China (33.4323°N, 108.4528°E, 1988 m). Genomic DNA was extracted from fresh leaves using DNAsecure Plant Kit (Tiangen Biotech, Beijing, China). Genomes were sequenced using a combination of paired-end Illumina and PacBio high fidelity (HIFI) sequencing. A high-throughput chromosome conformation capture sequencing (Hi-C) library was then constructed to facilitate chromosome-level assembly. For Illumina sequencing, short-read (~350 bp) libraries were prepared following standard protocols and paired-end reads (2 × 150 bp) were sequenced on the HiSeq X Ten platform (Illumina, NEB, USA). For PacBio sequencing, a 60 kb size-selected SMRTbell library was constructed and circular consensus sequencing was performed on the PacBio Sequel platform (Pacific Biosciences, Menlo Park, CA, USA). For Hi-C library construction, the leaf cells were digested with Dpnll endonuclease and sequenced on the HiSeq X Ten platform. RNA sequencing was used to assist genome assembly and annotation. RNAs of three tissues (leaves, stems, and roots) were isolated using the RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China). cDNA libraries were constructed using the NEBNext Ultra RNA Library Prep Kit (Illumina, NEB, USA) and sequenced on the HiSeq X Ten platform.
All raw sequencing data were filtered to eliminate duplicated reads, adaptors, and low-quality bases using different strategies depending on the platforms. For Illumina short-reads, RNA-seq reads, and Hi-C sequencing, raw data containing adapter sequences, 20% of low-quality bases (Q ≤ 10), and more than 10% unknown bases (N) were removed to generate high-quality sequences. For PacBio HiFi long reads, the subreads were filtered and corrected using the pbccs pipeline with default parameters.
2.2. Genome size and heterozygosity estimationWe conducted a genome survey based on 17 k-mer frequencies generated by Illumina short-read sequencing (Liu et al., 2013). The distribution of k-mer depends on the characteristics of the genome and follows a Poisson's distribution. The k-mer frequencies for multiple values of k were calculated using Jellyfish v.1.1.11 (Marçais and Kingsford, 2011). We estimated the genome size using the following formula: genome size = total k-mer num/k-mer depth (total k-mer num is the total number of k-mers from all reads, and k-mer depth is the peak depth). This method generated the necessary genome information, including genome size, heterozygosity rate, and repetitive elements (Fig. S1).
2.3. Genome assembly and quality assessmentPacBio HiFi long reads were used as a backbone for de novo genome assembly. After quality control, the high-quality HiFi reads were assembled using Hifiasm v.0.14-r312 (Cheng et al., 2021), and the redundant haplotigs were removed by Purge Haplotigs v.1.1.1 (Roach et al., 2018). The contig assemblies were scaffolded with the 3D de novo assembly pipeline v.190716 (Dudchenko et al., 2017). Then, Hi-C reads were mapped to the draft scaffold genome using BWA-MEM v.0.7.16 (Li, 2013), and only reads with unique alignment positions were extracted to construct a chromosome-scale assembly using Juicebox v.1.11.08 (Durand et al., 2016). Finally, Lachesis v.1.0.20 (Burton et al., 2013) was employed to cluster, order, and orient the scaffolds, which were finally anchored onto 11 chromosomes. To improve the assembly quality, artificial correction of orientation errors with discrete chromatin interaction patterns was conducted. The completeness of the assembled genome was evaluated by BUSCO v.5.3.2 (Simão et al., 2015) using gene content from the Embryophyta_odb10 database. To validate the alignment rates, the high-quality Illumina reads and assembled transcripts were remapped to the final assembly using HISAT2 v.2.1.0 (Kim et al., 2015).
2.4. Genome annotationTransposable elements in the Corylus chinensis genome were annotated using a combination of de novo and homology-based strategies. For de novo prediction, we used LTR_FINDER v.1.07 (Xu and Wang, 2007), RepeatScout (Price et al., 2005) and RepeatModeler v.1.0.4 (http://www.repeatmasker.org/RepeatModeler.html) to construct a de novo library, and then performed annotation using RepeatMasker v.4.0.7 (Tempel, 2012). For homologous alignment, RepeatMasker was employed to align the C. chinensis genome against the Repbase database (Bao et al., 2015), and RepeatProteinMask v.4.05 (Bergman and Quesneville, 2007) was used for homologs-based prediction (Jurka et al., 2005).
We applied an integrated pipeline to forecast protein-coding genes in the repeat masked genome, including AUGUSTUS v.3.3.3 (Stanke et al., 2006) for ab initio gene finding, GeneWise v.2.4.1 (Birney et al., 2004) and TBlastN v.2.6.0 (Camacho et al., 2009) for homologue-based prediction, Hmmer v.3.1b2 (Potter et al., 2018) and Transdecoder v.5.5.0 (https://github.com/TransDecoder/TransDecoder) for transcriptome-based prediction. Finally, all resulted gene models were merged into a consensus gene set using GETA v.2.4.6 (https://github.com/chenlianfu/geta). For RNA-seq evidence, sequencing data were mapped to the Corylus chinensis genome using HISAT2 v.2.1.0 (Kim et al., 2015). Trinity v.2.4.0 (Grabherr et al., 2011) was used to assemble de novo and genome-guided transcripts with clean reads. For homology-based prediction, we downloaded homologous proteomes of six plant species: Arabidopsis thaliana (GCA_000001735.2), Oryza sativa (GCA_009797565.1), Populus trichocarpa (GCA_000002775.4), Corylus avellana (GCA_901000735.2), Corylus heterophylla (GCA_016403345.1), and Corylus mandshurica (http://doi.org/10.6084/m9.figshare.12523124.v1). Functional annotation of coding genes was conducted via homologous searches against several public databases, including a NR database (NCBI), Swiss-Prot (Bairoch and Apweiler, 2000), GO, InterPro (Hunter et al., 2009), and KEGG pathway (Kanehisa et al., 2019).
2.5. Gene family and phylogenetic analysesProtein sequences of Corylus chinensis and the additional 14 species examined were retrieved to generate clusters of gene families. We filtered gene sets by selecting the longest transcript. Genes with internal stop codons, less than 30 amino acids, and incompatible reading frames were removed. We then conducted an all-vs-all comparison using BLASTP (E-value ≤ 1e-5) and OrthoFinder (Emms and Kelly, 2019) to identify orthologous and cluster gene families. The evolutionary relationships were inferred based on shared single-copy orthologous genes. The coding sequences of single-copy orthologues were aligned by MAFFT v.7.313 (Katoh and Standley, 2013) and trimmed by GBLOCKS v.0.91b (Talavera and Castresana, 2007). RAxML v.8.28 was used to construct a maximum-likelihood (ML) tree with 100 bootstrap replicates (Stamatakis, 2014). We used MCMCtree (Yang, 2007) to estimate divergence times based on the approximate likelihood method. Three calibrations were used: (ⅰ) 37–49 Ma for the crown node of Corylus–Ostryopsis–Ostrya–Carpinus (Pigg et al., 2003); (ⅱ) 47 Ma as the lower boundary for the Alnus–Betula split (http://www.timetree.org/); (ⅲ) 56.8–90.0 Ma for the split of Fagaceae (Castanea mollissima) from the order Fagales (http://www.timetree.org/).
CAFÉ v.5.1 (De Bie et al., 2006) was applied to examine expansion and contraction of orthologous gene families. The model was used to capture gain or loss of gene families for each species of the ML tree inferred from 13,481 shared gene families. A probabilistic graphical model was introduced to estimate the transition probability of gene family size from parent to child branches. The p-values were calculated in each species based on conditional likelihoods, and the species-specific gene families were determined according to the presence or absence of genes. To identify the macro-synteny blocks between Corylus chinensis and closely related species, we conducted a synteny analysis using MCScanX based on the protein sequences of five Corylus genomes, including C. chinensis, C. heterophylla, C. avellana, C. americana, and C. mandshurica.
2.6. Population resequencing, mapping, and genotypingFor genome resequencing, leaf materials of Corylus chinensis were collected from 12 natural populations (n = 47) across its main natural distribution region in subtropical China. One accession from Huoditang Forestry District was used for genome assembly. We also performed re-sequencing for the sympatric CKY complex (n = 44, pop = 10) to detect introgression and compare demographic history. To avoid re-sequencing cloned individuals, samples from each population were separated by at least 1000 m. Genomic DNA was extracted and then sequenced on the Illumina HiSeq X Ten platform. For raw resequencing data, we first used fastp (Chen et al., 2018) to filter adaptors and low-quality reads with default parameters. Then, the high-quality reads were mapped against the C. chinensis genome using BWA-MEM (Li, 2013). We employed SAMtools v.0.1.19 (Li et al., 2009) to sort BAM files and create index files. PCR duplications were removed using "MarkDuplicates" in Picard v.1.117 (https://broadinstitute.github.io/picard/). The coverage rate, sequencing depth, and mapping rate were calculated with the DepthOfCoverage program in GATK v.4.1.2.0 (McKenna et al., 2010). SNP calling was conducted using the HaplotypeCaller and GenotypeGVCFs tools. To minimize the influence of sequencing and mapping bias, hard filtration was performed with the following criteria: (ⅰ) sites with missing rates > 20% and minor allele frequency < 0.05; (ⅱ) sites with mapping quality and base quality < 20; (ⅲ) sites < 1/3 average depth and > 3-fold average depth.
2.7. Population structure and genomic diversityTo avoid the effect of linkage disequilibrium (LD) on population structure, we filtered excessive linkage sites using Plink v.1.9 (Purcell et al., 2007) with the parameter "–indep-pairwise 50 10 0.2". Admixture v.1.3.0 (Alexander et al., 2009) was used to investigate the population structure based on the LD-pruned SNPs, setting the assumed number of genetic clusters (K) from 2 to 7 and each running 10,000 iterations. The optimum K value was identified based on the lowest cross-validation (CV) error value. Principal component analysis (PCA) was conducted with GCTA v.1.93 (Yang et al., 2011), and the first two eigenvectors were plotted in two dimensions. A neighbor-joining (NJ) tree was constructed using PHYLIP v.3.69 (Felsenstein, 1989) based on p distance matrix calculated by VCF2Dis (https://github.com/BGI-shenzhen/VCF2Dis).
We evaluated genome-wide nucleotide diversity (π) based on individual BAM files using ANGSD v.0.937 (Korneliussen et al., 2014) over 100 kb non-overlapping windows. Genome heterozygosity of each sample was calculated using PLINK. Inbreeding coefficient (FROH) was used to evaluate the inbreeding level of samples, which was defined as the ratio of total runs of homozygosity (ROH) length to genome size. ROH was calculated using PLINK based on the SNP dataset without MAF and LD filtering. Only ROHs > 100 kb and containing < 20 SNPs were retained. LD decay pattern was inferred using PopLDdecay v.3.31 (Zhang et al., 2019), with decayed distance between SNPs measured by the maximum r2 dropped by half.
2.8. Inference of demographic historyTo investigate the demographic processes underlying the observed differentiation within and between Corylus chinensis and CKY, we used PSMC method (Li and Durbin, 2011) to infer changes in Ne, using the following parameters: -N25 -t15 -r5 -p "4 + 25 × 2 + 4 + 6". Because PSMC has high false negative rates at low sequencing depth, we chose four individuals sequenced at a high depth (> 30×) from each lineage for this analysis. PSMC analysis was conducted with a neutral nucleotide mutation rate of 3.75 × 10−8 per site per generation and a generation time of 15 years, which can be used to convert the scaled times and population sizes into real ones. Final results were visualized with the psmc_plot.pl function in PSMC.
2.9. Genetic load and deleterious mutationsThe ancestral sequence was constructed based on five genomes, Corylus heterophylla, C. mandshurica, C. avellana, C. americana, and C. chinensis (this study). Freebayes v.1.3.1 (Garrison and Marth, 2012) was used to call genotypes using the parameter "–report-monomorphic". Based on an empirical Bayesian strategy, IQ-tree v.1.6.12 (Nguyen et al., 2015) was then employed to reconstruct the ancestral state for each of the 11 chromosomes. The crown sequences from these related genomes were used to construct ancestral chromosome sequences. For genetic load analysis, we first employed SnpEff v.4.3 (Cingolani et al., 2012) to annotate our filtered VCF files. Then, deleterious mutations were identified using the Sorting Intolerant From Tolerant (SIFT) algorithm (Sim et al., 2012). A SIFT scoring database was created using ancestral chromosome sequences so that differences only occurred between ancestral and derived alleles, thus avoiding reference bias (Kono et al., 2016). The TrEMBL plant database was used to search for orthologous genes (Boeckmann et al., 2003). After polarization, SIFT4G (Vaser et al., 2016) was used to categorize coding sequence variants into synonymous (SYN), tolerated (TOL) (SIFT score ≥ 0.05), deleterious (DEL) (SIFT score < 0.05), and loss of function (LOF) (variants with the gain of a stop codon). The low-confidence and 'NA' sites were discarded. To evaluate genetic load within each lineage, we counted the heterozygous, homozygous, and derived alleles for SYN, TOL, DEL, and LOF in each sample.
2.10. Genomic signatures of selection and local adaptationTo detect genomic signatures potentially under selection in Corylus chinensis and CKY, we performed selective sweep analyses across all lineages (SW vs N) based on three genome-wide metrics, including diversity ratio (π ratio), differentiation index (FST), and cross-population composite likelihood ratio test (XP-CLR). We first scanned the genome for target regions under selection using π ratio and FST strategies (100-kb sliding windows and 10-kb steps), as implemented in VCFtools v.0.1.13 (Danecek et al., 2011). For XP-CLR analysis, we used a python module to compute the XP-CLR values of two lineages based on VCF files. The mean likelihood scores were calculated with the same sliding windows and step sizes as π ratio and FST. As recommended by Ma et al. (2021b), genomic regions in the top 5% of the three metrics were considered as putative selective sweeps, and selected genes shared by all the three methods were identified as candidate genes.
2.11. Introgression detection between Corylus chinensis and CKYTo test introgression patterns between Corylus chinensis and CKY and especially among different lineages, we implemented Patterson's D-statistic test and related admixture fraction estimates (f4-ratio statistics) using Dsuite v.0.5-r53 (Patterson et al., 2012). We performed multiple D-statistic tests using a four-taxon fixed model: (((P1, P2) P3) O), where P1 and P2 represented the sister lineages of C. chinensis or CKY, P3 indicated either the southwestern or northern lineage of two species, and C. mandshurica was used as an outgroup (O). Significant signal of introgression occurs between P1 and P3 with a D value < 0 and |Zscore| > 3, and between P2 and P3 with D value > 0 and |Zscore| > 3 (Durand et al., 2011). Because different trios of ((P1, P2), P3) share branches on the phylogeny and may lead to highly correlated f values, we hence estimated f-branch statistics for each phylogenetic branch using the Fbranch program.
For trios with significant D-statistics, we further calculated fd and its modified fdM statistics (Martin et al., 2015) using a sliding window method, which have been shown to be more useful in locating introgressive loci in small genomic regions compared to D-statistics. fd and fdM values were estimated with the python script ABBABABAwindows.py (https://github.com/simonhmartin/genomics_general) by setting a window size of 100 kb and a step size of 10 kb. For windows of D < 0, fd statistics will become noisy or meaningless, we thus defined introgressive regions based on fdM values as it is a modified index of fd and is symmetrically distributed near zero under the null hypothesis of no introgression. The windows with the 5% top fdM values were regarded as candidate introgressive regions.
3. Results 3.1. De novo assembly and genome assessmentA mature Corylus chinensis tree growing in the Huoditang Forestry District was selected for whole-genome sequencing (Fig. 1A–D). We obtained 24.36 Gb clean data (~87 million reads) for Illumina short reads (Table S1). Genome size was estimated to be approximately 365.64 Mb based on 17-mer analysis, with 1.16% heterozygosity and 68-fold coverage (Fig. S1 and Table S2). To obtain a high-quality genome, 18.83 Gb (50×) PacBio HiFi long reads (~1.5 million reads) were generated and used for genome assembly (Table S1). After deduplication, a contig-level assembly of 366.28 Mb spanning 367 contigs was produced with an N50 of 6.58 Mb (Table 1). Using Hi-C data (~44.06 Gb, 123×) (Fig. 1E and Table S1), up to 98.04% of the sequences were anchored onto 11 pseudo-chromosomes (Fig. 1f). The final assembly was 359.09 Mb and comprised 11 pseudo-chromosomes (Scaffold N50: 31.16 Mb) and 125 scaffolds (Table 1). Chromosomes were numbered from Chr01 to Chr11 based on their length, ranging from 47.04 to 19.56 Mb (Table S3).
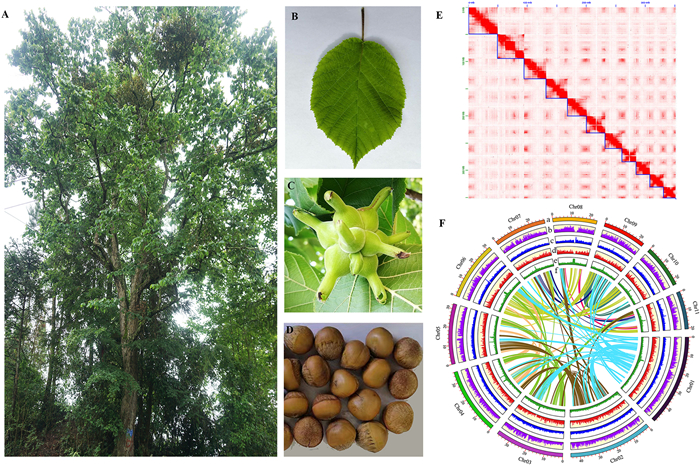
|
| Fig. 1 Morphological characteristics and genome information of Corylus chinensis. (A) Wild mature tree. (B) Mature leaf. (C) Fruits with bracted involucres. (D) Mature seeds. (E) Hi-C chromatin interaction heatmap for 11 pseudochromosomes in the C. chinensis genome, blue squares indicate chromosomes. (F) Circos plot showing the genomic features of C. chinensis from outer to inner (a–f): (a) 11 chromosomes featured in 10-Mb intervals across the genome; (b) SNP density; (c) repeat element density; (d) gene density; (e) GC content (f) intraspecific collinear blocks. |
| Assembly feature | Value |
| Total assembly size | 366.28 Mb |
| Total anchored size | 359.09 Mb |
| GC content | 37% |
| Total contig length | 384,074,399 bp |
| Number of contigs | 367 |
| Contig N50 size | 6,899,816 bp |
| Contig N90 size | 1,811,484 bp |
| Average contig length | 1,046,524 bp |
| Total scaffolds length | 376,540,788 bp |
| Number of scaffolds | 136 |
| Scaffold N50 size | 32,670,980 bp |
| Scaffold N90 size | 20,506,276 bp |
| Average scaffold length | 2,768,682 bp |
| Repetitive elements | 46.26% |
| BUSCO | C: 98.9% [S: 96.7%, D: 2.2%], F: 0.6%, M: 0.5%, n: 1614a |
| LAIb | 21.37 |
| a C: Complete BUSCO, S: Complete and single-copy BUSCO, D: Complete duplicated BUSCO, F: Fragmented BUSCO, M: Missing. n: Total BUSCO groups searched. b LAI, long terminal repeat (LTR) assembly index. |
|
BUSCO assessment showed that 98.9% of conserved genes (1614) could be completely annotated, of which 96.7% were single-copy, 2.2% were duplicated, 0.6% were fragmented, and 0.5% were missing (Tables 1 and S4). Genome completeness was also assessed by mapping all Illumina reads and transcriptome transcripts onto the Corylus chinensis genome, resulting in a mapping rate of 97.86% and 98.68%, respectively (Table S5). The genome had a high long terminal repeat (LTR) index (LAI = 21.37). These results collectively indicate that the genome is nearly complete and of high quality.
3.2. Repeat elements and gene annotationIntegrating de novo and homology-based approaches, we identified 166.09 Mb (46.26%) of the genome as transposable elements (Table S6). Among these repetitive sequences, long terminal retrotransposons (LTRs) emerged as the most prevalent, constituting 19.63% of the genome, with Gypsy and Copia elements making up 5.49% and 9.48%, respectively. Interspersed nuclear elements (LINEs and SINEs) comprised 0.12% of the genome. DNA transposons accounted for 22.75%, of which TIRs and Non-TIRs (Helitrons) represented 19.9% and 2.85%, respectively. A total of 25,965 genes were annotated using a combination of de novo, homology and transcriptome-based methods, with average lengths of transcripts, coding sequences, exons, and introns determined as 5745.19 bp, 1400.78 bp, 483.59 bp, and 728.14 bp, respectively. On average, there were 5.34 exons per gene (Table S7). Overall, 25,082 protein-coding genes (96.60%) were functionally annotated from six protein databases: GO (55.96%), KEGG (28.41%), KOG (79.94%), Nr (94.06%), Pfam (82.75%), and Swissprot (67.28%) (Table S8).
3.3. Comparative genomic analysesWe identified 27,751 orthologous groups, of which 5653 were species-specific and 47 were unique to Corylus chinensis. C. chinensis shared a large number of orthogroups with congeneric species, namely C. americana (14,689), C. mandshurica (14,669), C. avellana (14,443), and C. heterophylla (14,435), indicating their close genetic relatedness. Altogether, 13,027 orthogroups were shared among the five Corylus species while 192 orthogroups (611 genes) were exclusive to C. chinensis (Fig. 2A; Data S1). In C. chinensis, 371 gene families underwent substantial expansion, whereas 998 gene families experienced contraction (Fig. 2B). GO enrichment analysis demonstrated that the expanded gene families (2349 genes) primarily functioned in 'response to wounding,' 'response to stress and stimulus,' 'defense response,' and 'secondary metabolic processes' (Fig. 2C), which may be associated with the adaptation of C. chinensis to the subtropical alpine environment.
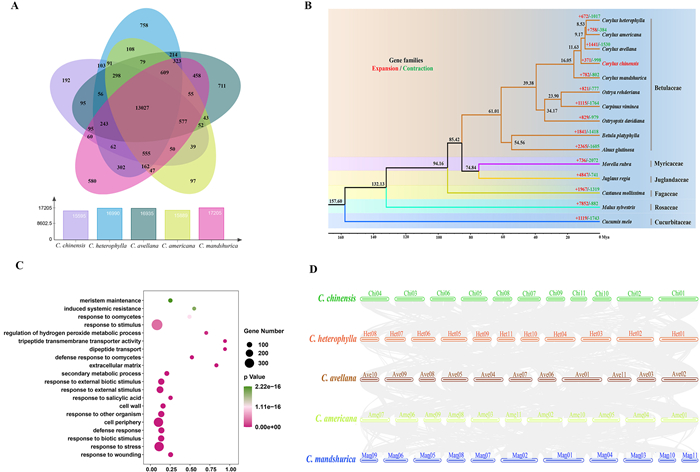
|
| Fig. 2 Genome evolution and comparative analysis of the Corylus chinensis genome and evolutionary analyses. (A) Venn diagram of C. chinensis and the other four Corylus genomes. Each number represents orthogroups shared between species. (B) Phylogenetic relationships, divergence time, and gene family expansions and contractions. The number above each branch represents the divergence time of each species in millions of years. The red and green values at the node represent the expanded and contracted gene families, respectively. (C) GO enrichment of significantly expanded gene families. (D) Macro-synteny patterns between C. chinensis and four closely related Corylus genomes. |
Based on 653 single copy orthologs shared by 15 species, the ML tree revealed that Betulaceae diverged from the common ancestor of Myricaceae and Juglandaceae at approximately 85.42 Ma. Within Betulaceae, Corylus occupied the basal position and formed sister to (Ostryopsis, (Carpinus, Ostrya)). Within Corylus, C. chinensis was located between C. manshurica and C. avellana, with the divergence occurring at around 11.63 Ma (Fig. 2B). Macro-synteny analysis identified 4457 intra-specific collinear genes within C. chinensis, which were relatively few and disorderly arranged, covering 17.17% of all genes. Inter-specific synteny indicated that the percentage of collinear genes between C. chinensis and other four Corylus genomes was 64.37%, 64.17%, 69.87%, and 62.82%, respectively (Table S9). Despite the presence of extensive collinearity and conserved gene order across all genomes, C. chinensis exhibited evident rearrangements and inversions of syntenic blocks throughout most chromosomes (Fig. 2D).
3.4. Genome variants and population structureWe performed re-sequencing for 47 Corylus chinensis accessions from 12 natural populations and 44 CKY accessions from ten natural populations (Fig. 3A). Population resequencing yielded an average of 148,791,919 clean reads per accession for C. chinensis and 84,205,090 for per accession for the CKY. As expected, C. chinensis had a higher mapping rate than did the CKY, with about 96.41% (93.08–97.72%) of C. chinensis sequence reads and 88.38% (86.85–91.52%) of CKY sequence reads accurately aligned to the reference genome. The average sequencing depth was 58.2× and 31.3× in C. chinensis and CKY accessions, respectively. All accessions exhibited high coverage rates (99.93% for C. chinensis and 99.70% for CKY) (Table S10). Employing a stringent filtering criterion, we ultimately identified 8,968,580 and 7,715,928 high-quality SNPs across 47 C. chinensis and 44 CKY accessions, respectively. We further generated LD-pruned datasets for both species, which included 975,254 and 1,194,659 SNPs, respectively.
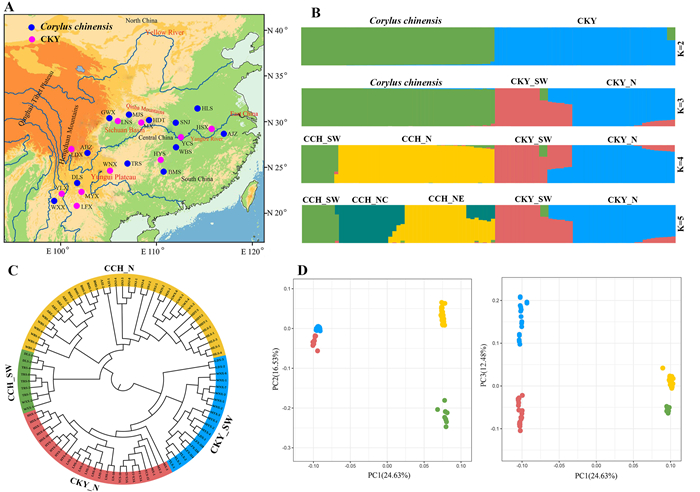
|
| Fig. 3 Spatial genetic structure of Corylus chinensis and the C. kwechowensis–C. yunnanensis complex (CKY). (A) Map showing the distribution of the 22 populations used in this study. C. chinensis and CKY are indicated as blue and pink, respectively. (B) Population structure inferred by admixture analysis for C. chinensis and CKY, with the number of clusters (K) ranging from 2 to 5. Different colors represent different genetic lineages derived from C. chinensis and CKY. (C) Phylogenetic analysis of C. chinensis and CKY. Color coding of the branches reflects the structure of genetic groups at K = 4. (D) PCA showing the first three components and the percentage of variation explained by each component. |
ADMIXTURE analyses revealed significant genetic segregation between Corylus chinensis and CKY when K = 2, although there was some degree of genetic admixture (Fig. 3B). At K = 3, CKY evolved into two subgroups, the southwestern and northern lineages (hereafter CKY_SW and CKY_N). At K = 4, ADMIXTURE determined four optimal clusters based on CV errors (Fig. S2), and C. chinensis was also divided into southwestern and northern lineages (hereafter CCH_SW and CCH_N). When K = 5, CCH_N further split into regional subclusters from Central and Northeast China (CCH_NC and CCH_NE), with adjacent populations showing genetic admixture to some extent. The NJ tree supported these patterns, dividing all accessions of C. chinensis and CKY into four clades: CCH_SW, CCH_N, CKY_SW, and CKY_N. Further population subclades were observed within each clade, corresponding to their geographical origin (Fig. 3C). PCA also confirmed the results above, with PC1 (24.63%) and PC2 (16.53%) distinguishing CKY, CCH_SW, and CCH_N, and PC1 (24.63%) and PC3 (12.48%) distinguishing C. chinensis, CKY_SW, and CKY_N (Fig. 3D).
3.5. Genome diversity, differentiation, and LD decayAcross the entire genomes, Corylus chinensis exhibited a mean nucleotide diversity (π) value of 8.87E-3, which was significantly lower than that of its widespread relative CKY (π = 11.68E-3). Among subgroups, CCH_SW (π = 7.16E-3) and CKY_SW (π = 9.89E-3) displayed slightly lower genome diversity than CCH_N (π = 7.58E-3) and CKY_N (π = 10.35E-3), respectively (Fig. 4A). When compared to an additional 18 threatened plants whose diversity information from genome resequencing was available, C. chinensis exhibited higher diversity than 15 species, and was similar to Litchi chinensis, but significantly lower than Pugionium dolabratum and Tetracentron sinense (Fig. 4B and Table S12). Among C. chinensis populations, the SNJ population showed the highest genetic diversity (π = 9.64E-3), whereas the TRS population displayed the lowest genetic diversity (π = 8.46E-3) (Table S11). The mean heterozygosity of C. chinensis was 0.0066 ± 0.0006, which was quite close to that of CKY (0.0067 ± 0.0005) (Table S10). Intraspecific differentiation revealed a mean FST value of 0.098 between CCH_SW and CCH_N, slightly lower than that between CKY_SW and CKY_N (0.102). The FST value between C. chinensis and CKY was 0.337, suggesting evident interspecific differentiation (Fig. 4A). The average Tajima's D value of CCH (1.06) was significantly higher than that of CKY (0.36), indicating a more severe population contraction in the former. The northern lineages of both species showed higher Tajima's D values (1.22, 1.23) compared to their southwestern counterparts (0.94, 1.06) (Fig. 4A). C. chinensis exhibited a lower linkage level and longer decay distance than CKY (2.4 kb vs 1.1 kb). The southwestern lineages of both species displayed extensively higher LD levels and slower decay rates, with 10.8 kb for C. chinensis and 2.9 kb for CKY, respectively. By contrast, the northern lineages showed similar LD levels and decay rates to their respective species (Fig. 4C).
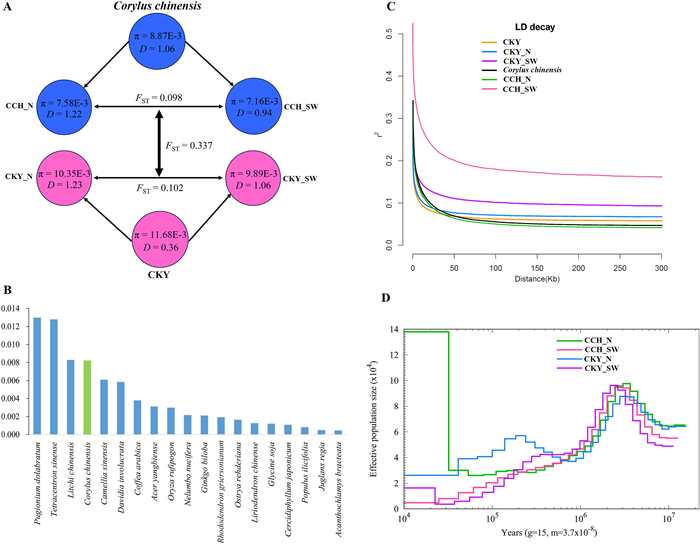
|
| Fig. 4 Comparison of genome diversity, linkage disequilibrium (LD) patterns and demographic histories between Corylus chinensis, the C. kwechowensis–C. yunnanensis complex (CKY), and their diverged lineages. (A) Nucleotide diversity (π), genetic divergence (FST) and Tajima's D (D) across C. chinensis, CKY, and four lineages. (B) Genome-wide sequence diversity for 18 tree species. (C) LD decay of C. chinensis, CKY, and four lineages, as measured by r2 against distance. (D) Demographic histories of C. chinensis, CKY, and four lineages, as indicated by changes in effective population size (Ne) through time inferred by PSMC. |
PSMC analysis revealed that the ancestral populations of both species experienced an expansion from 10 Ma to 3 Ma, followed by a dramatic bottleneck lasting until around 0.5 Ma, during which Ne decreased by approximately 66%. Thereafter, CCH_N maintained a stable population size until recently, whereas CKY_N underwent a population expansion (ca. 0.2 Ma) and subsequent decline. In contrast, both CCH_SW and CKY_SW experienced continuous population decline, with the Ne of CCH_SW decreasing to zero about 10,000 years ago (Fig. 4D).
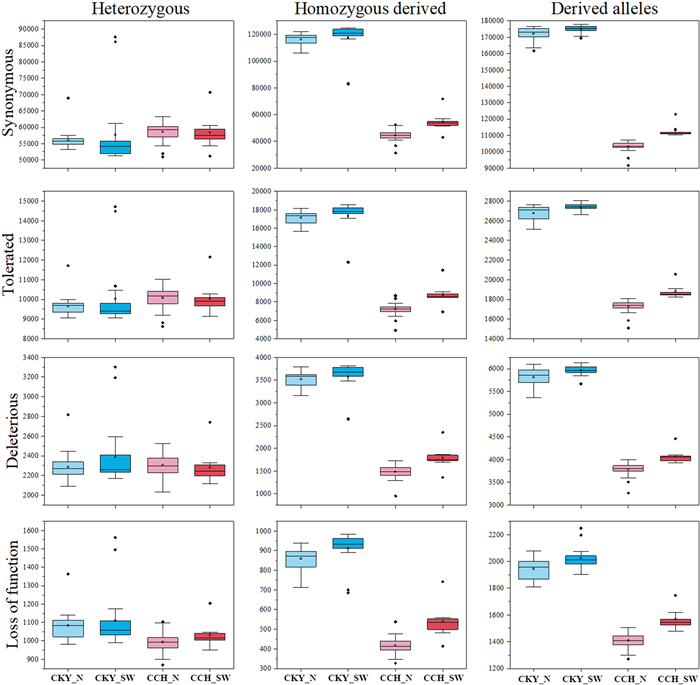
3.7. Genetic load, deleterious mutations, and inbreedingIn total, 11,127 genes (62,414 non-synonymous sites) were used to assess the functional effect of mutations (Data S2). As expected, the results demonstrated that CCH_N and CCH_SW carried equivalent levels of heterozygous mutations to CKY_N and CKY_SW for SYN, TOL, DEL, and LOF (Fig. 5 and Table S13), consistent with their similar heterozygosity (Table S10). However, CKY_N (mean = 34,406) and CKY_SW (mean = 34,796) accumulated significantly more homozygous sites than CCH_N (mean = 13,446) and CCH_SW (mean = 16,412) (Fig. 5 and Table S13). Consequently, the vast number of homozygous derived mutations resulted in extremely high number of derived alleles in CKY_N and CKY_SW. Specifically, CKY carried 30.5% more derived DEL alleles and 22.2% more derived LOF alleles than C. chinensis (Table S13). These results suggest that genetic load in C. chinensis is much lower compared to sympatric CKY. Functional annotation of these deleterious mutations suggested that the genes (952 genes) involved were mainly enriched in certain important biological processes, including macromolecule modification and metabolic processes, RNA modification, protein phosphorylation, and defense response (Data S3).
|
| Fig. 5 Comparison of genetic load between the four lineages of Corylus chinensis and the C. kwechowensis–C. yunnanensis complex (CKY). The number of derived alleles is based on counting each heterozygous genotype once and each homozygous-derived genotype twice. |
Inbreeding evaluation revealed that ROHs varied greatly across all accessions (Table S14). Corylus chinensis harbored significantly more short ROHs (100–500 kb, 94.93 Mb, 27.66% of the genome) and middle ROHs (500–1000 kb, 68.79 Mb, 20.04% of the genome) than CKY. Specifically, we detected long ROHs (> 1 Mb, 2.24 Mb, 0.65% of the genome) in C. chinensis, whereas this type of ROH was not found in CKY genome. Notably, CCH_SW showed the highest total number and length of ROHs, while CKY_N exhibited the lowest level in both indices. FROH was slightly higher in C. chinensis (0.13 ± 0.05) than in CKY (0.11 ± 0.04) (Fig. S3 and Table S14). The above results indicate that C. chinensis has a higher and more recent inbreeding level than CKY.
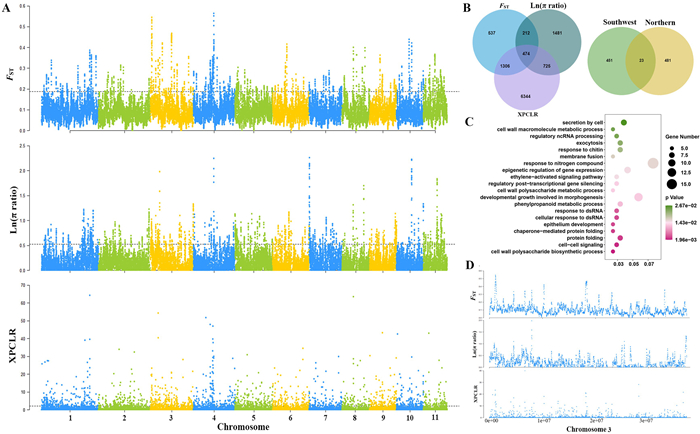
3.8. Genomic signatures of selection and local adaptationThree metrics (π ratio, FST, and XP-CLR) detected a total of 474 positively selected genes (PSGs) (108 genomic regions) in CCH_SW when compared CCH_N (Table S15). Among these PSGs, most were restricted to the selective sweeps of certain chromosomes, i.e., Chr11 (110 PSGs), Chr03 (106 PSGs), and Chr05 (87 PSGs) (Fig. 6A and B). GO enrichment indicated that these PSGs were mainly involved in cell wall polysaccharide biosynthetic process, cell–cell signaling, and protein folding (Fig. 6C; Data S4). In contrast, 504 PSGs (188 genomic regions) were identified in CCH_N (Fig. S4A and B; Table S15), and these PSGs were unevenly distributed from Chr03 (87 PSGs) to Chr10 (20 PSGs). The significantly enriched GO terms were related to tissue pattern formation, organ morphogenesis, transport and biosynthetic processes (Fig. S4C; Data S5). These findings indicate that selection could play an important role in selective sweeps on Chr03 for both lineages (Fig. 6D). Notably, only 23 PSGs were shared by CCH_SW and CCH_N (Fig. 6B), signs of their unique adaptive patterns.
|
| Fig. 6 Selective sweep signals and candidate positively selected genes (PSGs) in the southwestern lineage of Corylus chinensis (CCH_SW), with the northern lineage (CCH_N) as background. (A) Manhattan plots of three genome-wide metrices (FST, π ratio, and XP-CLR). The horizontal dashed lines indicate the significance threshold (5%) for selection signals. (B) Venn diagram of PSGs: the left indicates PSGs within CCH_SW identified by FST, π ratio, and XP-CLR methods, and the right indicates PSGs shared between CCH_SW and CCH_N. (C) GO enrichment scatter plot of PSGs in CCH_SW. (D) Selection signatures specific to Chr03. |
In CKY_SW, as many as 1703 PSGs were identified in 467 genomic regions, when compared to CKY_N (Fig. S5A and B; Table S15). Among these, Chr03, Chr02, and Chr06 had the largest number of PSGs, with 357,259, and 234 genes detected in the three chromosomes, respectively. In total, 261 GO terms were significantly enriched in categories such as macromolecule biosynthetic processes, regulation of gene expression, and developmental and metabolic processes (Fig. S5C; Data S6). By comparison, only 154 PSGs (41 genomic regions) were detected in CKY_N (Fig. S6A and B; Table S15), which participated chiefly in defense response, immune effector processes, and development (Fig. S6C; Data S7).
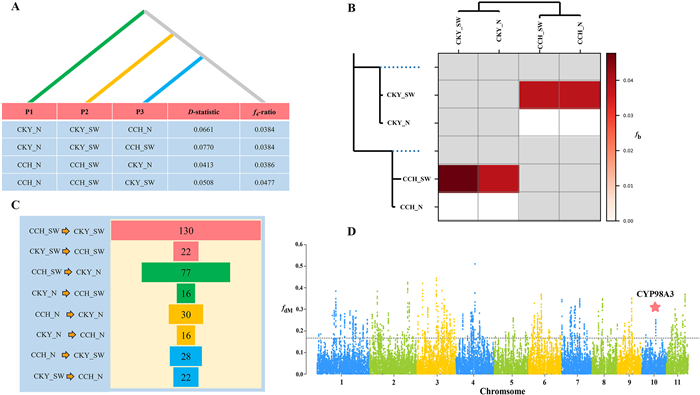
3.9. Genome-wide patterns of introgression between two speciesD-statistic and f4-ratio varied depending on the combination of trios, but all of them were significant with p < 0.001 and absolute Z-scores > 3 (Fig. 7A; Table S16), thus providing strong evidence of pervasive historical introgression between Corylus chinensis and CKY. Interestingly, we noted that interspecific introgression occurred mainly between the SW lineage of one species (C. chinensis or CKY) and two lineages (N and SW) of another species, whereas no significant introgression was detected between the N lineages of both species. To better illustrate introgression, we also estimated the f-branch statistic to assign introgression to specific internal branches on our phylogeny, which also revealed similar introgression patterns with those detected by D and f4 statistics. Particularly, CCH_SW and CKY_SW showed the highest introgression extent among all branches, when the trio was specified as ((CCH_N, CCH_SW), CKY_SW, O) (Fig. 7B).
|
| Fig. 7 Genome-wide introgression patterns between Corylus chinensis and the C. kwechowensis–C. yunnanensis complex (CKY). (A) Introgression detection measured by ABBA–BABA statistics. (B) The f-branch (fb) statistic identifies possible gene flow from the branch of the tree on the y axis to the species or lineages on the x axis. (C) The number and direction of adaptive introgression genes (AIGs) between pairwise lineages of C. chinensis and CKY. (D) Manhattan plot of fdM values calculated through a sliding windows analysis across genome. The horizontal dashed lines indicate the significance threshold (5%) for selection signals. The red star indicates two linked AIGs that are closely homologous to Arabidopsis CYP98A3, which is involved in secondary metabolite biosynthetic processes. |
We further calculated fd and its modified fdM statistics using a sliding window method, which has been shown to be more useful in locating introgressive loci in small genomic regions compared to D-statistics. Overall, the extent and proportion of genome-wide introgression estimated by fd and fdM varied greatly among different trios, ranging from 1.33% (688 genes) between CKY_N and CCH_SW to 4.34% (2150 genes) between CCH_SW and CKY_SW (Table S17). Furthermore, we found that CCH_SW and CKY_SW contributed dominantly to introgression in all trios, which can be indicated by the weakest introgression between CCH_N and CKY_N.
3.10. Adaptive introgression and functional significanceAdaptive introgressive regions were defined as those introgressive regions that overlap with the selective sweeps of the top 5% π ratio, FST, and XP-CLR (Table S15). Adaptive introgressive mainly occurred from CKY to Corylus chinensis whether in sympatric or allopatric lineages, with the largest number of adaptive introgressive genes (AIGs) detected from CKY_SW to CCH_SW (130 AIGs) (Fig. 7C). We also identified 77 AIGs that transferred from CKY_N to CCH_SW, of which 71 AIGs were commonly donated from CKY_SW and CKY_N to CCH_SW (Table S18). GO enrichment showed that the AIGs in CCH_SW were significantly enriched for secondary metabolite biosynthetic processes, regulation of response to water deprivation, and developmental growth (Data S8). Two linked AIGs, Hazelnut023377 and Hazelnut023378, that are closely homologous to Arabidopsis CYP98A3 (Fig. 7D) participated in secondary metabolite biosynthetic processes, such as coumarin, phenylpropanoid, lignin, and flavonoid metabolism and biosynthesis. These two AIGs may have promoted adaptive developmental growth in CCH_SW by regulating these secondary biological processes. Moreover, 28 and 30 AIGs were found to introgress from CKY_SW and CKY_N to CCH_N, respectively, and 28 AIGs were commonly shared (Table S18). These AIGs were mainly enriched for macromolecule biosynthetic processes, anatomical structure morphogenesis, and phosphorylation (Data S9). In contrast, adaptive introgression from C. chinensis to CKY was relatively weak, with only 22 and 16 AIGs determined to be donated from CCH to CKY_SW and CKY_N, respectively (Table S18).
4. Discussion 4.1. Phylogenetic status, genome evolution, and lineage divergenceCorylus chinensis is the sole macrophanerophyte species within Corylus. Due to the almost complete lack of genetic information on C. chinensis to date, its phylogenetic position has been debated for decades (Bassil et al., 2013; Erdogan and Mehlenbacher, 2000; Whitcher and Wen, 2001). Prior to this study, the only available phylogenetic information was from research based on nuclear ITS sequences that indicated C. chinensis was sister to C. colurna (Whitcher and Wen, 2001). However, a subsequent study based on plastome data revealed that C. chinensis was closely related to C. fargesii (Zhao et al., 2020). This cytonuclear discordance highlights the necessity of using whole genome information to accurately infer phylogenetic relationships within Corylus. Our phylogenomic analysis based on the newly sequenced C. chinensis genome and four published Corylus genomes confirms for the first time that C. chinensis is located between two shrub species complexes: the C. cornuta complex (represented by C. mandshurica) and the C. heterophylla complex (represented by C. heterophylla, C. americana, and C. avellana) (Fig. 2B). Phylogeny and morphological traits imply that C. chinensis has undergone a distinct evolutionary process compared to its close relatives. Here we found that 371 gene families have undergone significant expansion in C. chinensis. Furthermore, these gene families were functionally enriched to response to stress, stimulus, and defense, as well as function in secondary metabolic processes (Fig. 2C). Together, these biological functions may have contributed to the adaptive evolution and speciation of C. chinensis in subtropical alpine environments.
Population genomic analyses highlighted the genetic divergence between southwestern and northern lineages, which corroborates previous population structure assessments that used microsatellite markers (Lu, 2017). Similar differentiation patterns were also observed in the sympatric CKY. The distribution ranges of the two lineages correspond to the "Sino-Himalayan" and the "Sino-Japanese" floristic subkingdoms, respectively. The two subkingdoms are characterized by geographic barriers, environmental heterogeneity, and available connectivity, thus indicating the importance of isolation and selection in shaping genomic diversity in Corylus species. The finding that adaptive divergence is mediated by divergent selection has been reported in many plants, especially between the biogeographical boundaries of two floristic subkingdoms with relatively large ecological differences (Fan et al., 2013; Zhao et al., 2016). In our study, effective gene flow and local adaptation were detected between the southwestern and northern lineages for both C. chinensis and CKY (Fig. 3, Fig. 6, and S4–S6). Thus, divergent selection is more likely to be responsible for the divergence of these lineages in the face of gene flow. We also captured additional signals of subtle structure within CCH_N, i.e., CCH_NC and CCH_NE, consistent with local adaptation within this broader habitat. The subtle variations detected between these areas are crucial for the future survival of the species.
4.2. Maintenance of diversity and purification of deleterious variationsCompared to widespread species, the genetic diversity of endangered species is likely to be lower due to genetic drift and inbreeding depression (Allendorf et al., 2010; Setoguchi et al., 2011). In our study, the endangered Corylus chinensis did have lower genome diversity than its widespread relative CKY (Fig. 4A), but it also showed a much higher diversity than that of most other endangered plants (Fig. 4B). Moreover, the genome diversity of C. chinensis was similar to that of Betula pendula (π = 8.84E-3) (Salojärvi et al., 2017), although it was significantly higher than that of Ostrya chinensis (π = 2.79E-3) (Yang et al., 2018), which are two widespread species in the same family. Previous research used SSR and SRAP markers to show that the genetic diversity of C. chinensis was comparable to that of CKY (Chen, 2019). Our results generally support the point that some rare or endangered species can maintain high-level genetic diversity (Li et al., 2015; Stone et al., 2019; Liu et al., 2020).
Several factors might account for the maintenance of high levels of genetic diversity in endangered plant species such as Corylus chinensis. First, numerous studies have shown that genetic diversity decreases substantially due to temporary but severe reductions in Ne caused by reduced efficacy of natural selection and enhanced genetic drift (Jangjoo et al., 2016). This also seems plausible in C. chinensis and CKY, as the most severe bottleneck effect occurred at about 0.5 Ma (Fig. 4D), resulting in dramatic reductions in both Ne and genetic diversity. However, unlike the narrowly distributed southwestern lineages, the Ne of widespread northern lineages remained stable or even recovered through the LGM and the Holocene. Niche modeling also demonstrated that the distribution of C. chinensis has expanded continuously since the Holocene (He et al., 2022). Therefore, it is likely that the retained Ne during that time has guaranteed a baseline of genetic diversity. Second, genetic diversity is generally higher in outcrossing species than in selfing species (Nybom, 2004). Corylus species show relatively high self-incompatibility, which can effectively inhibit inbreeding and promote outcrossing. Thus, C. chinensis seems to have maintained high levels of heterozygosity and genetic diversity as a function of its breeding system. Third, research has shown that the genetic diversity Corylus species has increased due to incomplete reproductive isolation, which has led to widespread natural hybridization via the introduction of exogeneous genes into populations of related species (Zhao et al., 2020). In our study, we detected significant introgression signals, especially numerous AIGs from CKY to C. chinensis (Fig. 7 and Table S18). This finding indicates that adaptive introgression may play an important role in maintaining the stability and diversity of C. chinensis, as widely evidenced by population genomics of other plant species (Suarez-Gonzalez et al., 2017; Ma et al., 2019; Fu et al., 2022).
Endangered plants are generally more susceptible to inbreeding depression, as the probability of mating between relatives in small populations is high (Bortoluzzi et al., 2020). We compared the inbreeding levels of Corylus chinensis and CKY using ROH, with long and short ROHs accounting for recent and long-term inbreeding, respectively (Kirin et al., 2010; Hu et al., 2020; Yang et al., 2022b). In line with expectations, the results showed that C. chinensis had more ROHs than CKY at all three sizes (Fig. S3A), suggesting that inbreeding was much higher in C. chinensis than CKY in both historical and recent periods. Furthermore, the total number/length of ROHs and FROH were higher in CCH_SW and CKY_SW than in CCH_N and CKY_N, respectively, suggesting that southwestern lineages experienced more severe inbreeding and deserved greater conservation attention. Inbreeding also increases the genetic load of populations by accumulating more homozygous deleterious alleles (Marsden et al., 2016; Bortoluzzi et al., 2020). In small populations, genetic drift can reduce the strength of purifying selection, allowing deleterious variations to persist and become fixed in the population, leading to higher genetic loads (Yang et al., 2018; Ma et al., 2021b). Hence, a key issue arising from this research is understanding how this genetically constrained species survives, thrives, and successfully competes with taxa occupying similar niches. To clarify this question, we assessed the genetic loads in populations of C. chinensis and CKY by identifying putative missense mutations and loss of function. The results showed that CCH_SW, CCH_N, CKY_SW, CKY_N harbored comparable levels of heterozygous variants for SYN, TOL, DEL, and LOF sites (Fig. 5 and Table S13), consistent with their similar genetic diversity (Fig. 4A). Remarkably, C. chinensis carried 30.5% fewer derived DEL alleles and 22.2% fewer derived LOF alleles in a homozygous state than did CKY (Table S13), indicating that genetic load in C. chinensis is much lower than that of CKY. This phenomenon is largely due to more effective purging of highly deleterious mutations in C. chinensis, which may lead to a gradual decrease in inbreeding depression during long-term population declines. Similar patterns of purifying selection of deleterious mutations have also been observed in other endangered plants, such as Ostrya chinensis (Yang et al., 2018), Populus ilicifolia (Chen et al., 2020), and Rhododendron griersonianum (Ma et al., 2021a). Given the low genetic load and moderate diversity of C. chinensis, we can forecast its adaptability and resilience under natural conditions, if anthropogenic activities can be eliminated.
4.3. Adaptive divergence and interspecific introgressionEnvironmental heterogeneity between "Sino-Himalayan" and "Sino-Japanese" floristic subkingdoms can exert divergent selection on different lineages, thus promoting local adaptation. Despite population decline and inbreeding in Corylus chinensis, selection signatures suggested that genetic divergence between CCH_SW and CCH_N were locally structured (Figs. 6 and S4). We detected similar numbers of PSGs (474 vs 504) within CCH_SW and CCH_N, but these PSGs played different functional roles (Figs. 6C and S4C; Tables S19 and S20). These PSGs could, therefore, have contributed to their local adaptation to alpine (CCH_SW) and hygrothermal (CCH_N) environments, respectively. We also found evident functional differences in PSGs between intraspecific lineages of CKY (i.e., CKY_SW and CKY_N), and between interspecific sympatric lineages of C. chinensis and CKY (i.e., CCH_SW and CKY_SW, CCH_N and CKY_N) (Tables S21 and S22), implying that the genetic mechanisms underlying lineage divergence varied greatly, even if they displayed similar patterns of geographic differentiation.
A growing number of studies have demonstrated that introgression from other species may act as an important source of new genetic variation to facilitate adaptation of species to various environments (Khodwekar and Gailing, 2017; Miao et al., 2017; Suarez-Gonzalez et al., 2017). In Corylus, introgressive hybridization has been reported as a trigger for chloroplast capture, a special mechanism causing cytonuclear phylogenetic conflict (Zhao et al., 2020). C. chinensis and CKY, which are sympatric species in subtropical China, have a high possibility of interspecific introgression. Here, we elucidated for the first time genome-wide introgression patterns between these species. Specifically, introgression occurred asymmetrically between their sympatric and allopatric lineages. For instance, the proportion of introgression across the genome from CCH_SW to CKY_SW (4.34%) was the highest among all trios, whereas the reverse introgression from CKY_SW to CCH_SW was relatively low (2.93%) (Table S17). Selective sweep and sliding-windows introgression analyses identified some AIGs that played an important role in facilitating local adaptation across different lineages. Unexpectedly, although a large number of introgressions and selected genes were found in CKY_SW and CKY_N, only a few AIGs were involved in adaptive introgression (Table S18), indicating a low selection efficacy. In contrast, even with low introgression proportions (Table S17), CCH_SW and CCH_N fixed more AIGs than did CKY_SW and CKY_N (Table S18), suggesting a relatively high selection efficacy. This finding consistent with the fact that C. chinensis reduced deleterious variants through more effective purifying selection whereas CKY suffered a higher genetic load.
4.4. Implications for conservation and managementOverall, our findings provide the first comprehensive look at the demographic history of population collapse and the potential for future recovery of this endangered species. Despite its small population sizes and fragmented habitat, Corylus chinensis maintains relatively high genetic diversity, moderate genetic differentiation, and obvious population structure.
Given the current status, we recommend several conservation measures. First, the long-term survival of Corylus chinensis should be ensured through in situ conservation. In natural populations, saplings and seedlings are rarely found, and their habitats are frequently disturbed and destroyed by various factors, especially human activities and wild animals. Hence, it is imperative to establish conservation plots to protect the original habitats and natural ecosystem of C. chinensis. These plots will maintain and restore populations that can renew themselves naturally in their natural environments. Some representative populations of northern lineages (SNJ, AJZ, MJS, HLS) stem from national nature reserves, and their diversity is higher than the average level of C. chinensis. This indicates that in situ conservation can play an important role in protecting the genetic diversity of endangered C. chinensis populations.
Another important means for conservation of Corylus chinensis diversity is ex situ conservation. Ex situ conservation is especially important for populations whose habitats have been severely damaged or those that are unable to adapt to local climate changes. Our analysis of genome diversity, population stratification, and accumulation of deleterious alleles indicates that the narrow-ranged southwestern lineage (TRS, DLS, and WXX) represents a unique genetic resource that should be prioritized for protection and collection. Furthermore, we recommend establishing mini-reserve for the TRS population because it shows the lowest genetic diversity and does not belong to any nature reserve. Moreover, we found that the northern lineage evolved into two sub-lineages (central and northeast), which should be collected separately.
Finally, despite the conservation state of Corylus chinensis populations in several national nature reserves, inbreeding still occurs at a relatively high level. To avoid further inbreeding, habitat connectivity between different populations needs to be improved to enable gene exchange. If habitat connectivity and population linkages resulting from natural dispersal cannot be achieved quickly enough, other strategies need to be taken. In such cases, a translocation program should be adopted, especially those with high levels of inbreeding. We suggest adopting the rule of one-migrant-per-generation for conservation and management until stable gene flow is established (Wang, 2004). Notably, translocation between genetically similar populations should be avoided to minimize the negative effects of further inbreeding.
AcknowledgmentsWe sincerely thank Guowei Chen (Institute of Microbiology, Chinese Academy of Sciences) and Liya Dou (Beijing University of Chemical Technology) for their helpful discussions and guidance. This work was supported by the National Natural Science Foundation of China (Grant No. 32101541) and the National Key R&D Program of China (Grant No. 2022YFD2200400).
Data availability
All raw sequencing data from Illumina, PacBio HiFi, Hi-C, and RNA-seq have been deposited in the NCBI Genome and Sequence Read Archive, respectively, under BioProject PRJNA1039981, PRJNA1039955, PRJNA1039329, and PRJNA1039330. The genome assembly, annotation (gff), CDS and protein sequences were deposited in the figshare (10.6084/m9.figshare.24502669).
CRediT authorship contribution statement
Zhen Yang: Writing – review & editing, Writing – original draft, Visualization, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization. Lisong Liang: Writing – review & editing, Visualization, Resources, Formal analysis. Weibo Xiang: Writing – review & editing, Visualization, Methodology, Investigation. Lujun Wang: Writing – review & editing, Resources, Investigation. Qinghua Ma: Writing – review & editing, Supervision, Resources, Project administration, Investigation, Funding acquisition, Conceptualization. Zhaoshan Wang: Writing – original draft, Resources, Methodology, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2024.03.006.
Abascal, F., Corvelo, A., Cruz, F., et al., 2016. Extreme genomic erosion after recurrent demographic bottlenecks in the highly endangered Iberian lynx. Genome Biol., 17: 251. DOI:10.1186/s13059-016-1090-1 |
Alexander, D.H., Novembre, J., Lange, K., 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res., 19: 1655-1664. DOI:10.1101/gr.094052.109 |
Allendorf, F.W., Hohenlohe, P.A., Luikart, G., 2010. Genomics and the future of conservation genetics. Nat. Rev. Genet., 11: 697-709. DOI:10.1038/nrg2844 |
Bairoch, A., Apweiler, R., 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28: 45-48. DOI:10.1093/nar/28.1.45 |
Bao, W., Kojima, K., Kohany, 2015. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA, 6: 11. DOI:10.1186/s13100-015-0041-9 |
Bassil, N., Boccacci, P., Botta, R., et al., 2013. Nuclear and chloroplast microsatellite markers to assess genetic diversity and evolution in hazelnut species, hybrids and cultivars. Genet. Resour. Crop Evol., 60: 543-568. DOI:10.1007/s10722-012-9857-z |
Beech, E., 2018. Corylus chinensis. The IUCN Red List of Threatened Species. https://doi.org/10.2305/IUCN.UK.2018-1.RLTS.T32394A2817504.en e.T32394A2817504.
|
Beichman, A.C., Huerta-Sanchez, E., Lohmueller, K.E., 2018. Using genomic data to infer historic population dynamics of nonmodel organisms. Annu. Rev. Ecol. Evol. Syst., 49: 433-456. DOI:10.1146/annurev-ecolsys-110617-062431 |
Benazzo, A., Trucchi, E., Cahill, J.A., et al., 2017. Survival and divergence in a small group: the extraordinary genomic history of the endangered Apennine brown bear stragglers. Proc. Natl. Acad. Sci. U.S.A., 114: E9589-E9597. |
Bergman, C.M., Quesneville, H., 2007. Discovering and detecting transposable elements in genome sequences. Brief. Bioinform., 8: 382-392. DOI:10.1093/bib/bbm048 |
Birney, E., Clamp, M., Durbin, R., 2004. GeneWise and GenomeWise. Genome Res., 14: 988-995. DOI:10.1101/gr.1865504 |
Boeckmann, B., Bairoch, A., Apweiler, R., et al., 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31: 365-370. DOI:10.1093/nar/gkg095 |
Bortoluzzi, C., Bosse, M., Derks, M.F.L., et al., 2020. The type of bottleneck matters: insights into the deleterious variation landscape of small managed populations. Evol. Appl., 13: 330-341. DOI:10.1111/eva.12872 |
Burton, J.N., Adey, A., Patwardhan, R.P., et al., 2013. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol., 31: 1119. DOI:10.1038/nbt.2727 |
Camacho, C., Coulouris, G., Avagyan, V., et al., 2009. BLAST+: architecture and applications. BMC Bioinformatics, 10: 421. DOI:10.1186/1471-2105-10-421 |
Cammen, K.M., Andrews, K.R., Carroll, E.L., et al., 2016. Genomic methods take the plunge: recent advances in high-throughput sequencing of marine mammals. J. Hered., 107: 481-495. DOI:10.1093/jhered/esw044 |
Chen, S., Zhou, Y., Chen, Y., et al., 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34: i884-i890. DOI:10.1093/bioinformatics/bty560 |
Chen, Y.J., 2019. Genetic Diversity and Relationship Analysis of Hazelnut Germplasm Resources Based on SSR, SRAP Marker (Master thesis). Shenyang Agricultural University, Shenyang.
|
Chen, Z., Ai, F., Zhang, J., et al., 2020. Survival in the Tropics despite isolation, inbreeding and asexual reproduction: insights from the genome of the world's southernmost poplar (Populus ilicifolia). Plant J., 103: 430-442. DOI:10.1111/tpj.14744 |
Cheng, H., Concepcion, G.T., Feng, X., et al., 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods, 18: 170-175. DOI:10.1038/s41592-020-01056-5 |
Cingolani, P., Platts, A., Wang, L.L., et al., 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin), 6: 80-92. DOI:10.4161/fly.19695 |
Danecek, P., Auton, A., Abecasis, G., et al., 2011. The variant call format and VCFtools. Bioinformatics, 27: 2156-2158. DOI:10.1093/bioinformatics/btr330 |
De Bie, T., Cristianini, N., Demuth, J.P., et al., 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics, 22: 1269-1271. DOI:10.1093/bioinformatics/btl097 |
Dedato, M.N., Robert, C., Taillon, J., et al., 2022. Demographic history and conservation genomics of caribou (Rangifer tarandus) in Québec. Evol. Appl., 15: 2043-2053. DOI:10.1111/eva.13495 |
Dudchenko, O., Batra, S.S., Omer, A.D., et al., 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science, 356: 92. DOI:10.1126/science.aal3327 |
Durand, E.Y., Patterson, N., Reich, D., et al., 2011. Testing for ancient admixture between closely related populations. Mol. Biol. Evol., 28: 2239-2252. DOI:10.1093/molbev/msr048 |
Durand, N.C., Robinson, J.T., Shamim, M.S., et al., 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst., 3: 99-101. DOI:10.1016/j.cels.2015.07.012 |
Emms, D.M., Kelly, S., 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol., 20: 238. DOI:10.1186/s13059-019-1832-y |
Erdogan, V., Mehlenbacher, S.A., 2000. Phylogenetic relationships of Corylus species (Betulaceae) based on nuclear ribosomal DNA ITS region and chloroplast matK gene sequences. Syst. Bot., 25: 727-737. DOI:10.2307/2666730 |
Fan, D.M., Yue, J.P., Nie, Z.L., et al., 2013. Phylogeography of Sophora davidii (Leguminosae) across the 'Tanaka-Kaiyong Line', an important phytogeographic boundary in southwest China. Mol. Ecol., 22: 4270-4288. DOI:10.1111/mec.12388 |
Felsenstein, J., 1989. PHYLIP-Phylogeny Inference Package (version 3.2). Cladistics-Int. J. Willi Hennig Soc., 5: 164-166. |
Fu, R., Zhu, Y., Liu, Y., et al., 2022. Genome-wide analyses of introgression between two sympatric Asian oak species. Nat. Ecol. Evol., 6: 924-935. DOI:10.1038/s41559-022-01754-7 |
Funk, W.C., McKay, J.K., Hohenlohe, P.A., et al., 2012. Harnessing genomics for delineating conservation units. Trends Ecol. Evol., 27: 489-496. DOI:10.1016/j.tree.2012.05.012 |
Garrison, E., Marth, G., 2012. Haplotype-based variant detection from short-read sequencing. arXiv, 1207: 3907. |
Grabherr, M.G., Haas, B.J., Yassour, M., et al., 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol., 29: 644-652. DOI:10.1038/nbt.1883 |
He, X., Wenxu, M., Tiantian, Z., et al., 2023. Ecological differentiation and changes in historical distribution of Corylus heterophylla species complex since the last interglacial. J. Beijing For. Univ., 45: 11-23. |
He, X., Wenxu, M., Tiantian, Z., et al., 2022. Prediction of potential distribution of endangered species Corylus chinensis Franch. in climate change context. For. Res., 35: 104-114. |
Hohenlohe, P.A., Funk, W.C., Rajora, O.P., 2021. Population genomics for wildlife conservation and management. Mol. Ecol., 30: 62-82. DOI:10.1111/mec.15720 |
Hu, J.Y., Hao, Z.Q., Frantz, L., et al., 2020. Genomic consequences of population decline in critically endangered pangolins and their demographic histories. Natl. Sci. Rev., 7: 84-100. DOI:10.1093/nsr/nwz143 |
Hunter, S., Apweiler, R., Attwood, T.K., et al., 2009. InterPro: the integrative protein signature database. Nucleic Acids Res., 37: D211-D215. DOI:10.1093/nar/gkn785 |
Jangjoo, M., Matter, S.F., Roland, J., et al., 2016. Connectivity rescues genetic diversity after a demographic bottleneck in a butterfly population network. Proc. Natl. Acad. Sci. U.S.A., 113: 10914-10919. DOI:10.1073/pnas.1600865113 |
Jurka, J., Kapitonov, V.V., Pavlicek, A., et al., 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res., 110: 462-467. DOI:10.1159/000084979 |
Kanehisa, M., Sato, Y., Furumichi, M., et al., 2019. New approach for understanding genome variations in KEGG. Nucleic Acids Res., 47: D590-D595. DOI:10.1093/nar/gky962 |
Katoh, K., Standley, D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Khodwekar, S., Gailing, O., 2017. Evidence for environment-dependent introgression of adaptive genes between two red oak species with different drought adaptations. Am. J. Bot., 104: 1088-1098. DOI:10.3732/ajb.1700060 |
Kim, D., Langmead, B., Salzberg, S.L., 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods, 12: 357-360. DOI:10.1038/nmeth.3317 |
Kirin, M., McQuillan, R., Franklin, C.S., et al., 2010. Genomic runs of homozygosity record population history and consanguinity. PLoS One, 5: e13996. DOI:10.1371/journal.pone.0013996 |
Kono, T.J.Y., Fu, F., Mohammadi, M., et al., 2016. The role of deleterious substitutions in crop genomes. Mol. Biol. Evol., 33: 2307-2317. DOI:10.1093/molbev/msw102 |
Korneliussen, T.S., Albrechtsen, A., Nielsen, R., 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics, 15: 1-13. |
Li, H., 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv E-Prints. |
Li, H., Durbin, R., 2011. Inference of human population history from individual whole-genome sequences. Nature, 475: 493. DOI:10.1038/nature10231 |
Li, H., Handsaker, B., Wysoker, A., et al., 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25: 2078-2079. DOI:10.1093/bioinformatics/btp352 |
Li, M., Chen, S., Shi, S., et al., 2015. High genetic diversity and weak population structure of Rhododendron jinggangshanicum, a threatened endemic species in Mount Jinggangshan of China. Biochem. Syst. Ecol., 58: 178-186. DOI:10.3901/JME.2015.05.178 |
Lidgard, S., Love, A.C., 2018. Rethinking living fossils. BioScience, 68: 760-770. DOI:10.1093/biosci/biy084 |
Liu, B., Shi, Y., Yuan, J., et al., 2013. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Quant. Biol., 35: 62-67. |
Liu, D., Zhang, L., Wang, J., et al., 2020. Conservation genomics of a threatened Rhododendron: contrasting patterns of population structure revealed from neutral and selected SNPs. Front. Genet., 11: 757. DOI:10.1201/9781351187435-95 |
Lu, Z.Q., 2017. Species Delimitation in the Subfamily Coryloideae of Betulaceae in China (PhD thesis). Lanzhou University, Lanzhou.
|
Ma, H., Liu, Y., Liu, D., et al., 2021a. Chromosome-level genome assembly and population genetic analysis of a critically endangered rhododendron provide insights into its conservation. Plant J., 107: 1533-1545. DOI:10.1111/tpj.15399 |
Ma, Y., Chen, G., Edward Grumbine, R., et al., 2013. Conserving plant species with extremely small populations (PSESP) in China. Biodivers. Conserv., 22: 803-809. DOI:10.1007/s10531-013-0434-3 |
Ma, Y., Liu, D., Wariss, H.M., et al., 2022. Demographic history and identification of threats revealed by population genomic analysis provide insights into conservation for an endangered maple. Mol. Ecol., 31: 767-779. DOI:10.1111/mec.16289 |
Ma, Y., Wang, J., Hu, Q., et al., 2019. Ancient introgression drives adaptation to cooler and drier mountain habitats in a cypress species complex. Commun. Biol., 2: 213. DOI:10.1038/s42003-019-0445-z |
Ma, Y.P., Wariss, H.M., Liao, R.L., et al., 2021b. Genome-wide analysis of butterfly bush in three uplands provides insights into biogeography, demography and speciation. New Phytol., 232: 1463-1476. DOI:10.1111/nph.17637 |
Marçais, G., Kingsford, C., 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics, 27: 764-770. DOI:10.1093/bioinformatics/btr011 |
Marsden, C.D., Ortega-Del Vecchyo, D., O'Brien, D.P., et al., 2016. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl. Acad. Sci. U.S.A., 113: 152-157. DOI:10.1073/pnas.1512501113 |
Martin, S.H., Davey, J.W., Jiggins, C.D., 2015. Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol. Biol. Evol., 32: 244-257. DOI:10.1093/molbev/msu269 |
McKenna, A., Hanna, M., Banks, E., et al., 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20: 1297-1303. DOI:10.1101/gr.107524.110 |
Miao, B., Wang, Z., Li, Y., 2017. Genomic analysis reveals hypoxia adaptation in the Tibetan Mastiff by introgression of the gray wolf from the Tibetan Plateau. Mol. Biol. Evol., 34: 734-743. |
Nguyen, L.T., Schmidt, H.A., Arndt, V.H., et al., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol., 32: 268-274. DOI:10.1093/molbev/msu300 |
Nybom, H., 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol., 13: 1143-1155. DOI:10.1111/j.1365-294X.2004.02141.x |
Patterson, N., Moorjani, P., Luo, Y., et al., 2012. Ancient admixture in human history. Genetics, 192: 1065. DOI:10.1534/genetics.112.145037 |
Pigg, K.B., Manchester, S.R., Wehr, W.C., 2003. Corylus, Carpinus, and Palaeocarpinus (Betulaceae) from the middle Eocene Klondike Mountain and Allenby Formations of northwestern North America. Int. J. Plant Sci., 164: 807-822. DOI:10.1086/376816 |
Pimm, S.L., Jenkins, C.N., Abell, R., et al., 2014. The biodiversity of species and their rates of extinction, distribution, and protection. Science, 344: 1246752. DOI:10.1126/science.1246752 |
Potter, S.C., Luciani, A., Eddy, S.R., et al., 2018. HMMER web server: 2018 update. Nucleic Acids Res., 46: W200-W204. DOI:10.1093/nar/gky448 |
Price, A.L., Jones, N.C., Pevzner, P.A., 2005. De novo identification of repeat families in large genomes. Bioinformatics, 21: i351-i358. DOI:10.1093/bioinformatics/bti1018 |
Purcell, S., Neale, B., Todd-Brown, K., et al., 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81: 559-575. DOI:10.1086/519795 |
Roach, M.J., Schmidt, S.A., Borneman, A.R., 2018. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics, 19: 460. DOI:10.1186/s12859-018-2485-7 |
Salojärvi, J., Smolander, O.P., Nieminen, K., et al., 2017. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet., 49: 904-912. DOI:10.1038/ng.3862 |
Setoguchi, H., Mitsui, Y., Ikeda, H., et al., 2011. Genetic structure of the critically endangered plant Tricyrtis ishiiana (Convallariaceae) in relict populations of Japan. Conserv. Genet., 12: 491-501. DOI:10.1007/s10592-010-0156-y |
Sim, N.L., Kumar, P., Hu, J., et al., 2012. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res., 40: W452-W457. DOI:10.1093/nar/gks539 |
Simão, F.A., Waterhouse, R.M., Panagiotis, I., et al., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics: 3210-3212. DOI:10.1093/bioinformatics/btv351 |
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Stanke, M., Keller, O., Gunduz, I., et al., 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res., 34: W435-W439. DOI:10.1093/nar/gkl200 |
Stone, B.W., Ward, A., Farenwald, M., et al., 2019. Genetic diversity and population structure in Cary's Beardtongue Penstemon caryi (Plantaginaceae), a rare plant endemic to the eastern Rocky Mountains of Wyoming and Montana. Conserv. Genet., 20: 1149-1161. DOI:10.1007/s10592-019-01204-1 |
Suarez-Gonzalez, A., Hefer, C.A., Lexer, C., et al., 2017. Introgression from Populus balsamifera underlies adaptively significant variation and range boundaries in P. trichocarpa. New Phytol., 217: 416. |
Sun, W.B., Ma, Y.P., Blackmore, S., 2019. How a new conservation action concept has accelerated plant conservation in China. Trends Plant Sci., 24: 4-6. DOI:10.1016/j.tplants.2018.10.009 |
Talavera, G., Castresana, J., 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol., 56: 564-577. DOI:10.1080/10635150701472164 |
Tempel, S., 2012. Using and understanding repeat masker. In: Bigot, Y. (Ed.), Mobile Genetic Elements: Protocols and Genomic Applications Methods in Molecular Biology. Humana Press, Totowa, NJ, pp. 29–51.
|
Vaser, R., Adusumalli, S., Leng, S.N., et al., 2016. SIFT missense predictions for genomes. Nat. Protoc., 11: 1-9. DOI:10.1038/nprot.2015.123 |
Wang, J., 2004. Application of the one-migrant-per-generation rule to conservation and management. Conserv. Biol., 18: 332-343. DOI:10.1111/j.1523-1739.2004.00440.x |
Wei, X.Z., Dong, H.E., Jiang, M.X., et al., 2009. Characteristics of riparian rare plant communities on the Shennongjia Mountains, Central China. J. Wuhan Bot. Res., 27: 607-616. |
Werth, Alexander J., Shear, William A., 2014. The evolutionary truth about living fossils. Am. Sci., 102: 434-443. DOI:10.1511/2014.111.434 |
Whitcher, I.N., Wen, J., 2001. Phylogeny and biogeography of Corylus (Betulaceae): inferences from ITS sequences. Syst. Bot., 26: 283-298. |
Xu, Z., Wang, H., 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res., 35: W265-W268. DOI:10.1093/nar/gkm286 |
Yang, F., Cai, L., Dao, Z., et al., 2022a. Genomic data reveals population genetic and demographic history of Magnolia fistulosa (Magnoliaceae), a plant species with extremely small populations in Yunnan Province, China. Front. Plant Sci., 13: 811312. DOI:10.3389/fpls.2022.811312 |
Yang, J., Lee, S.H., Goddard, M.E., et al., 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet., 88: 76-82. DOI:10.5815/ijeme.2011.06.12 |
Yang, L., Wei, F., Zhan, X., et al., 2022b. Evolutionary conservation genomics reveals recent speciation and local adaptation in threatened Takins. Mol. Biol. Evol., 39: msac111. DOI:10.1093/molbev/msac111 |
Yang, Y., Ma, T., Wang, Z., et al., 2018. Genomic effects of population collapse in a critically endangered ironwood tree Ostrya rehderiana. Nat. Commun., 9: 5449. DOI:10.1038/s41467-018-07913-4 |
Yang, Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol., 24: 1586-1591. DOI:10.1093/molbev/msm088 |
Zhang, C., Dong, S.S., Xu, J.Y., et al., 2019. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics, 35: 1786-1788. DOI:10.1093/bioinformatics/bty875 |
Zhao, J.L., Gugger, P.F., Xia, Y.M., et al., 2016. Ecological divergence of two closely related Roscoea species associated with late Quaternary climate change. J. Biogeogr., 43: 1990-2001. DOI:10.1111/jbi.12809 |
Zhao, T., Wang, G., Ma, Q., et al., 2020. Multilocus data reveal deep phylogenetic relationships and intercontinental biogeography of the Eurasian-North American genus Corylus (Betulaceae). Mol. Phylogenet. Evol., 142: 106658. DOI:10.1016/j.ympev.2019.106658 |



