b. Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Science, Kunming, 650201, China;
c. Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, 650201, Yunnan, China;
d. Department of Export Agriculture, Faculty of Animal Science and Export Agriculture, Uva Wellassa University, Badulla, 90000, Sri Lanka;
e. Department of Life Sciences, School of Science and Computing, South Eastern Kenya University, 170-90200, Kitui, Kenya;
f. Department of Biology, Columbus State University, University System of Georgia, Columbus, GA, 31907-5645, USA;
g. Royal Botanic Garden Edinburgh, 20A Inverleith Row, Edinburgh, EH3 5LR, Scotland, UK;
h. Lijiang Forest Biodiversity National Observation and Research Station, Kunming Institute of Botany, Chinese Academy of Sciences, Lijiang, 674100, Yunnan, China
The tea plant (Camellia sinensis (L.) O. Kuntze) is the first documented tree crop in China, with more than 3000 years of domestication history (Yamanishi, 1995). The tea plant was initially used as medicine nearly 5000 years ago and later as a beverage (Yamanishi, 1995; Mondal, 2009). Recent discoveries provide clear evidence of China’s long history of using and cultivating tea plants. For example, the first evidence of tea use was found in the Han Yangling Mausoleum in Xi’an, Shaanxi Province (dating 2100 years ago) and the Guargyam Cemetery in the Ngari District, Xizang Autonomous Region (Tibet) (dating 1800 years ago) (Lu et al., 2016). Although it is generally accepted that the tea plant was first domesticated in China, details of its origin and domestication remain unclear and controversial (Meegahakumbura et al., 2018a, 2018b; Wambulwa et al., 2021; Zhang et al., 2021). Stuart (1919) speculated that the place of origin of the tea plant lies in the mountain range between Yunnan Province in southwest China and Assam in Northeast India, whereas Kingdon-Ward (1950) postulated the origin of the tea plant to be in the area where Indo-Burma, Xizang, and Yunnan meet. More recently, it was proposed that Yunnan Province, China could be a possible domestication center (Yu and Chen, 2001); this area encompasses the long, narrow region between Wenshan and Honghe (Yu, 1986), as well as the Wenshan (primary center) and Xishuangbanna (secondary center) regions (Muramatsu, 1991).
The systematic cultivation of Chinese Assam tea in Yunnan is thought to have started during the Tang Dynasty (618–907 AD) (Zhou, 2004). The Puer region of Yunnan became a world-renowned tea trading center during the Song (960–1279 AD) and Yuan (1279–1368 AD) dynasties (Chen, 1986). Trading of the fermented Puer tea along the Tea Horse Road from Yunnan to Xizang flourished during the Tang Dynasty (618–907 AD) (Freeman and Ahmed, 2010) and was followed by a boom in the tea industry during the Qing Dynasty (1644–1911 AD) (Zhou, 2004). There is evidence that the domestication history of Assam tea in Yunnan coincided with the use and domestication of wild non-tea species (e.g., Camellia taliensis and C. crassicolumna), and that the latter might have been used directly in the preparation of beverage tea (Ming, 2000), or became incorporated into the germplasm of Chinese Assam tea through natural and selective hybridization (Zhao et al., 2014; Li et al., 2015; Meegahakumbura et al., 2018a, 2018b; Zhang et al., 2021). Therefore, it is essential to include close wild relatives of the tea plant when exploring the domestication history of Chinese Assam tea. Given that previous studies were based on relatively few populations/individuals within the region (Ji et al., 2011; Yao et al., 2012; Li et al., 2015; Wang et al., 2020; Zhang et al., 2020, 2021; Lu et al., 2021), a more comprehensive assessment is required to determine the degree of genetic partitioning within and among populations of Chinese Assam tea and the extent of past hybridization/introgression with wild congeners.
Based on a large-scale sampling and genetic analysis of tea plants, three independent domestication centers have been suggested: 1) China tea (Camellia sinensis var. sinensis) in South China, 2) Chinese Assam tea in Yunnan, Southwest China, and 3) Indian Assam tea in Assam, India (Meegahakumbura et al., 2016; 2018a). The independent domestication of Chinese Assam tea, in particular, was recently supported by genomic analysis (Li et al., 2021; Rawal et al., 2021). In addition, there is evidence that Chinese Assam tea may represent a new genetic lineage distinct from the Assam tea of India (Indian Assam tea) based on SSR data (Meegahakumbura et al., 2016, 2018a, 2018b). Although genomic studies on Chinese Assam tea cultivars and close wild relatives (Xia et al., 2017; Wang et al., 2020) and genome resequencing (Wang et al., 2020; Lu et al., 2021; Zhang et al., 2021) have shed some light on the domestication and breeding history of Chinese Assam tea, the findings have also generated interesting new questions. For instance, no information exists so far on the genetic diversity, genetic relationships, and domestication/breeding history of ancient Chinese Assam tea germplasm of the Puer, Lincang, and Xishuangbanna areas (domestication center) in Yunnan province, China.
To explore the domestication/breeding history of Chinese Assam tea and its genetic relationships with close wild relatives, we genotyped 469 individuals (ancient tea plants and two wild species), including 381 Camellia sinensis var. assamica (26 populations), 72 C. taliensis (six populations), and 16 C. crassicolumna (three populations) individuals, based on 16 nuclear SSR loci. We aimed to address the following three questions: 1) What is the level of genetic diversity of the Chinese Assam tea and its two wild relatives? 2) What is the breeding history of Chinese Assam tea? 3) Is there evidence of past hybridization/introgression between Chinese Assam tea and its wild relatives? Our findings will provide new insights into the domestication history of Chinese Assam tea and will contribute baseline data to future tea breeding programs and germplasm conservation.
2. Materials and methods 2.1. Plant materialsA total of 381 ancient trees from 26 populations of Chinese Assam tea (Camellia sinensis var. assamica) were collected from old or abandoned tea gardens near villages across Southwest Yunnan, China (Fig. 1). For each population, we collected samples from 10 to 16 ancient trees that had a basal diameter of at least 20 cm. Based on historical records and traditional knowledge, most of the sampled individuals were estimated to be more than 100 years old, with some trees being estimated to be over 1000 years old (e.g., samples BW1, LBZ11 and NNS4). To explore the genetic relationships between the Chinese Assam tea and its wild relatives, 72 additional ancient C. taliensis trees (representing six populations) and 16 trees of C. crassicolumna (representing three populations) were also sampled in the region, bringing the total sample size to 469 individuals. Vouchers of each sampled tree were collected and deposited at the herbarium of the Kunming Institute of Botany (KUN), Chinese Academy of Sciences, Yunnan, China (Table S1; Fig. 1).
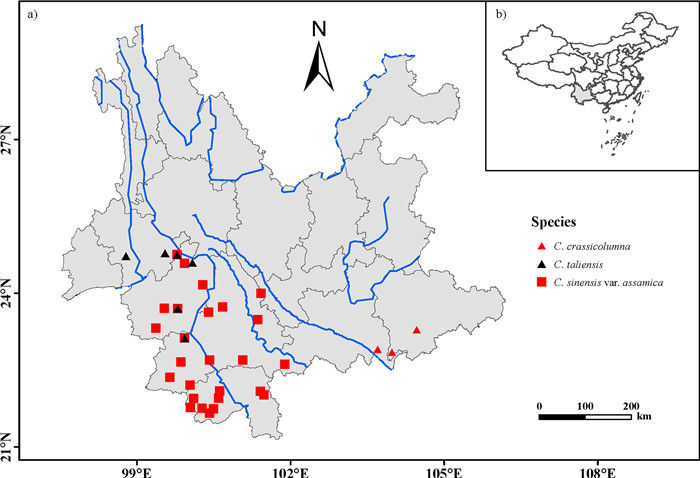
|
| Fig. 1 Geographical location of the 35 populations sampled here, including 26 populations of Camellia sinensis var. assamica (Chinese Assam type) ancient tea collected from Puer, Lincang, and Xishuangbanna, six C. taliensis wild tea populations collected from Puer and Lincang, and three C. crassicolumna wild tea populations collected from Honghe, Yunnan, China. Blue lines indicate the Mekong River and other rivers flowing through Yunnan province. |
Sixteen highly polymorphic microsatellite loci were selected from our previous study (Meegahakumbura et al., 2016) and used to genotype all 469 individuals. DNA extraction, PCR, fragment analysis, and data sorting protocols followed Wambulwa et al., 2016a, Wambulwa et al., 2016b. MICROCHECKER v.2.2.1 (Van Oosterhout et al., 2004) was used to examine the presence of null alleles and allele dropout. Indices of genetic diversity were estimated using GenAIEx v.6.5b4 (Peakall and Smouse, 2006), whereas gene diversity (HS) and inbreeding coefficients (FIS) were obtained using FSTAT v.2.9.3.2 (Goudet, 2002). The number of private alleles associated with each population (or region) was calculated in GenAIEx. An analysis of molecular variance (AMOVA) was carried out for all cultivated and wild populations using GenAIEx with 999 permutations.
To determine the population structure of Chinese Assam tea and its close wild relatives, we first performed Bayesian inference in STRUCTURE v.2.3.4 (Pritchard et al., 2000), as detailed in Meegahakumbura et al. (2016). The best K value was evaluated based on the delta K (ΔK) method (Evanno et al., 2005) and STRUCTURE Harvester v.0.6.94 (Earl and von Holdt, 2012). An individual was assigned to a particular cluster if its membership coefficient (Q value) for the cluster was greater than 0.8 (Q ≥ 0.8) (Diez et al., 2015), while individuals with Q values less than 0.8 were assigned to the admixture group. Further exploration of genetic structure was performed using Principal Co-ordinates Analysis (PCoA) on all 469 individuals in GenAIEx. The PCoA was first carried out on the 381 individuals belonging to the three genetic groups (Puer, Lincang, and Xishuangbanna) of Chinese Assam tea defined based on the STRUCTURE analysis results, the 72 individuals of C. taliensis, and the 16 individuals of C. crassicolumna. The analysis was then rerun at K = 4 with all admixture individuals excluded.
Genetic relationships among individuals of cultivated tea and their wild relatives were explored using the Nei’s distance measure (Nei et al., 1983) in UPGMA algorithm implemented in MSA v.4.05 (Dieringer and Schlӧ;tterer, 2003) and Phylip v.3.67 (Felsenstein, 2004). The UPGMA tree was visualized with FigTree v.1.3.1 (Rambaut, 2012), as described in Wambulwa et al., 2016a, Wambulwa et al., 2016b.
NewHybrids v.1.1 beta (Anderson and Thompson, 2002) was used to check the exchange of genetic material among individuals, as detailed in Wambulwa et al. (2016b). A total of 309 individuals selected from 26 ancient populations were used in the analysis. In the NewHybrids analysis, Lincang and Puer samples were used as parents, while all 165 individuals from Xishuangbanna were used as test samples. STRUCTURE results at K = 4 were used to identify pure individuals (Q ≥ 0.8) from these two gene pools (70 from Lincang and 74 from Puer), and these were used as parents in the NewHybrids analysis. Based on the NewHybrids analysis results, individuals were assigned to parental groups and distinct hybrid classes (F1, F2, BC1, and BC2) if their posterior probability was ≥ 0.8, and mixed hybrid (including genetic admixture of more than two gene pools) if their posterior probability < 0.8.
Additionally, a second NewHybrids analysis was carried out for 453 individuals belonging to the 31 ancient populations of both Chinese Assam tea and Camellia taliensis distributed in Puer, Lincang, and Xishuangbanna. As Chinese Assam tea and C. taliensis are sympatrically distributed in most of the tea gardens, the two taxa were used as parents for this NewHybrids analysis to determine the genetic exchange between the two species. Three populations of C. crassicolumna (16 individuals) were excluded from the analysis as its populations were collected from the Honghe area, far away from the main cultivating area (Puer, Lincang, and Xishuangbanna). Based on the results of this analysis, individuals were again assigned to parental, hybrid or backcross groups as explained above.
3. Results 3.1. Genetic diversity and partitioning of genetic variationIn our analysis of 469 ancient tea trees from 34 populations based on 16 nSSR markers, the proportion of missing data for the entire data set was less than 1.3% (Table S2). There were no null alleles and no allele dropouts detected in the nSSR data. The 16 primer pairs yielded 197 alleles with an average of 12.31 alleles per locus. Locus TUGMS2 135 had the highest number of alleles (19), while locus S87 had the lowest (7).
Overall, we found a moderately high gene diversity for ancient Chinese Assam tea (HS = 0.638), with the highest (HS = 0.673) and lowest (HS = 0.615) values being observed for the Puer and Lincang populations, respectively (Table 1). Camellia crassicolumna showed the highest gene diversity (HS = 0.658), while C. taliensis had the lowest (HS = 0.615). However, C. taliensis showed the highest indices of allelic diversity at the species level (Ar = 6.517; Ap = 8). Within the ancient Chinese Assam tea populations, Xishuangbanna populations showed the highest allelic diversity (Ar = 9.223; Np = 16), while populations from Lincang had the lowest (Ar = 8.1474; Np = 4) (Table 1).
| Group/Species | N | NA | NE | AR | NP | HO | HE | HS | F |
| CSAP | 121 | 9.313 | 4.07 | 9 | 9 | 0.604 | 0.705 | 0.673 | 0.154 |
| CSAX | 165 | 10.00 | 3.892 | 9.223 | 16 | 0.568 | 0.658 | 0.625 | 0.152 |
| CSAL | 95 | 8.19 | 3.448 | 8.147 | 4 | 0.561 | 0.632 | 0.617 | 0.150 |
| CSA | 381 | 11.063 | 4.139 | 6.5 | 33 | 0.578 | 0.688 | 0.638 | 0.178 |
| CT | 72 | 9.063 | 3.868 | 6.517 | 8 | 0.564 | 0.668 | 0.615 | 0.164 |
| CC | 16 | 6.313 | 3.996 | 6.217 | 7 | 0.565 | 0.718 | 0.658 | 0.216 |
| CSAP: Camellia sinensis var. assamica samples collected from Puer; CSAL: Camellia sinensis var. assamica samples collected from Lincang; CSAX: Camellia sinensis var. assamica samples collected from Xishuangbanna; CSA: C. sinensis var. assamica; CT: C. taliensis; CC: C. crassicolumna; N: Number of samples; NA: Number of alleles; NE: Number of effective alleles; AR: Allelic richness; NP: Private alleles; HO: Observed heterozygosity; HE: Expected heterozygosity; HS: Gene diversity; F: Inbreeding coefficient. | |||||||||
AMOVA based on the entire data set of C. sinensis var. assamica, C. taliensis and C. crassicolumna showed that 29.81% of the genetic variation was partitioned among species, 9.66% among populations, and 60.53% resided within populations (Table 2). Furthermore, the AMOVA results for ancient Chinese Assam tea populations in Puer, Lincang, and Xishuangbanna revealed that 6.4% of the genetic variation was partitioned among the three regions, 8.5% among populations, while most of the genetic variation (85.1%) resided within populations (Table 3).
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation |
| Among species | 2 | 875.39 | 5.54 | 29.81 |
| Among populations | 32 | 1141.1 | 1.79 | 9.66 |
| Within populations | 434 | 4877.87 | 11.24 | 60.53 |
| Total | 468 | 6894.36 | 18.57 | 100 |
| df, degrees of freedom. | ||||
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation |
| Among Regions | 2 | 263.88 | 0.84 | 6.4 |
| Among Populations | 23 | 638.23 | 1.13 | 8.5 |
| Within Populations | 355 | 3988.56 | 11.24 | 85.1 |
| Total | 380 | 4890.67 | 13.21 | 100 |
| df, degrees of freedom. | ||||
The best K value for our data in STRUCTURE was K = 2 (Fig. S1), at which there was clear genetic clustering of populations of Chinese Assam tea from those of its wild relatives (Camellia taliensis and C. crassicolumna). There was some genetic admixture between the two clusters for some Chinese Assam tea trees, indicating introgression from the two wild gene pools into Chinese Assam tea (Table 1; Fig. 2). At K = 3, populations of C. taliensis and C. crassicolumna, Chinese Assam tea from the Puer region (CSAP), and Chinese Assam tea from Lincang (CSAL) separately formed three relatively clean genetic clusters, but with some exceptions (Fig. 2). The Chinese Assam populations in the Xishuangbanna region, however, showed an admixture between the Chinese Assam tea gene pools from Puer and Lincang. At K = 4, the 26 Chinese Assam tea populations were separated into three genetic clusters, consistent with their geographical origins (i.e., Puer, Lincang and Xishuangbanna); specifically, 54.3% of the populations grouped into distinct geographical clusters based on the threshold of Q ≥ 0.8 (Fig. 3A), where 45.7% showed various genetic admixtures. Although we found the best K value to be K = 2, a more biologically meaningful clustering was shown at K = 4. When the 469 accessions of Chinese Assam tea and the two wild species were regrouped based on an admixture coefficient of Q ≥ 0.8, at K = 4, four ‘pure’ groups (three geographical groups and wild) and an admixture group were defined.

|
| Fig. 2 Results of the structure analysis at K = 2 to 4 based on nSSR data for a total of 469 individuals, comprising 26 populations of Camellia sinensis var. assamica (Chinese Assam type tea) from Puer (CSAP), Xishuangbanna (CSAX), and Lincang (CSAL), six populations of C. taliensis, and three populations of C. crassicolumna. |
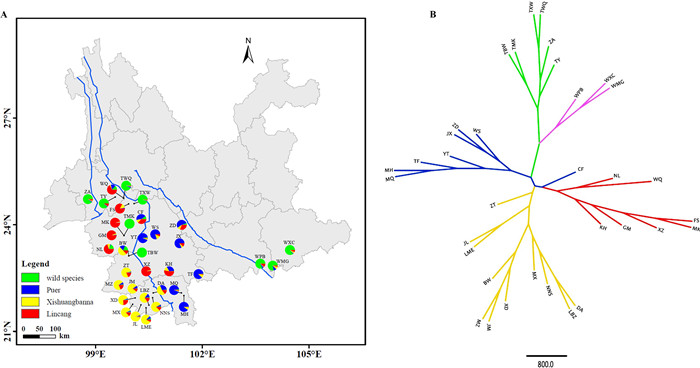
|
| Fig. 3 Geographic distribution of genetic clusters defined by STRUCTURE analysis at K = 4 (A), and Unrooted UPGMA tree with colors corresponding to the genetic clusters at K = 4 in STRUCTURE (B). |
The tree-based UPGMA analysis supported three distinct gene pools of Chinese Assam tea in Southwest Yunnan, with the twowild species (Camellia taliensis and C. crassicolumna) clearly separate from these and from each other (Fig. 3B). All populations whose individuals had Q ≥ 0.8 in STRUCTURE clustered into their respective clades, except for one population (CF), which was placed in the Puer group based on STRUCTURE analysis but clustered within the Lincang group in the UPGMA tree (Fig. 3B).
The first three principal coordinates of the PCoA plot explained 72% of the total genetic variation and separated Chinese Assam tea from the two wild species (C. taliensis and C. crassicolumna) (Fig. 4). The PCoA, however, failed to delineate the three genetic groups of Chinese Assam tea that were defined in STRUCTURE (Puer, Lincang and Xishuangbanna (Fig. 2). When the PCoA was repeated while excluding admixture individuals at K = 4, the first three principal coordinates explained 76% of the total genetic variation. Here, Chinese Assam tea accessions from Puer and Lincang were separated, though a considerable degree of overlap was evident (Fig. 4A). Additionally, accessions from Xishuangbanna clustered between those from Puer and Lincang in Axis 3, but also showed some overlap (Fig. 4B).
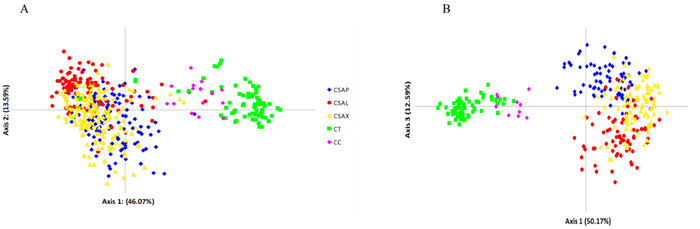
|
| Fig. 4 Principal Co-ordinates Analysis (PCoA) for Axis 1 and 2 (A) and 1 and 3 (B) of Chinese Assam type tea samples and their close wild relatives after re-grouping of individuals in STRUCTURE based on a cut-off membership coefficient (Q value) of 0.8 and excluding the admixture samples. |
The first NewHybrids analysis included 309 Chinese Assam tea accessions, out of which 74 accessions from Puer and 70 from Lincang were used as parents due to their ‘pure’ genetic constitution; the remaining 165 from Xishuangbanna were used as test samples. We found that 52 individuals from Xishuangbanna were parental types, 42 (25.4%) of which were genetically similar to the “Lincang” parent, and 10 accessions (6.1%) were assigned to the “Puer” parent. A total of 55 individuals (33.3%) were assigned F2 hybrid status, while the remaining 58 (35.2%) were found to be mixed hybrids (Table 4; Fig. 5). Posterior probability values of the NewHybrids analysis showed that four accessions from Puer and two accessions from Lincang were also mixed hybrids (Table 4).
| Genetic lineage | P1 | P2 | F1 | F2 | BC1 | BC2 | Mixed |
| CSAP | 70 | 4 | |||||
| CSAL | 68 | 2 | |||||
| CSAX | 10 | 42 | 55 | 58 | |||
| Total | 80 | 110 | 55 | 64 | |||
| P1: Parent 1; P2: Parent 2; F1: First filial generation; F2: Second filial generation; BC1: Backcross to parent 1; BC2: Backcross to parent 2; Mixed: Hybrids with posterior probability value less than 0.8 were assigned into mixed hybrid group. | |||||||
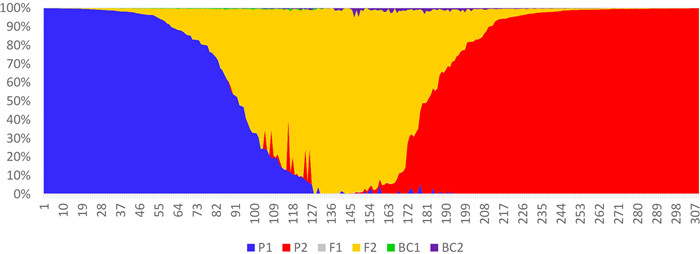
|
| Fig. 5 NewHybrids analysis of 309 samples representing Chinese Assam type tea pure samples collected from Puer and Lincang, and test samples collected from Xishuangbanna. |
In the second NewHybrids analysis, out of the 453 samples used (including 381 and 72 accessions of Chinese Assam tea and C. taliensis, respectively), 38 (10%) were found to be of hybrid origin (Table S3). Among the 72 C. taliensis trees, 9 (12.5%) were of hybrid origin, while one individual (TF5) was identified as Chinese Assam tea, possibly due to a misidentification. Of the 46 accessions of hybrid origin, 7 (15.2%) were BC2 (backcross) hybrids to Chinese Assam tea, and 4 (8.7%) were BC1 hybrids to C. taliensis. The remaining 35 (76.1%) accessions were mixed hybrids (Table S3; Fig. S2). Interestingly, we did not observe any first generation (F1), or second generation (F2) hybrids based on the NewHybrids analysis.
4. Discussion 4.1. Genetic diversity, molecular variation, and population structureOur overall estimate of gene diversity for the ancient Chinese Assam tea trees included in our study (HS = 0.638) was moderately high, consistent with the average gene diversity reported for Chinese tea germplasm collected from 14 provinces in China (H = 0.64; Yao et al., 2012). The genetic diversity estimation for Camellia taliensis (HS = 0.615) in the current study was also comparable to the diversity value reported by Zhao et al. (2014) (HS = 0.597) for the same species collected from the same region. Despite having the lowest sample size, C. crassicolumna had the highest gene diversity (HS = 0.658) and a relatively high number of private alleles (7) (Table 2). When comparing the ancient Chinese Assam tea populations collected from the three geographical areas, Puer (HS = 0.673) showed the highest gene diversity followed by Xishuangbanna (HS = 0.625), with Lincang showing the lowest (HS = 0.617). Trade activities along the Tea Horse Road in Yunnan (Freeman and Ahmed, 2010) may have played a role in the exchange of genetic material throughout Southwest China and the neighboring countries (Thailand, Laos, and Myanmar), considering that the starting point of the road (Yiwu, Mengla) was in ancient Puer. Trade activities in countries bordering Yunnan towards Puer flourished in this area around this time and might explain the relatively high genetic diversity of ancient Chinese Assam tea populations in Xishuangbanna and Puer populations. Zhao et al. (2021) also recently reported high genetic diversity (HE = 0.655–0.795) for ancient C. sinensis in Sandu County, Guizhou Province, China. In contrast, Fang et al. (2012) reported low genetic diversities for modern tea cultivars from China based on nSSR markers (HS = 0.286–0.53). Collectively, the findings suggest that a decrease in genetic diversity from wild species to landraces to modern cultivars has occurred in tea plant populations during domestication. Our results (particularly within the Chinese Assam tea category) may have been influenced by the disparities in sample sizes among the different groups. Moreover, the comparisons highlighted above should be treated with caution owing to differences in sample size, number and type of markers, as well as the specific statistical approaches used to estimate genetic diversity.
Analysis of Molecular Variance (AMOVA) in the current study revealed that 29.81% of the genetic variation was partitioned among species, 9.66% among populations, and 60.53% within the population (Table 2), as would be expected for a wild out-crossing species. This finding indicates considerable levels of gene flow among the analyzed populations. Ji et al. (2011) analyzed the genetic differentiation of 10 ancient tea populations from the same region as our study (Yunnan, SW China) using ISSR markers and reported similar results (39.7% among populations and 60.3% within population genetic variation). Similarly, an analysis of wild and ancient cultivated tea populations in the Wuyi mountains of China using SNPs found that 34% of the genetic variation was partitioned between populations and 66% within populations (Liu et al., 2022). Our AMOVA analysis on the ancient Chinese Assam tea populations grouped according to the three geographical regions (Puer, Lincang and Xishuangbanna) showed that 6.4% of the genetic variation was partitioned among regions, 8.5% among populations, and 85.1% within populations (Table 3). Similar results based on EST-SSR markers were reported in a study that analyzed 450 Chinese cultivated tea accessions from 14 tea-growing regions (Yao et al., 2012). Genotyping by sequencing of tea germplasm in Hunan province also showed that 18.62% of the genetic variation was partitioned between populations and 81.38% within populations (Huang et al., 2022). Yet, SSR analysis of the ancient tea germplasm of Sandu County, Guizhou Province of China indicated that 95% of the genetic diversity resided within populations (Zhao et al., 2021), while SSR analysis of Korean cultivated tea germplasm found 99% of the genetic diversity residing within populations (Lee et al., 2019). However, ancient Chinese Assam tea populations in the current study showed considerable regional and population differentiation.
The ancient Chinese Assam tea populations could be assigned to three distinct genetic groups, Puer, Lincang and Xishuangbanna (K = 4; Fig. 2), corresponding well with its geographic distribution range (Fig. 3A), although a considerable proportion of the alleles were shared among the three groups (K = 4; Figs. 2 and 3). The Lincang and Xishuangbanna gene pools were separated from the Puer gene pool by the Lancang (Mekong) River, while the former two genetic groups were only separated from each other by an administrative boundary (Fig. 3A), indicating initial local adaptation and spread of genotypes during consecutive artificial selection in the domestication of the tea plants. The overall lack of substantial population structure in our study may be explained by the long generation time, allogamy, and self-incompatibility (Fuchinour, 1979) of the tea plants, the relatively short domestication history of Chinese Assam tea and the local artificial dissemination of suitable genotypes (Meegahakumbura, 2016; Meegahakumbura et al., 2018a).
4.2. Breeding history and early dissemination of Chinese Assam teaOur results support the existence of three gene pools of Chinese Assam tea in southwest Yunnan, in Puer, Lincang, and Xishuangbanna (Figs. 2 and 3). Recently, Zhang et al. (2021) reported the presence of two subgroups (ancestral Chinese Assam tea and cultivated Chinese Assam tea) based on genome resequencing. Lu et al. (2021) demonstrated the differential clustering of two ancient tea populations collected from Lincang and Menghai (Xishuangbanna) in Yunnan. A recent genome re-sequencing study of Lincang ancient tea tree populations also revealed the presence of three subpopulations in the Lincang area (Lei et al., 2022). This collectively points to the existence of multiple gene pools of ancient Chinese Assam tea in Yunnan. Our earlier demographic modelling study indicated that Chinese Assam tea was possibly domesticated around 2770 years ago (Meegahakumbura et al., 2018a), consistent with the early records of tea usage in Yunnan (Zhao and Yin, 2008). Therefore, Chinese Assam tea was possibly domesticated locally in the three gene pools of Puer, Lincang, and Xishuangbanna in the Mekong River valley. The existence of some hybrid individuals was also evident in the NewHybrid analysis in the present study. Seeds of the domesticated tea trees may have been dispersed among the three regions along the Tea Horse Road (more likely for the Puer type Chinese Assam tea), and subsequently established hybrid tea landrace populations. During post-domestication breeding, ancient hybrid (F1) tea trees were possibly selected, and their seeds planted extensively in the mountains of Xishuangbanna during the Qing Dynasty (1644–1911 AD), which might explain the existence of F2 and mixed hybrid ancient tea trees in the area. Interestingly, the populations near regional boundaries (e.g., CF, NL, BW and DA) have more mixed genetic components (Fig. 3A), indicating unhindered germplasm exchange between the three gene pools. A sudden boom in local tea production during the Qing Dynasty (Zhou, 2004) likely explains the fixing of unique alleles within the Xishuangbanna populations observed in our current study (Fig. 3A; Table 1).
Our data also indicate that natural or human-mediated gene flow among wild tea populations may have played an important role in tea domestication in Yunnan, Southwest China. The tea plant has a wide array of phenotypes (Wight, 1959), which may result from hybridization between C. sinensis and C. taliensis (Wight and Barua, 1957). Ancient Chinese Assam tea and C. taliensis populations often grow sympatrically in the Mekong River basin in Yunnan, creating opportunities for natural hybridization. Indeed, our NewHybrids results indicated that interspecific hybrids exist within ancient Chinese Assam tea (9.7%) and C. taliensis (12.5%) gene pools (Table S3; Fig. 5). Among these, 76.1% and 15.2% are mixed and BC2 hybrids, respectively, indicating that advanced generation hybrids likely dominate the region. These findings support the view that C. taliensis might have contributed to the domestication of Chinese Assam tea in Yunnan, as earlier noted by Li et al. (2015), and that C. taliensis was ‘contaminated’ with the Ancient Chinese Assam tea gene pool. In this regard, our analysis was limited by the low number of wild relatives of the tea plant included in the study. Future investigations that incorporate more wild relatives will shed light not only on the domestication footprints of the tea plant, but also on the viable germplasm improvement pathways.
Trade activities along the Tea Horse Road in Yunnan are thought to have flourished during the Qing Dynasty (1644–1911 AD) (Freeman and Ahmed, 2010). Consequently, the road probably played a key role in the distribution of the domesticated tea germplasm throughout Southwest China, likely through the direct movement of plants or seeds, thus facilitating gene flow among previously isolated gene pools. For example, Puer Chinese Assam tea populations (XZ) showed a similar genetic composition to tea from Lincang. Furthermore, individuals from the Chinese Assam tea population CF, which is geographically located in the Lincang area separated by the Mekong River from the Puer area, appear to be of hybrid origin between Puer and Lincang gene pools. Additionally, the ancient tea populations analyzed in our study were domesticated before the inter-county boundary between Puer and Xishuangbanna regions was established in 1951, long after the beginning of tea domestication here. Therefore, the natural distribution of Chinese Assam tea is expected to not always agree with the geopolitical boundaries, upon which our sample collection was based. For instance, MH, a historically significant ancient Chinese Assam tea population located at the starting point of the Tea Horse Road, was assigned to Puer populations based on our genetic analysis but is currently located in the Xishuangbanna, indicating that the ancient Chinese Assam tea populations were selected during local domestication of previously locally adapted plants. To obtain a clearer picture of the domestication and dissemination dynamics of Chinese Assam tea in the region, future investigations should incorporate maternally-inherited genetic markers such as plastid DNA sequences, which may offer insights into the role of seed dispersal in the expansion of the tea industry. Although the domestication and breeding history of Chinese Assam tea in southwestern Yunnan is complex, the results presented here will help identify sources of unique and historically significant germplasm that may be valuable for future tea breeding and improvement programs.
4.3. Implications for germplasm conservationWe report a relatively high genetic variation within and among the ancient Chinese Assam tea populations in southwestern Yunnan, which is a valuable resource for future tea breeding. Our results highlight the need for in-situ conservation interventions to safeguard populations against overexploitation due to increasing demand for the sale and use of leaves from ancient tea trees (Ji et al., 2011; Zhao et al., 2014). We recommend that awareness programs be initiated to protect the recently defined distinct genetic lineages making up Chinese Assam tea that primarily occur only in this region (Meegahakumbura et al., 2016, 2018a), and which have not been fully used for tea breeding worldwide (Wambulwa et al., 2016b, 2017). Recent efforts to replace existing ancient tea populations with “improved” clones, which may be high-yielding and of immediate benefit to farmers, may represent an additional risk for the ancient Chinese Assam tea populations in southwestern Yunnan. Numerous biotic (e.g. pests and diseases) and abiotic (e.g. climate change) stressors make it desirable to maintain a genetically diverse tea germplasm, which will in turn guarantee a resilient and sustainable tea industry. In such cases, it may be helpful to compensate farmers who protect these ancient populations against over-harvesting or replacement with clonal plantations.
The present study identified three gene pools for Camellia sinensis var. assamica (Chinese Assam tea) populations in southwestern Yunnan that will serve as critical genetic resource centers for the further improvement of tea germplasm. Although we collected only 16 individuals from three populations of C. crassicolumna from the forest in Honghe of Yunnan, these populations exhibited a high genetic diversity and had seven private alleles. As our study indicated that wild tea populations have also contributed to the gene pool of domesticated Chinese Assam tea in Yunnan, it is essential to conserve the populations of wild species that are closely related to the tea plant.
AcknowledgementsWe are grateful to Drs. Lijun Yan, Jie Liu, Junbo Yang, Hongtao Li, Mr. Zhirong Zhang, and Guangfu Zhu for their help with sample collection, laboratory work and data analysis. This study was supported by funds from the National Natural Science Foundation of China (31970363, 31161140350), and the Key Basic Research Program of Yunnan Province, China (202101BC070003). The Royal Botanic Garden Edinburgh is supported by the Scottish Government’s Rural and Environment Science and Analytical Services division.
Author contributions
LMG and DZL designed the research and acquired funding. LMM, LMG, MKM, and MCW collected the materials, performed the experiments. LMM, MKM, MCW, and ZFS carried out data analysis. MKM, MCW, LMG, MM, and KSB wrote the first draft of the manuscript. All authors have contributed to interpretation of the results and editing of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.06.002.
Anderson, E.C., Thompson, E.A., 2002. A model-based method for identifying species hybrids using multilocus genotypic data. Genetics, 160: 1217-1229. DOI:10.1093/genetics/160.3.1217 |
Chen, X.Y., 1986. The Original Locality of Tea Plant-Yunnan. Kunming: Yunnan People Press: p. 156.
|
Dieringer, D., Schlӧtterer, C., 2003. Microsatellite analyzer (MSA): a flatform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes, 3: 167-169. DOI:10.1046/j.1471-8286.2003.00351.x |
Diez, C.M., Trujillo, I., Martinez-Urdiroz, N., et al., 2015. Olive domestication and diversification in mediterranean basin. New Phytol., 206: 436-447. DOI:10.1111/nph.13181 |
Earl, D.A., vonHoldt, B.A., 2012. Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour., 4: 359-361. DOI:10.1007/s12686-011-9548-7 |
Evanno, G., Regnaut, S., Goudet, J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol., 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Fang, W., Cheng, H., Duan, Y., et al., 2012. Genetic diversity and relationship of clonal tea (Camellia sinensis) cultivars in China as revealed by SSR markers. Plant Syst. Evol., 298: 469-483. DOI:10.1007/s00606-011-0559-3 |
Felsenstein, J., 2004. Inferring Phylogenies. Sunderland, Massachusetts: Sinauer Associates.
|
Freeman, M., Ahmed, S., 2010. Tea Horse Road: Chi's Ancient Trade Route to Tibet. Bangkok, Thailand: River Book Company.
|
Fuchinour, Y., 1979. Analysis of self-incompatibility alleles of major varieties of tea. Jpn. Agric. Res. Q., 13: 43-48. |
Goudet, J., 2002. FSTAT: a Programme to Estimate and Test Gene Diversities and Fixation Index, Version 2.9.3.2.
|
Huang, F., Duan, J., Lei, Y., Liu, Z., Kang, Y., et al., 2022. Genetic diversity, population structure and core collection analysis of Hunan tea plant germplasm through genotyping-by-sequencing. Beverage Plant Resear., 2: 5. |
Ji, P.Z., Li, H., Gao, L.Z., et al., 2011. ISSR Diversity and genetic differentiation of ancient tea C. sinensis. var. assamica plantations from China. Pakistan J. Bot., 43: 281-291. |
Kingdon-Ward, F., 1950. Does wild tea exists?. Nature, 165: 297-299. DOI:10.1038/165297a0 |
Lee, K.J., Lee, J.R., Sebastin, R., et al., 2019. Assessment of genetic diversity of tea germplasm for its management and sustainable use in Korea Genebank. Forests, 10: 780. DOI:10.3390/f10090780 |
Lei, Y., Yang, L., Duan, S., et al., 2022. Whole-genome resequencing reveals the origin of tea in Lincang. Front. Plant Sci., 10: 984422. |
Li, M.M., Meegahakumbura, M.K., Yan, L.J., et al., 2015. Genetic involvement of Camellia taliensis in the domestication of Camellia sinensis var. assamica (Assamica Tea) revealed by nuclear microsatellite markers. Plant Divers. Resour., 37: 29-37. |
Li, L., Hu, Y., He, M., et al., 2021. Comparative chloroplast genomes: insights into the evolution of the chloroplast genome of Camellia sinensis and the phylogeny of Camellia. BMC Genom., 22: 138. DOI:10.1186/s12864-021-07427-2 |
Liu, C., Yu, W., Cai, C., et al., 2022. Genetic diversity of tea plant (Camellia sinensis (L.) Kuntze) germplasm resources in Wuyi Mountain of China based on Single Nucleotide Polymorphism (SNP) markers. Horticulture, 8: 932. DOI:10.3390/horticulturae8100932 |
Lu, H.Y., Zhang, J.P., Yang, Y.M., et al., 2016. Earliest tea as evidence for one branch of the silk road across the Tibetan Plateau. Sci. Rep., 6: 18955. DOI:10.1038/srep18955 |
Lu, L., Chen, H., Wang, X., et al., 2021. Genome-level diversification of eight ancient tea populations in the Guizhou and Yunnan regions identifies candidate genes for core agronomic traits. Hortic. Res., 8: 190. DOI:10.1038/s41438-021-00617-9 |
Meegahakumbura, M.K., 2016. In: Kuntze, O. (Ed.), Genetic Assessments of Asian Tea Germplasm and Domestication History of the Tea Plant (Camellia sinensis (L.)). University of Chinese Academy of Sciences, Beijing, China.
|
Meegahakumbura, M.K., Wambulwa, M.C., Thapa, K.K., et al., 2016. Indications for three independent domestication events for tea plant (Camellia sinensis (L.) O. Kuntze) and new insights into the origin of tea germplasm in China and India revealed by nuclear microsatellites. PLoS One, 11: e0155369. DOI:10.1371/journal.pone.0155369 |
Meegahakumbura, M.K., Wambulwa, M.C., Li, M.M., et al., 2018a. Domestication origin and breeding history of the tea plant (Camellia sinensis) in China and India based on nuclear microsatellites and cpDNA sequence data. Front. Plant Sci., 8: 2270. DOI:10.3389/fpls.2017.02270 |
Meegahakumbura, M.K., Wambulwa, M.C., Li, D.Z., et al., 2018b. Preliminary investigations on the genetic relationships and origin of domestication of the tea plant (Camellia sinensis (L.)) using genotyping by sequencing. Trop. Agric. Res., 29: 230-240. DOI:10.4038/tar.v29i3.8263 |
Ming, T.L., 2000. Monograph of the Genus Camellia. Kunming, China: Yunnan Science and Technology Press: pp. 110-135.
|
Mondal, T., 2009. Tea breeding. In: Jain, S.M., Priyadarshan, P.M. (Eds.), Breeding Plantation Tree Crops: Tropical Species. Springer Science + Business Media, New York, pp. 545-587.
|
Muramatsu, K., 1991. The Science of Tea. Japan: Asakura Publications: pp. 6-7.
|
Nei, M., Tajima, F., Tateno, Y., 1983. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol., 19: 153-170. DOI:10.1007/BF02300753 |
Peakall, R., Smouse, P., 2006. GenALEx 6. Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes, 6: 288-295. DOI:10.1111/j.1471-8286.2005.01155.x |
Pritchard, J.K., Stephens, M., Donnelly, P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. DOI:10.1093/genetics/155.2.945 |
Rambaut, A., 2012. FigTree Version 1.4. http://tree.bio.ed.ac.uk/software/figtree/.
|
Rawal, H.C., Borchetia, S., Bera, B., et al., 2021. Comparative analysis of chloroplast genomes indicated different origin for Indian tea (Camellia assamica cv TV1) as compared to Chinese tea. Sci. Rep., 11: 110. DOI:10.1038/s41598-020-80431-w |
Stuart, C.P., 1919. A basis for tea selection. Bull. Jard. Bot. Buitenzorg, 1: 193-320. |
Van Oosterhout, C., Hutchinson, W.F., Wills, D.P., Shipley, P., 2004. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes, 4: 535-538. DOI:10.1111/j.1471-8286.2004.00684.x |
Wambulwa, M.C., Meegahakumbura, M.K., Chalo, R., et al., 2016a. Nuclear microsatellites reveal the genetic architecture and breeding history of tea germplasm of East Africa. Tree Genet. Genomes, 12: 11. DOI:10.1007/s11295-015-0963-x |
Wambulwa, M.C., Meegahakumbura, M.K., Kamunya, S., et al., 2016b. Insights into genetic relationships and breeding patterns of African tea germplasm based on nSSR and markers and cpDNA sequences. Front. Plant Sci., 7: 1244. |
Wambulwa, M.C., Meegahakumbura, M.K., Kamunya, S., et al., 2017. Multiple origins and narrow genepool characterize African tea germplasm; concordant patterns revealed by nuclear and plastid DNA markers. Sci. Rep., 7: 4053. DOI:10.1038/s41598-017-04228-0 |
Wambulwa, M.C., Meegahakumbura, M.K., Kamunya, S., et al., 2021. From the wild to cup: tracking the footprints of tea species in time and space. Front. Nutr., 8: 706770. DOI:10.3389/fnut.2021.706770 |
Wang, X., Feng, H., Chang, Y., et al., 2020. Population sequencing enhances the understanding of tea plant evolution. Nat. Commun., 11: 4447. DOI:10.1038/s41467-020-18228-8 |
Wight, W., 1959. Nomenclature and classification of tea plant. Nature, 183: 1726-1728. DOI:10.1038/1831726a0 |
Wight, W., Barua, P.K., 1957. What is tea?. Nature, 179: 506-507. DOI:10.1038/179506a0 |
Xia, E.H., Zhang, H.B., Sheng, S., et al., 2017. The tea tree genome provides insides into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant, 10: 866-877. DOI:10.1016/j.molp.2017.04.002 |
Yamanishi, T., 1995. Special issue on tea. Food Rev. Int., 11: 371-546. DOI:10.1080/87559129509541049 |
Yao, M.Z., Ma, C.L., Qiao, T.T., et al., 2012. Diversity distribution and population structure of tea germplasms in China revealed by EST-SSR markers. Tree Genet. Genomes, 8: 205-220. DOI:10.1007/s11295-011-0433-z |
Yu, F.L., 1986. Discussion on the originating place and the originating center of tea plants. J. Tea Sci., 6: 1-8. |
Yu, F., Chen, L., 2001. Indigenous wild tea Camellias in China. In: Proceedings of International Conference of O-Cha (Tea) Culture and Science Session Ⅱ, pp. J1-J4.
|
Zhang, W., Zhang, Y., Qiu, H., et al., 2020. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat. Commun., 11: 3719. DOI:10.1038/s41467-020-17498-6 |
Zhang, X., Chen, S., Shi, L., et al., 2021. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet., 53: 1250-1259. DOI:10.1038/s41588-021-00895-y |
Zhao, F.R., Yin, Q.Y., 2008. The Khmer Meng nationalities in China earliest domesticated cultivated tea. J. Simao Teacher’s College, 24: 28-34. |
Zhao, D.W., Yang, J.B., Yang, S.X., et al., 2014. Genetic diversity and domestication origin of tea plant Camellia taliensis (Theaceae) as revealed by microsatellite markers. BMC Plant Biol., 14: 14. DOI:10.1186/1471-2229-14-14 |
Zhao, Y.C., Wang, R.Y., Liu, Q., et al., 2021. Genetic diversity of ancient Camellia sinensis (L.) O. Kuntze in Sandu county of Guizhou province in China. Diversity, 13: 276. DOI:10.3390/d13060276 |
Zhou, H.J., 2004. Yunnan Puer Tea. Kunming: Yunnan Science and Technology Press: pp. 18-21.
|



