b. Yunnan Academy of Forestry and Grassland, Kunming 650201, China;
c. Gaoligong Mountain, Forest Ecosystem, Observation and Research Station of Yunnan Province, Kunming 650201, China;
d. Yunnan Key Laboratory of Biodiversity of Gaoligong Mountain, Kunming 650201, China;
e. Department of Plant Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA;
f. College of Modern Biomedical Industry, Kunming Medical University, Kunming 650500, China
Apiaceae (Umbelliferae) are a large family of flowering plants, with 442 genera and 3575 species recognized (Christenhusz and Byng, 2016). While most members of its largest subfamily Apioideae are readily identifiable because of their characteristic inflorescences and fruits, their higher-level relationships have traditionally been obscure (Constance, 1971). A modern classification of Apioideae based on phylogenetic analyses of molecular data has elucidated the higher-level subdivisions within the subfamily and provided a framework for its classification (reviewed in Downie et al., 2001, 2010). However, several major clades have not yet been treated formally, pending confirmation from further studies (Downie et al., 2010).
The East Asia (or Physospermopsis) clade is one of these problematic groups. Calviño et al. (2006) recognized species of Physospermopsis H. Wolff, Trachydium Lindl., Tongoloa H. Wolff, Sinolimprichtia H.Wolff, Notopterygium H.de Boissieu, Haplosphaera Hand.-Mazz., and Hansenia Turcz. as comprising the Physospermopsis clade. Included were species with an almost exclusively East Asian distribution. Zhou et al. (2008) thus suggested the alternative name “East Asia clade” and included species of Pimpinella L., Sinocarum H. Wolff ex R.H. Shan & F.T. Pu, and Vicatia DC. within. Subsequent molecular phylogenetic studies supported its monophyly and expanded its circumscription to include representatives of 16 genera (Zhou et al., 2009, 2020, 2021; Downie et al., 2010; Mousavi et al., 2021; Wen et al., 2021). The East Asia clade is a sister group to tribe Komarovieae (Zhou et al., 2009), or arises from within it based on limited sampling and using the universal Angiosperms353 probe set (Clarkson et al., 2021).
Taxonomic relationships within the East Asia clade are extraordinarily complex, having species-rich genera whose boundaries are poorly defined and several monotypic genera. Taxonomic challenges surround Pimpinella, Sinocarum, Tongoloa, and Trachydium. Pimpinella includes ~150 species that occur disjunctly in Africa, Europe, and Asia (Pimenov and Leonov, 1993; Pu and Watson, 2005). A few Pimpinella species are also distributed in South America, with one species found in western North America (Pimenov and Leonov, 1993). Members of Pimpinella can be recognized by their perennial (rarely biennial or annual) herbaceous habit, stem bases without fibrous remnant sheaths, slightly laterally compressed fruits constricted at the commissure, and filiform ribs. Authors have arranged the species of Pimpinella into various sections (de Candolle, 1827; Bentham and Hooker, 1867; Boissier, 1875; Wolff, 1910, 1927; Shan and Sheh, 1985a), and molecular phylogenetic studies have shown that the sectional classification of Pimpinella is highly artificial. Furthermore, Pimpinella is not resolved as a monophyletic group, with its members assigned to seven tribes and other major clades in Apioideae including the East Asia clade (Downie et al., 2010; Wang et al., 2014; Fereidounfar et al., 2016; Zhou et al., 2020). As the type species of Pimpinella (P. saxifraga L.) is placed within tribe Pimpinelleae, nomenclatural changes are required for those Pimpinella species falling outside of Pimpinelleae.
Sinocarum is a small genus of approximately 20 species and exhibits a typical Sino-Himalayan distribution ranging from Nepal to SW China (Pu et al., 2005). Tongoloa also occurs at high elevations in the Sino-Himalayan region, with 15 species recognized (Pan and Watson, 2005a; Pimenov, 2017). Both genera are poorly defined with diffuse generic boundaries, and relationship to putatively allied genera Acronema Edgew., Pimpinella, Physospermopsis, and Trachydium have been suggested. Sinocarum and Tongoloa were each determined to be non-monophyletic in their current circumscription (Downie et al., 2010; Zhou et al., 2020). Moreover, Sinocarum displayed conflicting placements in different phylogenetic studies (Zhou et al., 2008, 2009, 2020; Xiao et al., 2021). The proper circumscriptions of Sinocarum and Tongoloa need to be ascertained.
Trachydium is another taxonomically complex genus, and its generic delimitation has historically been the subject of considerable debate. Since its establishment, those high montane species from the Sino-Himalayan region having a prostrate habit or shortened stems were described in, or transferred to, Trachydium (Pimenov and Kljuykov, 2000). These include species previously attributable to Aulacospermum Ledeb., Chamaesciadium C.A. Mey., Chamaesium H. Wolff, Ligusticum L., Physospermopsis, Pleurospermum Hoffm., Pseudotrachydium (Kljuykov, Pimenov & V.N. Tikhom.) Pimenov & Kljuykov, Schulzia Spreng., and Sinocarum. However, Pimenov & Kljuykov (2000) considered Trachydium to be a monotypic genus (T. roylei Lindl.) and indicated that some of its species belong to Sinocarum or Tongoloa. Previous molecular results partly support such a viewpoint, as the four species examined to date occur throughout the East Asia clade and are allied with Sinocarum, Tongoloa, and Physospermopsis (Zhou et al., 2008, 2009, 2020).
No traditional classification of Apioideae has grouped together those genera that are presently included in the East Asia clade. The morphological diversity among its members is high, making it particularly difficult to identify clade-specific diagnostic characters. In this study, we (1) identify additional members of the East Asia clade by increasing the sampling of species whose distributions are in East Asia or genera having one or more species already included in the clade; (2) assess the phylogenetic relationships of those members comprising the East Asia clade, placing emphasis on Sinocarum, Tongoloa, and Trachydium; and (3) confirm the phylogenetic placements of several species that have been treated variously (and perhaps erroneously) in previous molecular phylogenetic studies. The results obtained will be used to carry out nomenclatural transfers of some species and further clarify relationships within this taxonomically problematic group.
2. Materials and methods 2.1. SamplingThe nrDNA internal transcribed spacer (ITS) region was used for phylogenetic inference, as the numerous ITS sequences available in GenBank for Apiaceae subfamily Apioideae enabled an immediate consideration of these accessions for their possible inclusion in the East Asia clade. Comparative data for any specific chloroplast DNA locus does not yet exist across such a broad sampling of Apioideae.
To ascertain the limits of the East Asia clade, ITS sequence data were procured from 150 accessions of Apioideae (Table S1). These accessions included representation of species whose distributions in East Asia suggested possible close affinities with those taxa already included in the clade, or those genera whose species have already been unambiguously placed in the clade in earlier studies. Sampling of the genera Tongoloa, Sinocarum, and Trachydium was more comprehensive than the other genera. For those non-monophyletic genera whose species were previously determined to occur in several major clades, such as Hymenidium Lindl. and Pimpinella, those species that fell into the East Asia clade were resampled to confirm their phylogenetic positions. For those species having different ITS sequences in GenBank suggesting that at least one may be in error (i.e., Physospermopsis rubrinervis (Franch.) C. Norman and P. cuneata H. Wolff), we collected new leaf material from the type localities. Where possible, each included species was represented by more than one accession to represent morphological, geographical, or possible infraspecific molecular variation. Apioideae ITS sequences were downloaded from GenBank and screened using the strategies described by Zhou et al. (2020) for possible inclusion in the East Asia clade. We only used those sequences released after March 2018, as sequences prior to that date were extracted in a previous study to ascertain the broad phylogenetic placements of Ligusticum species (Zhou et al., 2020). Thirty-eight separate ITS1 and ITS2 sequences from the previous matrix of Zhou et al. (2020) were replaced with the resequencing of the entire ITS region (ITS1-5.8S-ITS2). In summary, 655 ITS sequences newly obtained from GenBank plus the 150 ITS sequences generated specifically for this study resulted in 805 sequences altogether. These were added to the matrix of Zhou et al. (2020) for a total of 3678 Apioideae ITS sequences.
2.2. DNA extractionTotal genomic DNAs were isolated from silica-gel dried materials or from herbarium specimens using a Plant Genomic DNA Kit (Tiangen Biotech, China) according to the manufacturer's protocol. The entire ITS region was amplified using the primers ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) and ITS5 (5′-GGA AGT AAA AGT CGT AAC AAG G-3’; White et al., 1990) or N-nc18S10 (5′-AGG AGA AGT CGT AAC AAG-3′) and C26A (5′-GTT TCT TTT CCT CCG CT-3’; Wen and Zimmer, 1996). If the PCR amplification failed, the region was amplified in two parts, using ITS-u1 (5′-GGA AGK ARA AGT CGT AAC AAG G-3′) and ITS-u2 (5′- GCG TTC AAA GAY TCG ATG RTT C-3′) for ITS1, and ITS-u3 (5′- CAW CGA TGA AGA ACG YAG C-3′) and ITS-u4 (5′-RGT TTC TTT TCC TCC GCT TA-3′) for ITS2 (Cheng et al., 2016). Procedures for amplification are the same as described in Zhou et al. (2008, 2009). The PCR products were sequenced at the Sequencing Laboratory of the Institute of Botany using the same primers as for PCR on an ABI 3730 automated sequencer (Applied Biosystems, Foster City, California, U.S.A.). ITS sequence data acquired in this study were deposited in GenBank (Table S1).
2.3. Phylogenetic analysisThe sequences were assembled, aligned, and manually adjusted using the BioEdit sequence alignment editor (Hall, 1999). Initially, we performed maximum likelihood (ML) analyses using RAxML v.8.0.0 (Stamatakis, 2014) on XSEDE via the CIPRES Science Gateway (http://www.phylo.org/). The settings were rapid bootstrap analysis with 1000 replicates and utilizing the GTR + I + G substitution model. Three species of tribe Bupleureae were used to root the trees as the tribe occupies a sister-group relationship to all other apioid genera excluding its most early-diverging branches (Downie et al., 2001; Calviño et al., 2006). When the circumscription of the East Asia clade was delimited through this ML analysis, we selected only its accessions to conduct Bayesian inference (BI) with MrBayes v.3.2.5 (Ronquist and Huelsenbeck, 2003; Ronquist et al., 2012). The BI trees were rooted with three members of tribe Komarovieae, as the tribe constitutes an immediate sister group to the East Asia clade (Zhou et al., 2009, 2020). For the BI analyses, we determined the appropriate nucleotide substitution models using the corrected Akaike information criterion (AIC) in jModeltest v.2.1.7 (Posada, 2008; Darriba et al., 2012). We performed the BI analysis using two independent runs of the Markov chain Monte Carlo (MCMC) for 10 million generations each and sampling one tree every 1000 generations. Analyses were run until the standard deviation of split frequencies was less than 0.001. Tracer was also used to graphically assess the convergence of runs, and to determine that the effective sampling size values exceeded 200. We discarded the first 25 % generations as burn-in, and the remaining trees were summarized into a 50% majority-rule consensus tree with posterior probabilities (PP).
3. ResultsThe aligned data matrix of 3678 ITS sequences contained 736 positions, with none excluded due to alignment ambiguities. Of these positions, 63 were autapomorphic, 63 were constant, and 610 were parsimony informative. Phylogenetic analysis of this data matrix resulted in a significant expansion of the East Asia clade from any previous circumscription. Two representatives from the genus Acronema are newly recognized members of the clade and the numbers of species from Hymenidium, Tongoloa, and Trachydium have increased substantially. Except for the genera Hansenia, Hymenolaena DC., Keraymonia Farille (including one accession), and Sinolimprichtia (a monotypic genus), all other genera comprising the East Asia clade are polyphyletic and greatly intermingled. The nomenclatural types of Acronema, Hymenidium, Physospermopsis, Pimpinella, Sinocarum, and Trachydium fall outside of the East Asia clade. The ML tree containing all 3678 representatives of Apioideae is presented in Fig. S1, while a portion of this ML tree representing only the East Asia clade and its simplified phylogenetic tree showing relationships among groupings of taxa are presented in Figs.1 and 2A, respectively. In addition, we extracted portions of the Acronema and Sinodielsia clades that contain taxa of interest, as shown in Figs. 3 and 4, respectively.
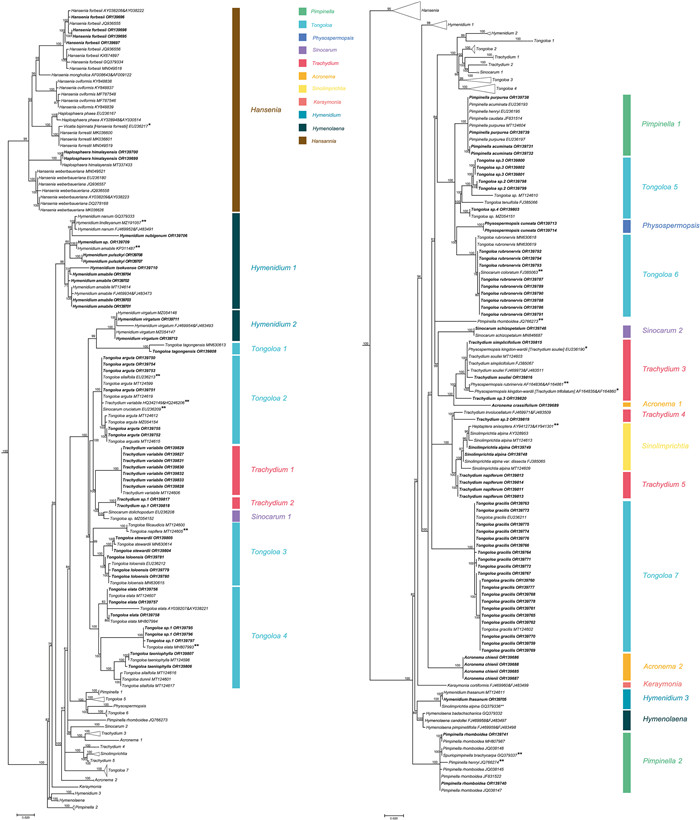
|
| Fig. 1 Portion of the large consensus tree (Fig. S1) obtained from maximum likelihood analysis of 3678 nrDNA internal transcribed spacer sequences from Apiaceae subfamily Apioideae showing only the East Asia clade (comprising 206 terminals). Support values ≥ 50 % are provided. Clade designations are those defined in the text. The 106 newly sampled accessions in this portion of the tree are shown in bold. GenBank accession numbers of all sequences are provided; two numbers indicate sequences from separate ITS1 and ITS2 regions. *Indicates that the identifications of original materials from these accessions were confirmed and that these sequences were used in the Bayesian inference analysis; names in [] are the correct ones. **Indicates that the identifications of these sequences are problematic and thus these taxa were excluded from the Bayesian analysis. |
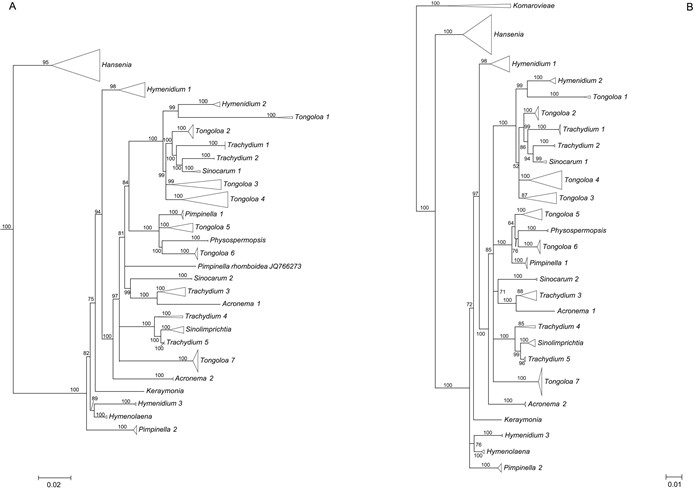
|
| Fig. 2 A. Simplified phylogenetic tree derived from maximum likelihood analysis of 3678 nrDNA internal transcribed spacer sequences from Apiaceae subfamily Apioideae showing the groupings of taxa in East Asia clade, with support values (≥ 50%) provided next to the branches. B. Simplified phylogenetic tree derived from Bayesian inference analysis of 195 nrDNA internal transcribed spacer sequences showing the group of taxa from the East Asia clade and outgroups, with Bayesian posterior probabilities (≥ 50%) provided next to the branches. |
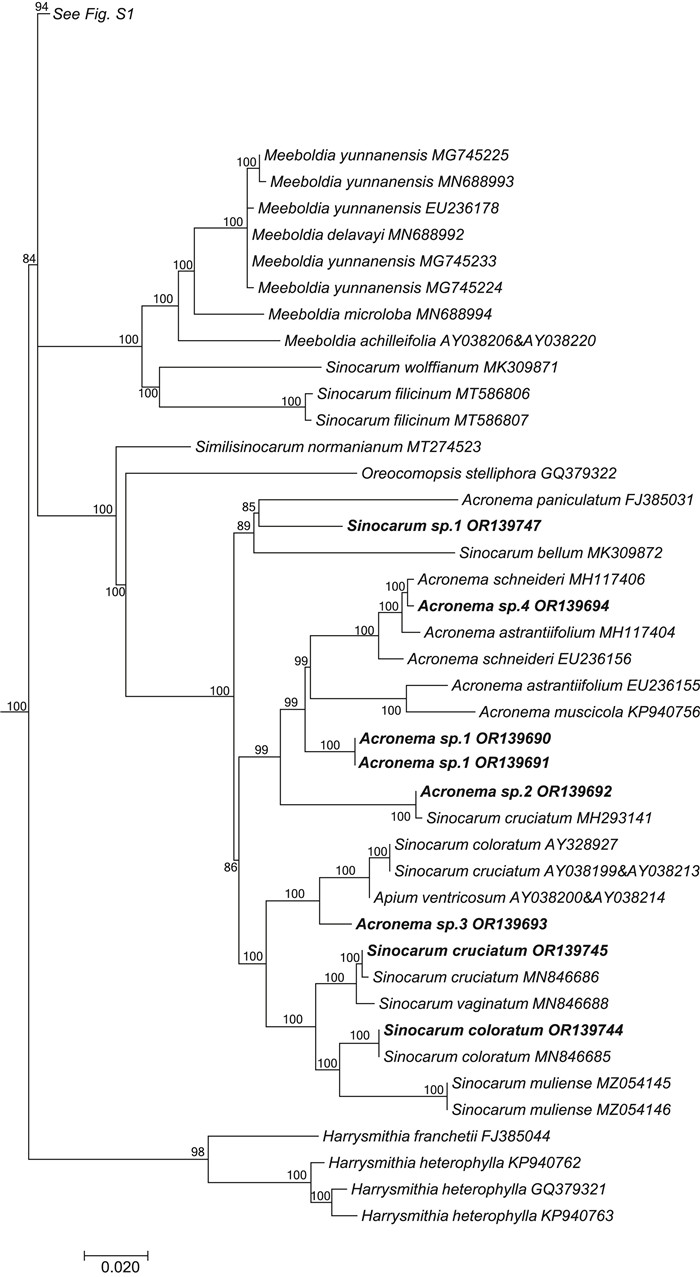
|
| Fig. 3 Portion of the large consensus tree (Fig. S1) obtained from maximum likelihood analysis of 3678 nrDNA internal transcribed spacer sequences from Apiaceae subfamily Apioideae showing only the accessions falling in the Acronema clade. Support values ≥ 50% are provided. The newly sampled accessions in this portion of the tree are shown in bold. GenBank accession numbers of all sequences are provided; two numbers indicate sequences from separate ITS1 and ITS2 regions. |
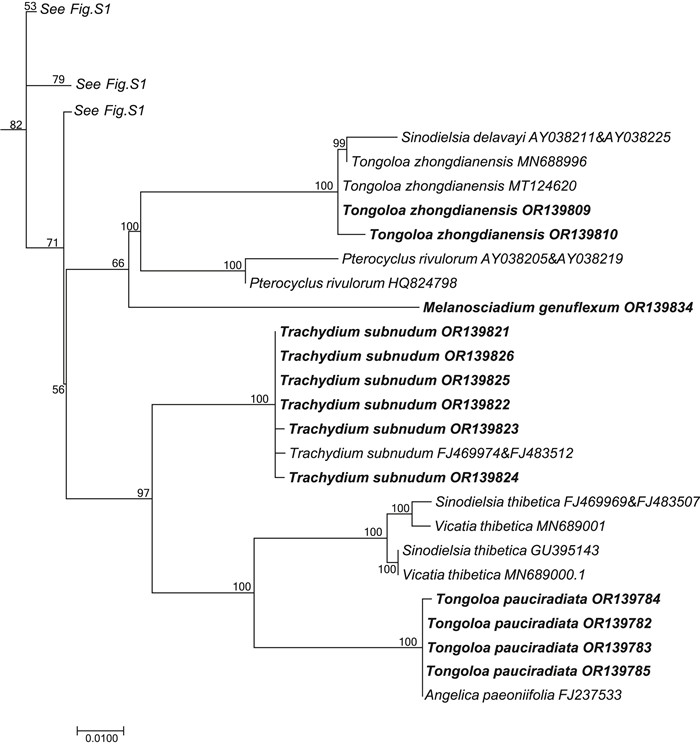
|
| Fig. 4 Portion of the large consensus tree (Fig. S1) obtained from maximum likelihood analysis of 3678 nrDNA internal transcribed spacer sequences from Apiaceae subfamily Apioideae showing only the accessions falling in the Sinodielsia clade. Support values ≥ 50% are provided. The newly sampled accessions in this portion of the tree are shown in bold. GenBank accession numbers of all sequences are provided; two numbers indicate sequences from separate ITS1 and ITS2 regions. |
Simultaneous analysis of multiple ITS sequences from the same species indicated that some may be either misidentifications or resulting from contamination or improper laboratory practices because they did not fall as monophyletic in the phylogenetic trees. Sequences of some conspecific taxa differed considerably and occurred in different tribes and other major clades of Apioideae. Physospermopsis cuneata is confirmed as belonging to the East Asia clade. The placement of its two previously examined accessions (GenBank accession nos. EU236188 and EU236189) in tribe Pimpinelleae (Zhou et al., 2008, 2009) must be attributable to identification errors. The accession of Physospermopsis rubrinervis collected from its type locality (Luoping Mt., Yunnan Province, China) allies with other representatives of the same species within the tribe Pleurospermeae; the accession of P. rubrinervis (AF164836 and AF164861), previously attributable to the East Asia clade, is also a misidentification. Similar situations also occur for Sinocarum coloratum (Diels) H. Wolff ex R.H. Shan & F.T. Pu and S. cruciatum (Franch.) H. Wolff ex R.H. Shan & F.T. Pu, where sequences of the same species fell into both the Acronema clade (Fig. 3) and the East Asia clade (Fig. 1). After examination of their voucher materials, we considered that Sinocarum cruciatum accession MN846686 and S. coloratum accession MN846685 are correctly identified, whereas S. coloratum FJ385063 and S. cruciatum EU236209 in the East Asia clade and S. coloratum AY328927 and S. cruciatum AY038199, AY038213 and MH293141 in the Acronema clade are not. Vicatia bipinnata R.H. Shan & F.T. Pu (EU236217) and its synonymous species Melanosciadium bipinnatum (R.H. Shan & F.T. Pu) Pimenov et Kljuykov (JX514721) fell into the East Asia clade (Fig. 1) and tribe Selineae (Fig. S1), respectively. The voucher specimen of V. bipinnata is actually Hansenia forrestii (H. Wolff) Pimenov & Kljuykov, while the ITS sequence of M. bipinnatum (JX514721) is identical to that of Pimpinella rhomboidea var. tenuiloba R.H. Shan & F.T. Pu. The newly obtained Melanosciadium genuflexum Pimenov & Kljuykov is placed in the Sinodielsia clade and is sister group to the clade of Tongoloa zhongdianensis S.L. Liou, Pterocyclus rivulorum (Diels) H. Wolff, and Sinodielsia delavayi (Franch.) Pimenov & Kljuykov (Fig. 4). Heptaptera anisoptera Tutin (AY941273 and AY941301) and Spuriopimpinella brachycarpa (Kom.) Kitagawa (GQ379337) arise from within accessions of Sinolimprichtia alpina H. Wolff and Pimpinella rhomboidea Diels, respectively. However, other sequences of H. anisoptera and S. brachycarpa are placed into the Opopanax and Acronema clades, so we excluded these two problematic genera from the East Asia clade. With nearly identical ITS sequences, Hymenidium lindleyanum (Klotzsch) Pimenov & Kljuykov (MZ191057) comprises a group with H. nanum (Rupr.) Pimenov & Kljuykov in the East Asia clade, while another accession of H. lindleyanum (FJ469949 and FJ483488) falls into tribe Pleurospermeae (Fig. S1).
The accessions of Physospermopsis kingdon-wardii (H. Wolff) C. Norman (AF164835, AF164860 and EU236190) do not ally. We further checked their voucher specimens (W810822 for EU236190, and E00052566 for AF164835 and AF164860), which should be identified as Trachydium souliei H.de Boissieu and T. trifoliatum H. Wolff, respectively. The accession Sinolimprichtia alpina (GQ379336) is sister group to the clade comprising two accessions of Hymenidium lhasanum Pimenov & Kljuykov, and distantly away from other accessions of the same species. With only one nucleotide difference between them, we consider that the species identified as S. alpina (GQ379336) is actually H. lhasanum. Three accessions of Pimpinella henryi Diels fall into both Pimpinelleae and the East Asia clade, with the two accessions in the East Asia clade not grouping together. Although we cannot confirm the identity of accessions with sequences EU236195 and MH710738, the one in the East Asia clade (JQ766274) arising from within Pimpinella rhomboidea is no doubt a misidentification. Furthermore, Tongoloa napifera (H. Wolff) C. Norman (MT124605) does not ally with its synonymous species Trachydium napiferum H. Wolff, and Pimpinella rhomboidea (JQ766273) forms an isolated lineage distantly away from its conspecific accessions. Other suspect sequences include Hymenidium amabile (Craib & W.W. Smith) Pimenov & Kljuykov (KP311487), Tongoloa elata H. Wolff (MH807993), Tongoloa silaifolia (H.de Boissieu) H. Wolff (EU236213), and Trachydium variabile H. Wolff (HQ342149 and HQ246206). In summary, 11 genera are recognized herein as constituting the East Asia clade (Table 1).
| Genus | Number of species falling into the East Asia clade/Total number of species in genus and reference | Distribution of species falling into the East Asia clade |
| Acronema* | 2/25a | China |
| Hansenia (including Notopterygium and Haplosphaera) | 7/8b,c,d,e | Bhutan, China, India, Mongolia, Nepal, Russia |
| Hymenidium* | 7/37f | Bhutan, China, India, Kazakhstan, Kirghizia, Sikkim, Tajikistan |
| Hymenolaena | 3/3g | Afghanistan, India, Kazakhstan, Kirghizia, Pakistan, Tadzhikostan |
| Keraymonia | 1/4g | China, Nepal |
| Physospermopsis* | 1/15a,g | China |
| Pimpinella* | 5/150a | China, India, Myanmar, Nepal, Pakistan |
| Sinocarum* | 2/20a,f | China, Myanmar |
| Sinolimprichtia | 1/1a | China |
| Tongoloa* | 12/17a,f,h,i | Bhutan, China, India, Nepal |
| Trachydium* | 6/10a,f | Bhutan, China, India, Nepal |
| a Sheh et al. (2005). b Pimenov et al. (2008). c Jia et al. (2019). d Tan et al. (2020). e Jiang et al. (2022). f Pimenov (2017). g Pimenov and Kljuykov (2000). h Gui et al. (2020a). i Gui et al. (2020b). | ||
The aligned 195 ITS sequences from taxa comprising the East Asia clade and three outgroup taxa (Komarovieae) resulted in a matrix of 620 positions, with none excluded because of alignment ambiguities. Of these positions, 249 were parsimony informative, 305 were constant, and 66 were autapomorphic. Gaps ranging in length from 1 to 3 bp were introduced to facilitate alignment. The mean percentage G + C content across all sequences was 54.8%. The BI results are generally congruent in topology with the portion of the ML tree containing the East Asia taxa; branch support values are indicated (Figs. 2B and 5).
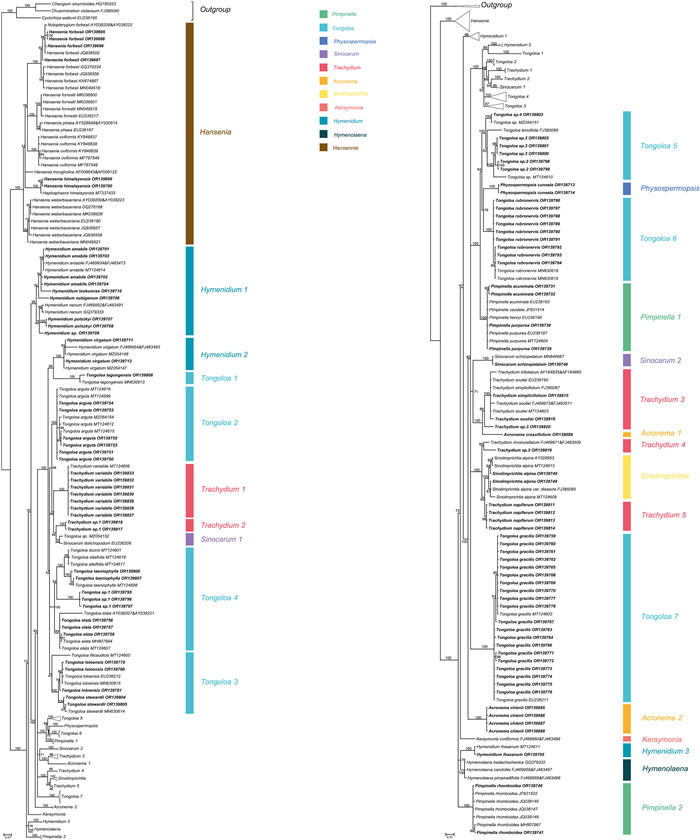
|
| Fig. 5 Majority-rule consensus tree for the East Asia clade and outgroups inferred from Bayesian inference analysis (195 terminals). Numbers above the branches are Bayesian posterior probabilities. The 106 newly sampled accessions are shown in bold. GenBank accession numbers of all sequences are provided; two numbers indicate sequences from separate ITS1 and ITS2 regions. Clades designations are described in the text. |
The seven species of Hansenia constituted a well-supported monophyletic group (BI = 100), with each of its species also arising as monophyletic. Two species of the former genus Haplosphaera, Haplosphaera phaea Hand.-Mazz. (≡Hansenia phaea (Hand.-Mazz.) J.B. Tan & X.G. Ma) and H. himalayensis Ludlow (≡Hansenia himalayensis (Ludlow) J.B. Tan & X.G. Ma), arise from within the Hansenia clade. The three species of Hymenolaena also allied as a strongly supported clade (BI = 100) and is sister group to Hymendium lhasanum. The four newly sampled species of Hymenidium [H. pulszkyi (Kanitz) Pimenov & Kljuykov, H. nubigenum (H. Wolff) Pimenov & Kljuykov, H. tsekuense (R.H. Shan) Pimenov & Kljuykov, and H. sp.] ally with their congeners H. amabile and H. nanum in clade Hymenidium 1 (BI = 98). Hymenidium virgatum Pimenov & Kljuykov (clade Hymenidium 2) and H. lhasanum (clade Hymenidium 3) are separately placed in the East Asia clade and do not cluster with the aforementioned congeners.
With the exception of Tongoloa zhongdianensis and T. pauciradiata H. Wolff that are placed into the Sinodielsia clade (Fig. 4), all other sampled accessions of Tongoloa fell into the East Asia clade, albeit in seven different groups (Tongoloa 1–7). Tongoloa tagongensis L.J. Gui & X.J. He (Tongoloa 1), T. arguta L.J. Gui & X.J. He (Tongoloa 2), T. rubronervis S.L. Liou (Tongoloa 6), and T. gracilis H. Wolff (Tongoloa 7) each comprises a separate, monophyletic group. Three species of Tongoloa [T. loloensis (Franch.) H. Wolff, T. stewardii H. Wolff, and T. filicaudicis K.T. Fu] comprised a well-supported group (Tongoloa 3; BI = 87). Five Chinese endemic species of Tongoloa [Tongoloa elata H. Wolff, T. dunnii (H.de Boissieu) H. Wolff, T. silaifolia, T. taeniophylla (H.de Boissieu) H. Wolff, and T. sp. 1] comprised the Tongoloa 4 group (BI = 100). Four unknown species of Tongoloa ally with T. tenuifolia H. Wolff in Tongoloa 5 (BI = 100).
The species of Trachydium also comprise multiple groups in the East Asia clade (Trachydium 1–5), while its nomenclatural type (T. roylei) and T. subnudum C.B. Clarke ex H. Wolff are included within tribe Pleurospermeae and the Sinodielsia clade, respectively (Figs. S1 and 4). All accessions of Trachydium variabile ally as monophyletic (Trachydium 1; BI = 100) and comprise a sister group to the clade of Trachydium 2 and Sinocarum 1. Trachydium 2 comprises two accessions of Trachydium having identical ITS sequences (BI = 100); their morphology is different from other species of Trachydium and therefore they likely represent a new species. Trachydium 3 contains Trachydium simplicifolium W.W. Smith, T. soulei H.de Boissieu, T. trifoliatum, and T. sp. 3, of which the included accessions of all but the latter comprise a large polytomy having sequence divergence values of identity to 0.50%. Trachydium napiferum (Trachydium 5) allies with the Chinese endemic genus Sinolimprichtia, comprising a clade sister group to the clade of T. involucellatum R.H. Shan & F.T. Pu and Trachydium sp. 2 (Trachydium 4).
The genus Sinocarum is also not monophyletic, with accessions of S. bellum Pimenov & Kljuykov, S. coloratum, S. cruciatum, S. filicinum H. Wolff, S. muliensis L.J. Gui, Y.P. Xiao & X.J. He, S. vaginatum H. Wolff, S. wolffianum (Fedde ex H. Wolff) P.K. Mukh. & Constance, and S. sp. 1 placed into the Acronema clade (Fig. 3) and away from S. schizopetalum (Franch.) H. Wolff ex R.H. Shan & F.T. Pu and S. dolichopodum (Diels) H. Wolff ex R.H. Shan & F.T. Pu in the East Asia clade. The two Sinocarum species in the East Asia clade are placed into two separate monophyletic groups (Sinocarum 1–2). Sinocarum dolichopodum allies with Tongoloa sp. MZ054152 (in Sinocarum 1), while the two included accessions of Sinocarum schizopetalum (Sinocarum 2) fall as a sister group to the clade of Trachydium 3 and Acronema 1 (BI = 71).
The genus Acronema is referred to both the Acronema clade and the East Asia clade. Two species occur in the East Asia clade. Acronema crassifolium H.C. Wang, X.M. Zhou & Y.H. Wang (Acronema 1) allies with Trachydium 3, and A. chienii R.H. Shan (Acronema 2) occupies a separate lineage. Five species of Pimpinella fall in the East Asia clade and are divided into two distantly related groups (Pimpinella 1 and 2). The first group comprises a large polytomy (with sequence divergence values ranging from identity to 0.17 %) and includes Pimpinella acuminata (Edgew.) C.B. Clarke, P. purpurea (Franch.) H.de Boissieu, P. caudata (Franch.) H. Wolff, and one accession of P. henryi. The second group consists solely of Pimpinella rhomboidea and comprises a trichotomy with all other species of the East Asia clade except for the genus Hansenia. Lastly, Keraymonia cortiformis Cauwet & S.B. Malla, the only included species of the genus, constitutes an isolated lineage without close relatives.
4. Discussion 4.1. Circumscription and distribution of the East Asia cladeThe East Asia clade has previously been considered as comprising representatives of 16 genera (Downie et al., 2010; Zhou et al., 2020, 2021). However, with the reduction of Haplosphaera, Notopterygium, and Ligusticum litangense F.T. Pu to Hansenia (Pimenov et al., 2008; Tan et al., 2020; Zhou et al., 2020), the exclusion of dubious sequences S. brachycarpa, H. anisoptera, and V. bipinnata from the clade, and the examination of additional sequences available in GenBank for possible inclusion, the East Asia clade is herein circumscribed to include 11 genera (Table 1). Seven of these genera (Acronema, Hymenidium, Physospermopsis, Pimpinella, Sinocarum, Tongoloa, and Trachydium) are each not monophyletic, and all have members falling outside of the East Asia clade. Within the East Asia clade each of these genera except Physospermopsis comprises two or more lineages with its species intermingled with other East Asia taxa, resulting in relationships more complex than previously realized. Excluding Tongoloa, the nomenclatural types of six genera fall into other tribes and major clades of Apioideae indicating that substantial taxonomic rearrangements and new combinations will be inevitable. Upon expanded sampling of species distributed in East Asia and genera in which species have already been unambiguously placed in the clade, the number of species belonging to Hymenidium, Tongoloa, and Trachydium in the East Asia clade has increased greatly.
When the East Asia clade was first identified (as Physospermopsis clade, Calviño et al., 2006), it included species almost exclusively distributed in East Asia. With the expansion of its circumscription herein, its distribution now extends to Central and South Asia, and even Asian Russia. Two species, namely S. schizopetalum and Pimpinella purpurea, have also been reported to occur in Southeast Asia (Pimenov, 2017). All genera are native to Asia (the monotypic genus Sinolimprichtia is endemic to China). With the exception of Hymenolaena, whose distribution is limited to Central and South Asia (Afghanistan, India, Kazakhstan, Kirghizia, Pakistan, and Tajikistan; Pimenov and Kljuykov, 2000), all other genera have members distributed in China. The single species of Keraymonia (K. cortiformis) is distributed in China and Nepal (Pimenov, 2017). Six species of Hansenia [H. forbesii (H.de Boissieu) Pimenov & Kljuykov, H. forrestii, H. oviformis (R.H. Shan) Pimenov & Kljuykov, H. phaea, H. weberbaueriana (Fedde ex H. Wolff) Pimenov & Kljuykov, and H. trifoliolata Q.P. Jiang & X.J. He (not included in present analysis)] are confined to China, H. himalayensis is distributed in Bhutan, China, India and Nepal, and the type H. mongolica Turcz. occurs in Mongolia and Siberia, Russia (Sheh and Watson, 2005; Pimenov et al., 2008; Pimenov, 2017). Members of the remaining East Asia clade genera are distributed in Bhutan, China, India, Kazakhstan, Kirghizia, Myanmar, Nepal, Pakistan, Sikkim, and Tajikistan, with each genus having a species occurring in or endemic to China (Sheh et al., 2005; Pimenov, 2017; Pimenov and Kljuykov, 2000).
4.2. Morphological diversification and relationships within the East Asia cladeUnlike the tribes of Apioideae inferred from molecular data that can also be delimited using morphological or anatomical synapomorphies (e.g., Bupleureae, Neves and Watson, 2004; Oenantheae, Hardway et al., 2004; Chamaesieae, Zhou et al., 2009), the East Asia clade is morphologically heterogeneous. The differences among its members are extreme, exhibiting a great diversity of habit, life form, flower morphology, umbel structures, and fruit morphology and anatomy. The fruit presents variation in shape, degree and direction of mericarp compression (laterally compressed or slightly dorsally compressed), surface of mericarp (tuberculate or glabrous), modifications of ribs (winged or filiform), and shape of the ventral endosperm (flat or concave). We are not aware of any morphological features that can be used to unambiguously define the East Asia clade.
Of the many distinct groupings of taxa within the East Asia clade, four are equivalent to generally recognized genera (Hansenia, Sinolimprichtia, Hymenolaena, and Keraymonia). With the incorporation of species from Notopterygium and Haplosphaera, as well as the recognition of a new species, Hansenia is now represented by eight species (Pimenov et al., 2008; Tan et al., 2020; Jiang et al., 2022). It is monophyletic and diagnostic morphologically, with its (sometimes somewhat unequally) broad wings on all ribs. Sinolimprichtia is a monotypic genus endemic to China and is characterized by scaphoid petals and dark purple anthers (Pan and Watson, 2005b). It falls as a sister group to Trachydium napiferum within the East Asia clade, which in turn is sister group to T. involucellatum and Trachydium sp. 2. These three species of Trachydium may be referred to Sinolimprichtia, but this remains to be confirmed. The genus Hymenolaena consists of three distinctive species distributed in western Himalaya, Afghanistan, and Middle Asia. Its pinnate leaves and large, conspicuous, and almost entirely membranous bracteoles well delimit the genus (Pimenov and Kljuykov, 2000); the group is also monophyletic in trees presented herein. Keraymonia currently comprises four species distributed in Nepal, Bhutan, and Xizang of China (Pimenov and Kljuykov, 2000). The only species included in this study, K. cortiformis, forms an isolated lineage in the East Asia clade. The type species, Keraymonia nipaulensis Cauwet-Marc & Farill, allies closely with members of tribe Scandiceae in analyses of chloroplast gene rbcL sequences (Saslis-Lagoudakis et al., 2012). The generic status of K. cortiformis must be reconsidered.
Tongoloa is highly polyphyletic, however. Two of its species (Tongoloa zhongdianensis and T. pauciradiata) ally with the Sinodielsia clade, while the remainder form seven separate clades within the East Asia clade (Table 2). Some authors have suggested that T. zhongdianensis is conspecific with T. loloensis (Pan and Watson, 2005a). In our analysis, T. zhongdianensis is paraphyletic, with Sinodielsia delavayi arising from within. S. delavayi is the type of Sinodielsia H. Wolff, which is conspecific with Meeboldia yunnanensis (H. Wolff) Constance & F.T. Pu; thus, it has been renamed as Meeboldia delavayi (Franch.) W. Gou & X.J. He (Gou et al., 2021). All species of Meeboldia cluster together in the Acronema clade. Tongoloa loloensis, which has been variously designated as a species of Carum, Pimpinella or Trachydium (Franch, 1894; Boissieu, 1902; Hiroe, 1958), allies strongly with Tongoloa stewardii. Tongoloa pauciradiata is a poorly known species endemic to China and is not recorded in the Flora of China (Pan and Watson, 2005a); Hiroe (1979) considered it to be a species of Pimpinella. In the present study, T. pauciradiata allies with Angelica paeoniifolia R.H. Shan & C.Q. Yuan, both of which constitute a clade sister group to Vicatia thibetica H.de Boissieu (≡Sinodielsia thibetica (H.de Boissieu) Kljuykov & P.K. Mukh.). The ITS sequences of four accessions of T. pauciradiata are identical to that of A. paeoniifolia (FJ237533), while the sequences of our own accessions of A. paeoniifolia (Zhou et al., unpublished data), all falling in the Sinodielsia clade, do not match those of other conspecific members. A comparison of our material of A. paeoniifolia (Z21-117 and Z21-124) with the types (PE00935513 and PE00617248) confirm that our collection is consistent with the types and protologue presented by Shan et al. (1980). Except for groups Tongoloa 3, Tongoloa 4, and Tongoloa 5, which each include two or more species of Tongoloa, the other Tongoloa species (including the type, T. gracilis) each constitute a separate clade. The morphological features characterizing the species comprising Tongoloa 4 are subtle and include ultimate segments (length vs. width ratio), plant height (< 30 cm vs. > 30 cm), and rays (thick vs. slender). These features make species identifications extremely difficult. In addition, the morphological characters of five undescribed species (i.e., T. sp. 1, T. sp. 2, T. sp. 3, T. sp. 4, and T. sp. MT124610) prevent them from being assigned to existing Tongoloa species and describing them now as new species would only add to the confusion, given the polyphyletic nature of the genus. Overall, Tongoloa is a rather critical genus, and its proper circumscription and taxonomy will be among the most difficult challenges remaining in Apioideae.
| Species | Clade |
| Sinocarum bellum, S. coloratum, S. cruciatum, S. muliensis, S. vaginatum, S. sp. 1 | Acronema clade |
| Sinocarum dolichopodum, S. schizopetalum; Tongoloa arguta, T. dunnii, T. elata, T. filicaudicis, T. gracilis, T. loloensis, T. rubronervis, T. silaifolia, T. sp. 1, T. sp. 2, T. sp. 3, T. sp. 4, T. sp. MZ054152, T. sp. MT124610, T. taeniophylla, T. tenuifolia, T. tagongensis, T. stewardii; Trachydium involucellatum, T. napiferum, T. simplicifolium, T. souliei, T. sp. 1, T. sp. 2, T. sp. 3, T. trifoliatum, T. variabile | East Asia clade |
| Tongoloa pauciradiata, T. zhongdianensis; Trachydium subnudum | Sinodielsia clade |
| Trachydium roylei | Pleurospermeae |
Sinocarum is closely related to, and difficult to distinguish from, Acronema because the characters used to delimit these genera are variable and overlapping (Pu et al., 2005). Acronema is characterized by long-linear or long-aristate petals; however, some species have acute or obtuse petals (e.g., A. yadongense S.L. Liou and A. chinense H. Wolff) that can also be seen in Sinocarum. Both genera have species with ultimate linear segments making them indistinguishable (e.g., Acronema schneideri H. Wolff, A. graminifolium (W.W.Smith) S.L. Liou & R.H. Shan, and Sinocarum vaginatum). In the present study, species of Sinocarum and Acronema fall into both the Acronema and East Asia clades (Table 2). In the Acronema clade, their species are intermingled, indicative of their close relationship. The generic type of Sinocarum (S. coloratum) was synonymized with S. cruciatum by Pimenov (2017). However, S. coloratum allies with S. muliense, both of which comprise a clade sister group to the clade of S. cruciatum and S. vaginatum. Upon examination of these specimens, we believe that S. coloratum and S. cruciatum should each be maintained as distinct species. Sinocarum bellum does not ally with its congeners. In a previous study (Xiao et al., 2021), S. bellum fell as sister group to all Acronema species. Two other species of Sinocarum in the Acronema clade, S. filicinum and S. wolffianum, form a sister group with three species of Meeboldia. Their fruits are similar, being ovoid with filiform ribs, tapering toward the apex, slightly laterally compressed, and having a concave seed face (Wolff, 1930; Xiao et al., 2021); all have recurved styles too (Wolff, 1930; Shan and Sheh, 1985b, Table 3). As a result, we transfer S. filicinum and S. wolffianum into the genus Meeboldia (see Taxonomic treatment section). The morphological characters of S. dolichopodum, such as its long rhizome, leaf morphology, and divided bracteoles, are rather uncharacteristic of Sinocarum, suggesting that the species may be better placed elsewhere (Pu et al., 2005). The present results support such a treatment, as the species allies with several species of Tongoloa and Trachydium. The most characteristic feature of S. schizopetalum (Franch.) H. Wolff is its petal apex being palmately 3–4-lobed. This species is sister group to the Trachydium 3 group plus A. crassifolium. Its broad-ovoid mature fruits and sub-rhomboidal pollen are also different from those Sinocarum species in the Acronema clade (Xiao et al., 2021). A. crassifolium is a newly described species characterized by thickly papery, ternate, abaxially dark purple leaves, terminal umbels with 8–13 rays, and no calyx teeth (Wang et al., 2013). Because the apex of its petals is linear (ca. 1.7 mm long), it was described under the genus Acronema. However, the species falls within the East Asia clade, distantly away from most of its congeners in the Acronema clade. With its roots conic or tuberous, ultimate segments linear or linear-lanceolate, and petal bases long clawed and with an acute apex, Acronema chienii is also separated from congeners and occupies an isolated position in the East Asia clade, further adding to the taxonomic complexities of the genus and the clade as a whole.
| Character | S. filicinum | S. wolffianum | Meeboldia |
| Roots | Taproot elongate, often branched | Taproot fusiform | Taproot fusiform |
| Basal leaves | 2-pinnate | Bipinnatisect | 3-pinnate/pinnatifid |
| Bracteoles | Linear-lanceolate | Linear | Linear |
| Styles | Recurved | Recurved | Recurved |
| Calyx | Subulate | Subulate | Lanceolate-subulate |
| Fruits | Ovoid or oblong-ovate, slightly laterally compressed, tapering toward the apex, seed face concave | Ovate-oblong, slightly laterally compressed, tapering toward the apex, seed face concave | Narrowly ovoid, slightly laterally compressed, tapering toward the apex, seed face concave |
| Ribs | Filiform | Filiform | Filiform |
Of the 37 species of Hymenidium recognized by Pimenov (2017), seven species (and one unknown species) occur in the East Asia clade as three separate lineages. H. amabile is widely distributed in China (Sichuan, Yunnan, and Xizang Provinces), Bhutan, and India (Pimenov, 2017), and is easily identified by its showy leaf sheaths and bracteoles having its membranous and main veins dark purple. Hymenidium tsekuense is a narrowly endemic species of northwestern China (Qinghai Province) and is part of the Pleurospermum hookeri C.B. Clarke [ = H. hookeri (C.B. Clarke) Pimenov & Kljuykov] complex (Pan and Watson, 2005c). The main morphological differences between H. amabile and H. tsekuense are that the latter is slender in habit and has fewer rays (Shan and Sheh, 1979). In the present study, H. amabile is paraphyletic with H. tsekuense contained within, whereas H. hookeri falls into the Acronema clade. Thus, H. tsekuense and H. amabile may be regarded as conspecific or possibly close relatives having geographical variants. Hymenidium nubigenum is another poorly known taxon recorded from only a few collections, with its bracteoles margins white-membranous and ribs crisped-winged (Pan and Watson, 2005c). The sample collected from its type locality (Que'er Mtn, Sichuan Province) yielded only a portion of its ITS sequence and this was enough to inform its phylogenetic position. H. nubigenum allies with H. nanum, as either a geographically distant species in the ML analysis (Fig. 1) or a poorly supported sister group to H. tsekuense and H. amabile in the BI analysis (Fig. 5). Both H. virgatum and H. lhasanum are newly described species endemic to China (Pimenov and Kljuykov, 2004). In the protologue, the authors considered H. virgatum to be closely related to the genera Sinolimprichtia and Tongoloa. In the present study, it is a sister group to Tongoloa tagongensis. However, the morphological features of H. virgatum and T. tagongensis are distinctly different. Hymenidium lhasanum forms a sister group to three species of Hymenolaena. Both taxa have clearly different morphologies, especially in the leaf blade (1-pinnate vs. bipinnatisect) and bracteoles (completely membranous vs. membranous-margined).
Trachydium has been described as a storage-box genus, holding all taxa with shortened stems from the alpine Sino-Himalaya. Besides the two clades to which Trachydium has been referred to previously (East Asia clade and Pleurospermeae; Downie et al., 2010), one species (Trachydium subnudum) is allied with the Sinodielsia clade (Table 2 and Fig. 4). Trachydium subnudum is characterized by its scabrous leaf surfaces and densely tuberculate fruit, especially on its ribs. It occurs as a sister group to the clade of Tongoloa pauciradiata and V. thibetica. These three species share several characteristics, such as long rhizomes, carrot-like roots, and scabrous leaves, and all are distant from their respective generic types and congeners. The type species of Trachydium (T. roylei) is clustered with several species of Hymenidium in the tribe Pleurospermeae. Whether these Hymenidium species are transferred into Trachydium or separated as a distinct genus remains to be seen. Species of Trachydium in the East Asia clade each comprise separate groups. The exception is Trachydium 3, which includes four species of Trachydium. Trachydium souliei, T. simplicifolium, and T. trifoliatum are morphologically distinct, but have almost identical ITS sequences. These species are distributed primarily in the Himalayas and their origins and low sequence divergence could be the result of rapid species radiation.
Pimpinella rhomboidea is endemic to China and is characterized by its terminal leaflets being broad-ovate or rhombic. It has two varieties (P. rhomboidea and P. rhomboidea var. tenuiloba; Pu and Watson, 2005) that do not cluster together. In the protologue, the authors considered the main morphological differences between the varieties to refer to the basal leaves (ternate-2-pinnate vs. 2-ternate) and ultimate segments (1–1.5 × 0.5–1 cm vs. 5–8 × 2–5 cm); fruit characters were not used (Shan and Pu, 1989) for there are no ripe fruits on the type specimen (CDBI0172296). Pimpinella rhomboidea var. tenuiloba was synonymized with M. bipinnatum (≡ V. bipinnata, Pimenov and Kljuykov, 2006; Tan et al., 2015), which has close relationships to both Angelica angelicifolia (Franch.) Kljuykov and A. yanyuanensis (F.T. Pu) J. Zhou of tribe Selineae. The generic type of Melanosciadium, M. pimpinelloideum H.de Boissieu also belongs to Selineae, and it arises from within Angelica species. Thus, we consider that both M. bipinnatum (=Pimpinella rhomboidea var. tenuiloba) and M. pimpinelloideum will be transferred to Angelica pending confirmation from fruit characters. While M. genuflexum is away from congeners and falls into the Sinodielsia clade. Pimpinella rhomboidea occupies an isolated position in the East Asia clade. The other four species of Pimpinella in the East Asia clade comprise a polytomy. Considering that members of the genus Pimpinella are scattered in no less than seven major clades of Apioideae, there is still a long way to go to resolve its nomenclatural and taxonomic problems.
4.3. Three species with ambiguous phylogenetic placementsPhysospermopsis cuneata is a little-known species endemic to China. It is unusual within the genus because of its lack of conspicuous bracts and bracteoles (Pan and Watson, 2005d). Based on fruit structure, general habit, and leaf dissection, Pimenov and Kljuykov (1999) transferred it into the genus Sinodielsia, as S. cuneata (H. Wolff) Pimenov & Kljuykov. The results presented herein are consistent with our findings from prior studies in supporting its placement in the East Asia clade (Zhou et al., 2009, 2020). In this clade, it comprises a sister group to Tongoloa rubronervis, which in turn forms either a trichotomy with Tongoloa 5 and Pimpinella 1 (Fig. 1) or is a poorly supported sister group to Tongoloa 5 (Fig. 5). The type of Sinodielsia [S. yunnanensis H. Wolff (≡M. yunnanensis)] falls into the Acronema clade. Upon the results presented herein, the species may be better placed in Tongoloa, or a new genus awaits description.
Physospermopsis kingdon-wardii was originally described as a species of Trachydium under the name of T. kingdon-wardii H. Wolff (1929). It was later transferred into the genus Pleurospermum as P. kingdon-wardii (H. Wolff) Hiroe (1979). Based on anatomical and micromorphological fruit characters, Pu and Liu (2005) considered it more appropriately placed in Trachydium. Pimenov (2017) classified it as synonymous with Physospermopsis obtusiuscula (DC.) C. Norman, while other authors have argued that P. kingdon-wardii differs from P. obtusiuscula in its reduced stem, dwarf plants, and small fruits with prominent and sinuate ribs (Xu et al., 2021). After careful comparison of the type specimens of Physospermopsis kingdon-wardii (E00000221) and P. obtusiuscula (K000697363), we believe that the treatment of Pimenov (2017) is justified. It is also supported by the present study, as accessions of P. kingdon-wardii, with its entire or divided bracteoles and shortened stems (OR139727, OR139725, OR139724 and OR139726), and P. obtusiuscula (OR139722, OR139723 and OR139728), with its stems not shortened, comprised a polytomy. Herein, the generic attribution of P. kingdon-wardii is confirmed in the genus Physospermopsis.
Tongoloa stewardii is disjunctly distributed in Central and Southwest China and is distinguished from other species of the genus by its ovate to lanceolate-ovate ultimate segments and margins irregularly pinnate or coarsely serrate. Hiroe (1958) transferred the species to Pimpinella as P. stewardii (H. Wolff) Hiroe. Some authors considered it to be synonymous with Tongoloa fortunatii (H.de Boissieu) Pimenov & Kljuykov (Pimenov and Kljuykov, 1999; Pimenov, 2017). We have examined type materials of both species and conclude that T. stewardii and T. fortunatii cannot be separated from each other. Herein, Tongoloa stewardii is a sister group to T. loloensis, both of which fall into a large clade consisting of Trachydium, Tongoloa, and Sinocarum.
5. ConclusionThis is the most comprehensive study of the East Asia clade (Apiaceae subfamily Apioideae) carried out to date, specifically with regard to the number of new accessions examined, many of which are now included within the group. The clade is circumscribed to include representatives of 11 genera, seven (Acronema, Hymenidium, Physospermopsis, Pimpinella, Sinocarum, Tongoloa, and Trachydium) of which are not monophyletic and with some having their nomenclatural types falling outside of the group. Within the clade, the species comprising the polyphyletic genera are seriously intermingled, resulting in more confusing relationships than previously realized. The most perplexing taxonomic problems remaining in East Asia clade surround these polyphyletic genera, which are notoriously difficult to define. Thus, despite our attempt to clarify phylogenetic relationships of all genera comprising the East Asia clade, additional taxonomic study is warranted for the problematic species highlighted here.
6. Taxonomic treatmentMeeboldia filicina (H. Wolff) J. Zhou, comb. nov.
≡Sinocarum filicinum H. Wolff, 1929, Repert. Spec. Nov. Regni Veg. 27: 182, nom. inval. (Art. 35.1).
≡Carum chinense M. Hiroe, 1979, Umbelliferae World: 872.
Type: CHINA. “Yunnan, eastern flank of the Tali Range, 2540 m a.s.l., Forrest 6863” (lectotype E! –barcode K00000440, designated by M. Farille in E; isolectotype K! –barcode K000685663).
Meeboldia wolffiana (Fedde ex H. Wolff) J. Zhou, comb. nov.
≡Acronema wolffianum Fedde ex H. Wolff, Repert. Spec. Nov. Regni Veg. 27: 328. 1930.
≡Sinocarum wolffianum (Fedde ex H. Wolff) P.K. Mukh. & Constance, 1991, Edinburgh J. Bot. 48(1): 42.
≡Pimpinella feddeana M. Hiroe, 1979, Umbelliferae World: 832.
Type: INDIA. “Himalaya, Sikkim, Jongri, 12,000 ft, 15. 10. 1875, Clarke 25796” (lectotype K! –barcode K000075372, designated by Mukherjee, Constance, 1991: 42).
AcknowledgementsThis work was supported by the National Natural Science Foundation of China (no. 31960048 and 31872649), Yunnan Revitalization Talent Support Program (no. YNWRQNBJ-2019-208), the Department of Science and Technology of Yunnan Province (no. 202201AT070118), the Hundred Talents Program of Kunming Medical University (no. 60118260127), and the Gaoligong Mountain, Forest Ecosystem, Observation and Research Station of Yunnan Province (no. 202205AM070006). We thank three anonymous reviewers for valuable, constructive comments that helped improve the manuscript.
Author contributions
Jing Zhou: Writing – review & editing, Writing – original draft, Funding acquisition, Conceptualization. Zhenwen Liu: Writing – original draft, Conceptualization. Jia-Rui Yue: Investigation. Jun-Mei Niu: Formal analysis. Shi-Lin Zhou: Investigation. Xin-Yue Wang: Visualization, Data curation. Stephen R. Downie: Writing – review & editing
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.11.002.
Bentham, G., Hooker, J.D., 1867. Umbelliferae. Genera Plantarum, 1: 859-931. |
Boissier, E., 1875. Flora Orientalis. In: Calyciflorae, Gamopetalae. Geneve & Basileae: apud H. Georg, et Lugduni: apud eumdem, 3.
|
Boissieu, H.D., 1902. Note sur quelques Ombellifères de Chine d’après les collections du Muséum d’Histoire naturelle de Paris. Bulletin de l’Herbier Boissier, 2: 801-810. |
Clarkson, J.J., Zuntini, A.R., Maurin, O., et al., 2021. A higher-level nuclear phylogenomic study of the carrot family (Apiaceae). Am. J. Bot., 108: 1252-1269. DOI:10.1002/ajb2.1701 |
Calviño, C.I., Tilney, P.M., Van Wyk, B.E., et al., 2006. A molecular phylogenetic study of southern African Apiaceae. Am. J. Bot., 93: 1828-1847. DOI:10.3732/ajb.93.12.1828 |
Cheng, T., Xu, C., Lei, L., et al., 2016. Barcoding the kingdom plantae: new PCR primers for ITS regions of plants with improved universality and specificity. Mol. Ecol. Resour., 16: 138-149. DOI:10.1111/1755-0998.12438 |
Christenhusz, M., Byng, J.W., 2016. The number of known plant species in the world and its annual increase. Phytotaxa, 261: 201-217. DOI:10.11646/phytotaxa.261.3.1 |
Constance, L., 1971. History of the classification of Umbelliferae (Apiaceae). In: Heywood, V.H. (Ed.), The Biology and Chemistry of the Umbelliferae. Academic Press, pp. 1-8.
|
Darriba, D., Taboada, G.L., Doallo, R., et al., 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods, 9: 772. DOI:10.1038/nmeth.2109 |
De Candole, A.P., 1827. Dictionnaire classique d’histoire naturelle 11. Rey et Gravier, ed. 406. Libraries-Editeurs.
|
Downie, S.R., Plunkett, G.M., Watson, M.F., et al., 2001. Tribes and clades within Apiaceae subfamily Apioideae: the contribution of molecular data. Edinb. J. Bot., 58: 301-330. DOI:10.1017/s0960428601000658 |
Downie, S.R., Spalik, K., Katz-Downie, D.S., et al., 2010. Major clades within Apiaceae subfamily Apioideae as inferred by phylogenetic analysis of nrDNA ITS sequences. Plant Divers. Evol., 128: 111-136. DOI:10.1127/1869-6155/2010/0128-0005 |
Fereidounfar, S., Ghahremaninejad, F., Khajehpiri, M., 2016. Phylogeny of the Southwest Asian Pimpinella and related genera based on nuclear and plastid sequences. Genet. Mol. Res., 15: gmr15048767. |
Franchet, A., 1894. Notes sur quelques Ombellifères du Yunnan. Bulletin de la Société Philomathique de Paris, 86: 106-146. |
Gou, W., Guo, X.L., Zhou, S.D., et al., 2021. Phylogeny and taxonomy of Meeboldia, Sinodielsia and their relatives (Apiaceae: Apioideae) inferred from nrDNA ITS, plastid DNA intron (rpl16 and rps16) sequences and morphological characters. Phytotaxa, 482: 121-142. DOI:10.11646/phytotaxa.482.2.2 |
Gui, L.J., Jia, S.B., Guo, X.L., Price, M., Zhou, S.D., He, X.J., 2020a. Tongoloa tagongensis (Apiaceae), a new species from the hengduan mountains, China. Phytotaxa, 461(1): 17-25. |
Gui, L.J., Wen, J., Xiao, Y.P., Ren, T., Zheng, H.Y., He, X.J., 2020b. Tongoloa arguta (Apiaceae), a new species from southwest China. PhytoKeys, 164: 11-19. DOI:10.3897/phytokeys.164.54927 |
Hall, T., 1999. BioEdit: Biological Sequence Alignment Editor for Win95/98/NT/2K/XP version 6.0.7. http://www.mbio.ncsu.edu/BioEdit/bioedit.html.
|
Hardway, T.M., Spalik, K., Watson, M.F., et al., 2004. Circumscription of Apiaceae tribe Oenantheae. South Afr. J. Bot., 70: 393-406. DOI:10.1016/S0254-6299(15)30222-2 |
Hiroe, M., 1958. Umbelliferae of Asia (Excluding Japan). Kyoto: Eicodo (Akira Imagawa). Maruzen Ltd: pp. 1-219.
|
Hiroe, M., 1979. Umbelliferae of World. Tokyo: Ariake Book Company: pp. 741-747.
|
Jia, S.B., Guo, X.L., Zhou, S.D., et al., 2019. Hansenia pinnatiinvolucellata is conspecific with H. weberbaueriana (Apiaceae) based on morphology and molecular data. Phytotaxa, 418: 203-210. DOI:10.11646/phytotaxa.418.2.5 |
Jiang, Q.P., Price, M., Zhang, X.Y., et al., 2022. Hansenia trifoliolata, a new species (Apiaceae) from Shaanxi, China. PhytoKeys, 213: 79-93. DOI:10.3897/phytokeys.213.83632 |
Mousavi, S., Mozaffarian, V., Mummenhoff, K., et al., 2021. An updated lineage-based tribal classification of Apiaceae subfamily Apioideae with special focus on Iranian genera. Syst. Biodivers., 19: 89-109. DOI:10.1080/14772000.2020.1834002 |
Neves, S.S., Watson, M.F., 2004. Phylogenetic relationships in Bupleurum (Apiaceae) based on nuclear ribosomal DNA ITS sequence data. Ann. Bot., 93: 379-398. DOI:10.1093/aob/mch052 |
Pan, Z.H., Watson, M.F., 2005a. Tongoloa H. Wolff. In: Flora of China Editorial Committee Eds. Flora of China. Beijing: Science Press, 14. Missouri Botanical Garden Press, St.Louis, pp. 34-37.
|
Pan, Z.H., Watson, M.F., 2005b. Sinolimprichtia H.Wolff. In: Flora of China Editorial Committee Eds. Flora of China. Beijing: Science Press, 14. Missouri Botanical Garden Press, St.Louis, pp. 55-56.
|
Pan, Z.H., Watson, M.F., 2005c. Pleurospermum Hoffm. In: Flora of China Editorial Committee Eds. Flora of China. Beijing: Science Press, 14. Missouri Botanical Garden Press, St.Louis, pp. 158-169.
|
Pan, Z.H., Watson, M.F., 2005d. Physospermopsis H.Wolff. In: Flora of China Editorial Committee Eds. Flora of China. Beijing: Science Press, 14. Missouri Botanical Garden Press, St.Louis, pp. 31-33.
|
Pimenov, M.G., 2017. Updated checklist of Chinese Umbelliferae: nomenclature, synonymy, typification, distribution. Turczaninowia, 20: 106-239. DOI:10.14258/turczaninowia.20.2.9 |
Pimenov, M.G., Kljuykov, E.V., 1999. New nomenclatural combinations for Chinese Umbelliferae. Feddes Repert., 110: 481-491. DOI:10.1002/fedr.19991100705 |
Pimenov, M.G., Kljuykov, E.V., 2000. Taxonomic revision of Pleurospermum Hoffm. And related genera of Umbelliferae. Ⅱ: the genera Pleurospermum, Pterocyclus, Trachydium, Keraymonia, Pseudotrachydium, Aulacospermum, and Hymenolaena. Feddes Repert., 111: 517-534. DOI:10.1002/fedr.20001110718 |
Pimenov, M.G., Kljuykov, E.V., 2004. Hymenidium (Umbelliferae) in China and India: the new species, a new combination and some corrections. Bot. Zh. (St. Petersbg.), 89: 1652-1665. |
Pimenov, M.G., Kljuykov, E.V., 2006. Generic disposition of Vicatia bipinnata in Melanosciadium and a new species (Umbelliferae). Feddes Repert., 117(7–8): 471-472. |
Pimenov, M.G., Kljuykov, E.V., Ostroumova, T.A., 2008. Reduction of Notopterygium to Hansenia (Umbelliferae). Willdenowia, 38: 155-172. DOI:10.3372/wi.38.38110 |
Pimenov, M.G., Leonov, M.V., 1993. The Genera of the Umbelliferae: A Nomenclator. London: Royal Botanic Gardens.
|
Posada, D., 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol., 25: 1253-1256. DOI:10.1093/molbev/msn083 |
Pu, G.Z., Liu, Q.X., 2005. Comparative anatomical study on the genus Physospermopsis fruit from China and its systematic significance. J. Plant Resour. Environ., 14: 1-6. |
Pu, F.D., Watson, M.F., 2005. Pimpinella L. In: Flora of China Editorial Committee Eds. Flora of China. Beijing: Science Press, 14. Missouri Botanical Garden Press, St.Louis, pp. 93-104.
|
Pu, F.D., Watson, M.F., Holmes-Smith, I., 2005. Sinocarum H. Wolff ex R.H.Shan & F.T.Pu. In: Flora of China Editorial Committee Eds. Flora of China. Beijing: Science Press, 14. Missouri Botanical Garden Press, St.Louis, pp. 82-85.
|
Ronquist, F., Huelsenbeck, J.P., 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572-1574. DOI:10.1093/bioinformatics/btg180 |
Ronquist, F., Teslenko, M., Van der Mark, P., et al., 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol., 61: 539-542. DOI:10.1093/sysbio/sys029 |
Saslis-Lagoudakis, C.H., Savolainen, V., Williamson, E.M., et al., 2012. Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proc. Natl. Acad. Sci. U.S.A., 109: 15835-15840. DOI:10.1073/pnas.1202242109 |
Shan, R.H., Pu, F.D., 1989. New taxa of the Chinese Umbelliferae (3). Acta Phytotaxon. Sin., 27: 62-67. |
Shan, R.H., Sheh, M.L., 1979. Pleurospermum Hoffm. In: Flora Reipublicae Popularis Sinicae Editorial Committee Eds. Flora Reipublicae Popularis Sinicae, 55 (1). Science Press, Beijing, pp. 133-184.
|
Shan, R.H., Sheh, M.L., 1985a. Pimpinella L. In: Flora Reipublicae Popularis Sinicae Editorial Committee Eds. Flora Reipublicae Popularis Sinicae, 55 (2). Science Press, Beijing, pp. 67-113.
|
Shan, R.H., Sheh, M.L., 1985b. Sinocarum Wolff ex R.H.Shan & F.T.Pu. In: Flora Reipublicae Popularis Sinicae Editorial Committee Eds. Flora Reipublicae Popularis Sinicae, 55 (2). Science Press, Beijing, pp. 30-38.
|
Shan, R.H., Sheh, M.L., Yuan, C.Q., et al., 1980. New taxa of the Umbelliferae from Xizang (Tibet). Acta Phytotaxon. Sin., 18: 374-379. |
Sheh, M.L., Pu, F.D., Pan, Z.H., et al., 2005. Apiaceae. In: Flora of China Editorial Committee Eds. Flora of China. Beijing: Science Press, 14. Missouri Botanical Garden Press, St.Louis, pp. 1–205.
|
Sheh, M.L., Watson, M.F., 2005. Haplosphaera Hand.-Mazz. In: Flora of China Editorial Committee Eds. Flora of China. Beijing: Science Press, 14. Missouri Botanical Garden Press, St.Louis, pp. 37-38.
|
Stamatakis, A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Tan, J.B., Jia, S.B., He, X.J., et al., 2020. Accommodating Haplosphaera in Hansenia (Apiaceae) based on morphological and molecular evidence. Phytotaxa, 464: 207-216. DOI:10.11646/phytotaxa.464.3.2 |
Tan, J.B., Wang, Z.X., Xie, D.F., et al., 2015. Pimpinella rhomboidea var. tenuiloba is a synonym of Melanosciadium bipinnatum (Apiaceae). Nord. J. Bot., 33: 659-661. DOI:10.1111/njb.00748 |
Wang, H.C., Zhou, X.M., Sun, H., et al., 2013. Acronema crassifolium sp. nov. (Apiaceae), a distinct new species from Yunnan, southwest China. Phytotaxa, 87: 39-44. DOI:10.11646/phytotaxa.87.3.1 |
Wang, Z.X., Downie, S.R., Tan, J.B., et al., 2014. Molecular phylogenetics of Pimpinella and allied genera (Apiaceae), with emphasis on Chinese native species, inferred from nrDNA ITS and cpDNA intron sequence data. Nord. J. Bot., 32: 642-657. DOI:10.1111/j.1756-1051.2013.00343.x |
Wen, J., Xie, D.F., Price, M., et al., 2021. Backbone phylogeny and evolution of Apioideae (Apiaceae): new insights from phylogenomic analyses of plastome data. Mol. Phylogenet. Evol., 161: 107183. DOI:10.1016/j.ympev.2021.107183 |
Wen, J., Zimmer, E.A., 1996. Phylogeny and biogeography of Panax L. (the ginseng genus, Araliaceae): inferences from ITS sequences of nuclear ribosomal DNA. Mol. Phylogenet. Evol., 6: 167-177. DOI:10.1006/mpev.1996.0069 |
White, T.J., Bruns, T., Lee, S., et al., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J. (Eds.), PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, pp. 315-322.
|
Wolff, H., 1910. Umbelliferae-Apioideae-Bupleurum, Trinia et reliquae Ammineae heteroclitae. In: Engler, A. (Ed.), Das Pflanzenreich Ⅳ, 43. Wilhelm Engelmann, Berlin, pp. 1-214.
|
Wolff, H., 1927. Umbelliferae-Apioideae-Ammineae-Carinae, Ammineaenovemjugatae et genuinae. In: Engler, A. (Ed.), Das Pflanzenreich Ⅳ, 90. Wilhelm Engelmann, Berlin, pp. 1-398.
|
Wolff, H., 1929. Umbelliferae Asiatiae novae relictae. Ⅱ. Repertorium specierum novarum regni vegetabilis, 27: 179-192. DOI:10.1002/fedr.4870270909 |
Wolff, H., 1930. Umbelliferae asiaticae novae relictae. Ⅲ. Repertorium specierum novarum regni vegetabilis, 27: 301-335. DOI:10.1002/fedr.4870271612 |
Xiao, Y.P., Guo, X.L., Price, M., et al., 2021. New insights into the phylogeny of Sinocarum (Apiaceae, Apioideae) based on morphological and molecular data. PhytoKeys, 175: 13-32. DOI:10.3897/phytokeys.175.60592 |
Xu, X.R., Guo, X.L., Price, M., et al., 2021. New insights into the phylogeny and taxonomy of Chinese Physospermopsis (Apiaceae). PhytoKeys, 175: 67-78. DOI:10.3897/phytokeys.175.57681 |
Zhou, J., Gao, Y.Z., Wei, J., et al., 2020. Molecular phylogenetics of Ligusticum (Apiaceae) based on nrDNA ITS sequences: rampant polyphyly, placement of the Chinese endemic species, and a much-reduced circumscription of the genus. Int. J. Plant Sci., 181: 306-323. DOI:10.1086/706851 |
Zhou, J., Gong, X., Downie, S.R., et al., 2009. Towards a more robust molecular phylogeny of Chinese Apiaceae subfamily Apioideae: additional evidence from nrDNA ITS and cpDNA intron (rpl16 and rps16) sequences. Mol. Phylogenet. Evol., 53: 56-68. DOI:10.1016/j.ympev.2009.05.029 |
Zhou, J., Guo, M.J., Liu, Z.W., 2021. Advances in the study of systematics of “East-Asia clade” in Apiaceae. Guihaia, 41: 21-30. |
Zhou, J., Peng, H., Downie, S.R., et al., 2008. A molecular phylogeny of Chinese Apiaceae subfamily Apioideae inferred from nuclear ribosomal DNA internal transcribed spacer sequences. Taxon, 57: 402-416. DOI:10.2307/25066012 |



