b. College of Information Science and Technology, Nanjing Forestry University, Nanjing 210037, China;
c. Research Institute of Subtropical Forestry, Chinese Academy of Forestry, Hangzhou 311400, China;
d. Co-Innovation Center for Sustainable Forestry in Southern China, College of Life Sciences, Nanjing Forestry University, Nanjing 210037, China;
e. Department of Paleontology, University of Vienna, Vienna, Austria
There are three distinct genetic systems in higher plants, the dominant nuclear genome and the semi-autonomous organelle genomes (plastids and mitochondria). In contrast to the conserved plastid genome (plastome), the plant mitochondrial genome (mitogenome) is characterized by an intriguing “evolutionary paradox” distinguished by a remarkably low mutation rate but with a significantly high rearrangement rate (Palmer and Herbon, 1988; Lai et al., 2022). Plant mitochondria are considered as an important genetic system for studying the evolution of genome structure and functional content due to their extremely low sequence mutation rate, but frequent genomic recombination (Wu et al., 2022). They are also an ideal resource for studying the mechanisms of plant genetic diversity formation and maintenance (Wang et al., 2022). However, plant mitogenomes contain a large number of repetitive sequences and foreign DNA transfers, which lead to the fragmentation of mitogenome assembly and makes it difficult to obtain a complete mitogenome. So far, the total number of released plant mitogenomes is less than 5% of the plastomes, most of which are from eudicots and bryophytes, while the remainder belong to other land plants (e.g., ferns, gymnosperms, magnoliids, and monocots).
Plant mitogenomes have many unique evolutionary features, as compared to the compact and conservative animal mitogenomes. The most notable difference is that animal mitogenomes are very limited in size (15–18 kb), whereas plant mitogenomes span a wide range of sizes (66 kb–12 Mb). Comparative genomic analyses have revealed that frequent mitogenomic recombination and foreign DNA transfers could integrate large amounts of foreign DNA during evolution, ultimately leading to dramatic variations in the size and structure of the plant mitogenomes (Rice et al., 2013; Wu et al., 2022). A plant mitogenome is conventionally depicted as a single master circle (MC), like a plastome and animal mitogenome. With recent advances in DNA sequencing technology, the increased number of plant mitogenomes suggests that the real structure of the plant mitogenome is far more complicated than a single MC model would represent (Sloan, 2013). In addition to inducing the variations in mitogenomic structure and size, recombination and foreign DNA transfers also modify the gene and intron contents of the mitogenome. In terms of intron contents among land plant mitogenomes, the number of introns, including group Ⅰ and Ⅱ introns, ranges from only 4 in Viscum scurruloideum to 45 in Anthoceros angustus. The content of group Ⅱ introns is highly conserved in vascular plants, although less so relative to nonvascular plants (Mower, 2020).
The largest number of mitogenomes reported so far are those of angiosperms, which are characterized by a variety of unusual properties, including variable sizes, frequent posttranscriptional modifications, low gene densities, and extensive foreign DNA transfers (Knoop et al., 2011). Investigations of the mitogenomes of primitive angiosperms will facilitate the comprehensive insight into the evolutionary patterns of plant mitogenomes. The family Lauraceae is the largest family in the order Laurales of primitive angiosperms and comprises more than 3000 species in ca. 55 genera, which are widely distributed in tropical and subtropical regions (Yang et al., 2022b). Characterizing the genetic information of the Lauraceae is thus of great significance for understanding the evolution, phylogeny, and sustainable utilization of the family. Cinnamomum belongs to the family Lauraceae, and is now known to be restricted to the Old World. The genus possesses an unusual combination of morphological characters, e.g., evergreen trees, tripliveined leaves opposite or subopposite, paniculate inflorescences with ultimate cymes having strictly opposite lateral flowers, tepals mostly partially persistent in fruits. Trees of the genus are economically important because they have been widely used for their chemical components either as spices, or traditional Chinese medicines, or essential oils.
Mitogenomic data are uniparental and have been used for phylogenetic studies in many groups of higher plants. However, it remains unclear if mitochondrial sequences or genomic data are useful for the phylogeny of the family Lauraceae or not. There is not any mitogenome data of Lauraceae available in the public nucleotide database of NCBI, although it is known that the mitogenomes of the family have experienced an unusual and complicated evolutionary history (e.g., Cassytha) (Zhang et al., 2020a, 2022). With the development of sequencing technologies, the PacBio HiFi sequencing method has yielded highly accurate long-read sequencing datasets (Hon et al., 2020), which have great application potential in genome assembly and complex structure detection. This advancement has made it possible to reveal the complicated metagenomes. Here, we assembled the mitogenome of Cinnamomum chekiangense into a single master circle using HiFi sequencing data to provide the first mitogenome reference for the family Lauraceae. We further annotated and characterized the mitogenome and conducted a phylogenetic study to better understand its evolutionary significance.
Using the Revio sequencing platform, we obtained a total of 448,389 HiFi sequencing reads, with the maximum and average lengths of 32,397 bp and 14,109 bp, respectively. With the highly accurate long-read sequencing data, the complete mitogenome of C. chekiangense was assembled into a single circular molecule with a total length of 750,457 bp (GenBank accession number: ncbi-n:NC_082065) (Fig. 1A), which is the average size of mitogenomes in magnoliids. In fact, the mitogenome sizes vary considerably among the magnoliid clade, ranging from 535,805 bp in Hernandia nymphaeifolia to 967,100 bp in Magnolia biondii. The mitogenome structure of magnoliids is also variable, most being assembled into a single circular molecule (e.g., M. biondii, M. officinalis, M. figo, Liriodendron tulipifera, and H. nymphaeifolia), while some were assembled into multi-circular molecules (e.g., Saururus chinensis and Machilus pauhoi; Yu et al., 2023).
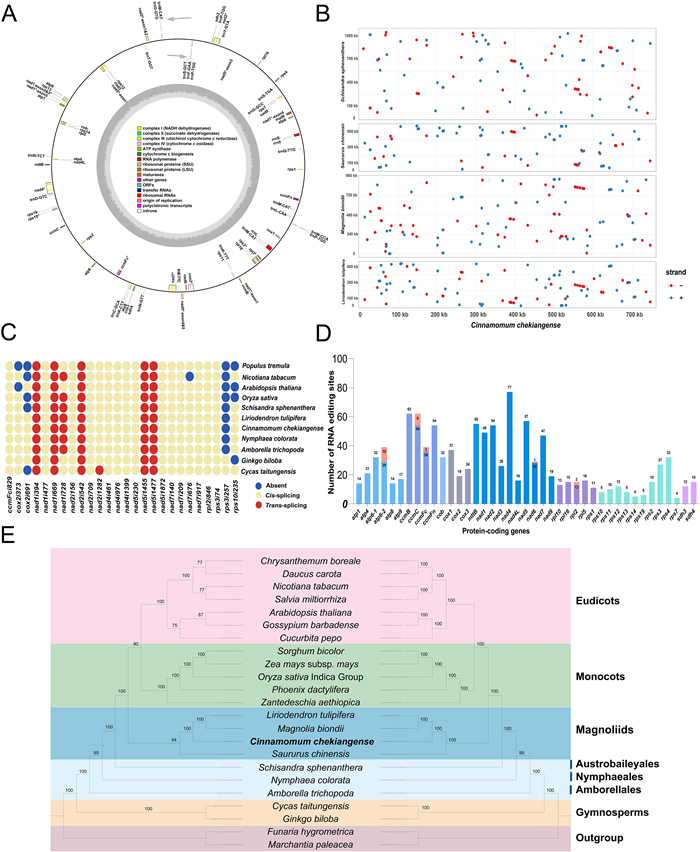
|
| Fig. 1 (A) The Circular mitogenome map of Cinnamomum chekiangense. Asterisks beside genes represent intron-containing genes. Genes with different functions are depicted using different colors. (B) Schematic representation of the collinearity among five magnoliid mitogenomes. The blue and red dots represent the direct and inverted syntenic regions, respectively. (C) Intron content among 11 seed plant mitogenomes. Blue, intron is absent; Yellow, intron is cis-splicing; Red, intron is trans-splicing. (D) Characteristic of RNA editing sites across all PCGs in the mitogenome of C. chekiangense. PCGs with different functions are depicted using different colors. (E) The phylogenetic relationships of 24 plant species based on mitochondrial PCGs (left) and complete plastome sequences (right). Funaria hygrometrica and Marchantia paleacea were used as outgroup. Numbers on each branch are bootstrap support values. Colors indicate the groups for each species. |
Frequent rearrangement is the major driver of plant mitogenome evolution. Using the nucmer program of MUMmer v.3.23 (Kurtz et al., 2004), we compared the mitogenome of C. chekiangense with another four closely related mitogenomes, i.e., L. tulipifera, M. biondii, Saururus chinensis, and S. sphenanthera. As clearly illustrated in Fig. 1B, the five mitogenomes exhibit very poor collinearity, with numerous regions lacking homology between these mitogenomes. The highest collinearity (~37%) was found between the mitogenomes of C. chekiangense and L. tulipifera, while the collinearity between other species and C. chekiangense was less than 25% (Table S1). The results of collinearity analysis showed that the mitogenome of Lauraceae may have experienced frequent genomic rearrangements during evolution, making it difficult to establish the ancestral mitogenomic structure.
Despite the substantial loss or transfer of mitochondrial protein-coding genes (PCGs) to the nucleus during the endosymbiosis of mitochondria, the plant mitogenomes still retain some unique PCGs (Zardoya, 2020). These retained PCGs of land plant mitogenomes include 24 core genes (atp1, 4, 6, 8, and 9; ccmB, C, Fc, and Fn; cob, cox1–3, nad1–7, 9, and 4L; mttB, and matR) and 19 variable genes (sdh3, sdh4, rpl2, 5, 6, 10, and 16; rps1–4, 7, 8, 10–14, and 19). In this study, most of the PCGs were identified in the C. chekiangense mitogenome, and only one core PCG (matR) and two variable PCGs (rpl6 and rps8) were lost during evolution (Fig. 1A and Table S2). Almost all of the ancestral PCGs have also been found in other mitogenomes of magnoliids (Fig. S1), such as M. biondii, S. chinensis and L. tulipifera. Additionally, the basally-diverging groups of angiosperms (ANA: Amborellales, Nymphaeales and Austrobaileyales) also retained almost all mitochondrial PCGs. In contrast to the mitogenomes of magnoliids and ANA clade, which have retained almost all of the ancestral repertoire, some gymnosperms (e.g., Welwitschia mirabilis), hornworts (e.g., Phaeoceros laevis and Anthoceros angustus), and the hemiparasitic angiosperm Viscum scurruloideum have dispensed with half or more of these PCGs (Mower, 2020).
Most eukaryotic mitochondrial introns are of two types, group Ⅰ and group Ⅱ, which differ in their splicing mechanisms and secondary structure (Mower, 2020). A total of 19 cis-spliced and 6 trans-spliced introns were identified in 13 PCGs (Fig. 1C). All introns in the C. chekiangense mitogenome belong to group Ⅱ. Using the naming scheme proposed by Dombrovska and Qiu (2004), each intron was named according to its position relative to the reference gene in the Marchantia polymorpha mitogenome. Comparison of cis- and trans-splicing intron content in seed plant mitogenomes revealed a relatively conservative pattern of intron evolution. Most of the introns were shared among seed plants (Fig. 1C), with the exception of nad1i728, rps3i257, cox2i691, cox2i373, and rps10i235. Among these, rps3i257 was completely lost during the divergence of angiosperms and gymnosperms, whereas nad1i728 was retained in some angiosperms (e.g., Arabidopsis thaliana and Populus tremula). Additionally, nad7i676 was lost from the mitogenome of Nicotiana tabacum, but was retained in other seed plants.
RNA editing is a common phenomenon in plant mitogenomic transcripts and may lead to massive diversity in post-transcriptional gene sequences. The number of RNA editing sites varies substantially across plant lineages. RNA editing is rare in mosses and liverworts (Rüdinger et al., 2009), but it is abundant in lycophytes (Zhang et al., 2020b). In gymnosperms, the number of RNA editing sites varies from as few as 99 sites in Welwitschia mirabilis to 1405 sites in Ginkgo biloba (Fan et al., 2019). RNA editing frequency is generally lower in angiosperms, especially in monocots and eudicots. The number of RNA editing sites in the mitogenomes of angiosperms is between 400 and 500 (Bi et al., 2016). In this study, we utilized GATK (https://github.com/broadinstitute/gatk), Bcftools (Danecek et al., 2021) and REDItools (Picardi and Pesole, 2013) to identify RNA editing sites. The thresholds to define an RNA editing site are QUAL > 30, depth > 100 ×, and P-value > 0.1. We identified a total of 1119 RNA editing sites in 41 PCGs of the C. chekiangense mitogenome based on RNA sequencing data (Fig. 1D and Table S3), which is the highest number in angiosperms to date. The above results suggest that the decreasing number of RNA-editing sites may be caused by gene loss from liverworts, mosses, gymnosperms to angiosperms. Although we have manually checked all RNA editing sites using IGV (Thorvaldsdottir et al., 2012), PCR experiment and Sanger sequencing are also required to obtain a more accurate result.
The advent of high-throughput sequencing (HTS) has allowed plant systematists to address long-standing phylogenetic issues at different taxonomic levels. In plant phylogenomic studies, plastomes have been widely used to infer phylogenetic relationships at different taxonomic levels due to their easily assembled genomes (Twyford and Ness, 2016; Yang et al., 2022a). In contrast, the mitogenomes have been largely neglected in plant phylogenies due to the difficulty of obtaining complete mitogenomes and generally low rates of nucleotide substitution (Sloan et al., 2009). Despite extensive studies on the early diversification of five major lineages in Mesangiospermae (Ceratophyllales, Chloranthales, eudicots, magnoliids, and monocots), their phylogenetic relationships remain elusive. In recent phylogenetic trees inferred from organellar genomes, monocots have been considered to be more closely related to eudicots than to magnoliids (Li et al., 2019, 2021; Xue et al., 2022). However, the phylogenetic relationships inferred from nuclear genes are more chaotic and unstable among the five Mesangiospermae clades (Zhang et al., 2019; Guo et al., 2021; Ma et al., 2021), implying possible hybridization and incomplete lineage sorting in the early history of angiosperms.
To investigate the phylogenetic position of C. chekiangense and the magnoliids relative to the monocots and eudicots, our study utilized the whole plastome sequences and 23 conserved mitochondrial PCGs to reconstruct the phylogenetic maximum likelihood (ML) trees of 24 plant species, respectively. These conserved mitochondrial PCGs were extracted to perform the multiple sequence alignment in MAFFT v.7.407 (Katoh and Standley, 2013). The aligned sequences were subsequently concatenated to construct the ML tree in IQ-TREE v.2.0.3 with 1000 bootstrap replicates (Minh et al., 2020). Both of the phylogenetic trees emphasized the magnoliids as a sister group to the clade comprising monocots and eudicots (Fig. 1E), which is consistent with the APG IV botanical classification system (Angiosperm Phylogeny Group Ⅳ et al., 2016). Previous study has used complete set of mitochondrial genes from 18 angiosperms to elucidate the phylogenetic relationships among the five Mesangiospermae clades of angiosperms (Xue et al., 2022). The results provided valuable information and alternative hypotheses to investigate the early evolution of angiosperms. However, it remains unclear whether mitogenomes contribute to the phylogeny of the family Lauraceae or not, as no mitogenomes have been published in this family. With the improvement of HTS technology and the development of effective genome assembly methodology for complex plant mitogenomes, we will be able to further investigate the large-scale phylogenetic relationships based on mitogenome sequences. In due course, some of the complex phylogenetic issues may be resolved based on the genomic data from nuclear, plastid, and mitochondrial genomes.
As the first reported mitogenome in the Lauraceae family, this study provides a valuable reference for mitogenome analysis in Lauraceae. Simultaneously, it provides important insights into RNA editing, mitogenome evolution, and phylogeny in angiosperms.
AcknowledgmentsThe work is supported by the Natural Science Foundation of Jiangsu Province (BK20220414) and the Natural Science Foundation of the Higher Education Institutions of Jiangsu Province (22KJB220003).
Data availability
The mitochondrial and plastid genomes supporting this study are available at GenBank with accession numbers: ncbi-n:NC_082065 and OR360835, respectively. The HiFi and RNA sequencing data of C. chekiangense are deposited in the SRA repository under SRR26158200 and SRR26157632, respectively.
Declaration of competing interest
The authors declare no conflicts of interest.
Author contributions
CB and YY planned and designed the research. CB, NS, and FH analyzed the data and prepared the figures. CB and KX provided the materials and conducted experiments. CB wrote the initial version of the manuscript. YY and DKF revised this and provided comments. All authors read and approved the manuscript.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.11.001.
Angiosperm Phylogeny Group Ⅳ, Chase, M.W., Christenhusz, M.J., et al., 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc., 181: 1-20. DOI:10.1111/boj.12385 |
Bi, C., Paterson, A.H., Wang, X., et al., 2016. Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches. BioMed Res. Int., 2016: 1-18. DOI:10.1155/2016/5040598 |
Danecek, P., Bonfield, J.K., Liddle, J., et al., 2021. Twelve years of SAMtools and BCFtools. GigaScience, 10: giab008. DOI:10.1093/gigascience/giab008 |
Dombrovska, O., Qiu, Y.L., 2004. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol. Phylogenet. Evol., 32: 246-263. DOI:10.1016/j.ympev.2003.12.013 |
Fan, W., Guo, W., Funk, L., et al., 2019. Complete loss of RNA editing from the plastid genome and most highly expressed mitochondrial genes of Welwitschia mirabilis. Sci. China Life Sci., 62: 498-506. DOI:10.1007/s11427-018-9450-1 |
Guo, C., Ma, P.-F., Yang, G.-Q., et al., 2021. Parallel ddRAD and genome skimming analyses reveal a radiative and reticulate evolutionary history of the temperate bamboos. Syst. Biol., 70: 756-773. DOI:10.1093/sysbio/syaa076 |
Hon, T., Mars, K., Young, G., et al., 2020. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci. Data, 7: 399. DOI:10.1038/s41597-020-00743-4 |
Katoh, K., Standley, D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol., 30: 772-780. DOI:10.1093/molbev/mst010 |
Knoop, V., Volkmar, U., Hecht, J., et al., 2011. Mitochondrial genome evolution in the plant lineage. In: Kempken, F. (Ed.), Plant Mitochondria. Springer, New York, NY, pp. 3–29.
|
Kurtz, S., Phillippy, A., Delcher, A.L., et al., 2004. Versatile and open software for comparing large genomes. Genome Biol., 5: R12. DOI:10.1186/gb-2004-5-2-r12 |
Lai, C., Wang, J., Kan, S., et al., 2022. Comparative analysis of mitochondrial genomes of Broussonetia spp. (Moraceae) reveals heterogeneity in structure, synteny, intercellular gene transfer, and RNA editing. Front. Plant Sci., 13: 1052151. DOI:10.3389/fpls.2022.1052151 |
Li, H.-T., Luo, Y., Gan, L., et al., 2021. Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biology, 19: 232. DOI:10.1186/s12915-021-01166-2 |
Li, H.-T., Yi, T.-S., Gao, L.-M., et al., 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants, 5: 461-470. DOI:10.1038/s41477-019-0421-0 |
Ma, J., Sun, P., Wang, D., et al., 2021. The Chloranthus sessilifolius genome provides insight into early diversification of angiosperms. Nat. Commun., 12: 6929. DOI:10.1038/s41467-021-26931-3 |
Minh, B.Q., Schmidt, H.A., Chernomor, O., et al., 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol., 37: 1530-1534. DOI:10.1093/molbev/msaa015 |
Mower, J.P., 2020. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion, 53: 203-213. DOI:10.1016/j.mito.2020.06.002 |
Palmer, J.D., Herbon, L.A., 1988. Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J. Mol. Evol., 28: 87-97. DOI:10.1007/BF02143500 |
Picardi, E., Pesole, G., 2013. REDItools: high-throughput RNA editing detection made easy. Bioinformatics, 29: 1813-1814. DOI:10.1093/bioinformatics/btt287 |
Rice, D.W., Alverson, A.J., Richardson, A.O., et al., 2013. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science, 342: 1468-1473. DOI:10.1126/science.1246275 |
Rüdinger, M., Funk, H.T., Rensing, S.A., et al., 2009. RNA editing: only eleven sites are present in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol. Genet. Genom., 281: 473-481. DOI:10.1007/s00438-009-0424-z |
Sloan, D.B., 2013. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol., 200: 978-985. DOI:10.1111/nph.12395 |
Sloan, D.B., Oxelman, B., Rautenberg, A., et al., 2009. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol. Biol., 9: 12. DOI:10.1186/1471-2148-9-12 |
Thorvaldsdottir, H., Robinson, J.T., Mesirov, J.P., 2012. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings Bioinf., 14: 178-192. |
Twyford, A.D., Ness, R.W., 2016. Strategies for complete plastid genome sequencing. Mol. Ecol. Resour., 17: 858-868. |
Wang, N., Li, C.C., Kuang, L.H., et al., 2022. Pan-mitogenomics reveals the genetic basis of cytonuclear conflicts in citrus hybridization, domestication, and diversification. Proc. Natl. Acad. Sci. U.S.A., 119: e2206076119. DOI:10.1073/pnas.2206076119 |
Wu, Z.Q., Liao, X.Z., Zhang, X.N., et al., 2022. Genomic architectural variation of plant mitochondria-A review of multichromosomal structuring. J. Syst. Evol., 60: 160-168. DOI:10.1111/jse.12655 |
Xue, J.Y., Dong, S.S., Wang, M.Q., et al., 2022. Mitochondrial genes from 18 angiosperms fill sampling gaps for phylogenomic inferences of the early diversification of flowering plants. J. Syst. Evol., 60: 773-788. DOI:10.1111/jse.12708 |
Yang, Z., Deng, C., Wang, L., et al., 2022a. A new species of Cinnamomum (Lauraceae) from southwestern China. PhytoKeys, 202: 35-44. DOI:10.3897/phytokeys.202.76344 |
Yang, Z., Liu, B., Yang, Y., et al., 2022b. Phylogeny and taxonomy of Cinnamomum (Lauraceae). Ecol. Evol., 12: e9378. |
Yu, R., Chen, X., Long, L., et al., 2023. De novo assembly and comparative analyses of mitochondrial genomes in Piperales. Genome Biol. Evol., 15: evad041. |
Zardoya, R., 2020. Recent advances in understanding mitochondrial genome diversity. F1000Research, 9: F1000. |
Zhang, C., Ma, H., Sanchez-Puerta, M.V., et al., 2020a. Horizontal gene transfer has impacted cox1 gene evolution in Cassytha filiformis. J. Mol. Evol., 88: 361-371. DOI:10.1007/s00239-020-09937-1 |
Zhang, H., Florentine, S., Tennakoon, K.U., 2022. The angiosperm stem hemiparasitic genus Cassytha (Lauraceae) and its host interactions: a review. Front. Plant Sci., 13: 864110. |
Zhang, J., Fu, X.-X., Li, R.-Q., et al., 2020b. The hornwort genome and early land plant evolution. Nat. Plants, 6: 107-118. |
Zhang, L., Chen, F., Zhang, X., et al., 2019. The water lily genome and the early evolution of flowering plants. Nature, 577: 79-84. |



