b. CAS Center for Excellence in Biotic Interactions, University of Chinese Academy of Sciences, Beijing 100049, China;
c. State Key Laboratory of Conservation and Utilization of Bio-Resources in Yunnan and Center for Life Science, School of Life Sciences, Yunnan University, Kunming 650500, China
The plant vascular system transmits not only water and nutrients, but also various molecules, including proteins, peptides, RNAs, and metabolites, from different tissues/organs (named local tissues/organs) to other (systemic) parts of a plant. Intensive studies on plant systemic signaling have revealed that some of these mobile molecules may be signals that convey important information to systemic tissues/organs (Gilroy et al., 2016) and thus impact plant growth and development and adaptation to environmental stresses (Turnbull and Lopez-Cobollo, 2013).
Nitrogen (N) and phosphorus (P) are essential mineral elements for plant growth and development, which are absorbed by roots and transported to shoots by the vascular system. Due to the limited contents and heterogenous distribution of N and P in soils, plants have evolved elaborate local and systemic signaling systems to cope with such nutrient stresses (Ham et al., 2018; Vega et al., 2019). N and P nutrients are sensed by roots and then certain mobile signals are translocated to systemic tissues, including other parts of the roots, as well as stems and leaves to synchronize N and P uptake and metabolism (Ruffel et al., 2016; Li et al., 2022). In Arabidopsis, Ruffel et al. (2011) used a split-root system and showed that N starvation-induced systemic signals, which are cytokinin dependent, travel in a root-shoot-root route, although the identities of these systemic signals remain unclear. Mobile RNAs are implicated in local-systemic communications during plant adaptation to N or P starvation (Thieme et al., 2015). A grafting experiment using Arabidopsis indicated that in response to P deficiency, the shoot-derived small RNA miR399 is transported through the phloem into the root to mediate PHO2 mRNA degradation, thereby enhancing phosphate uptake and translocation (Huang et al., 2013). Moreover, Zhang et al. (2016) used a cucumber-watermelon grafting system to study early P starvation-induced movement of mRNAs. In the watermelon sink tissues, 1786 and 2949 cucumber mobile mRNAs were identified under control and P starvation conditions, respectively (Zhang et al., 2016). Similarly, bidirectional movement of mRNAs was also identified in a heterograft between the Arabidopsis ecotype Col-0 and Ped-0 under nutrient-limited conditions: when the nutrients were replete, 684 and 456 mRNAs were transported from shoot to root and from root to shoot, respectively, while P and N starvation stress resulted in 258 and 264 mobile mRNAs being transferred from root to shoot, respectively (Thieme et al., 2015). Proteins have long been known to act as mobile signals in plants. For example, the leaf-produced florigen FLOWERING LOCUS T (FT) protein is translocated to the apical meristem to induce flowering (Turck et al., 2008), and the transcription factor HY5 moves from shoot to root, where it modulates whole-plant carbon and N acquisition (Chen et al., 2016). Whether proteins can move systemically on large scales, whether plants reconfigure mobile proteins between different organs in response to environmental stresses, and the biological functions of mobile proteins in systemic signaling remain far from being fully understood.
Approximately 1% of angiosperms are parasitic plants, which extract nutrients and water from their hosts via a specialized organ called the haustorium (Nickrent, 2020). Cuscuta spp. (dodder, Convolvulaceae) are stem parasitic plants that use haustoria to establish vascular (phloem and xylem) connections with host plants, allowing the transfer of various molecules, including nutrients, proteins, mRNAs, and signals, between dodder and hosts (Shen et al., 2023). Previous studies have shown that aphid (Myzus persicae) feeding-induced systemic signals were translocated from dodder to host soybean (Glycine max) and activated defenses (Zhuang et al., 2018). When two hosts were connected by a dodder parasite, herbivory- or salt stress-induced systemic signals are transferred between the two host plants (Hettenhausen et al., 2017; Li et al., 2020; Lei et al., 2021). Moreover, secondary metabolites (Smith et al., 2016), microRNAs (Shahid et al., 2018), siRNAs (Jhu et al., 2021), and mRNAs (Kim et al., 2014; Thieme et al., 2015; Song et al., 2022) travel from dodder to hosts and/or from hosts to dodder. Hundreds to more than one thousand proteins were found to move between dodder and hosts and between different dodder-connected hosts (Liu et al., 2020). Biochemical assays suggested that the mobile proteins may still retain biochemical activity (Liu et al., 2020), among which, host-derived flowering-inducer FT protein was found to translocate to dodder (Cuscuta australis) where it activated the flowering, allowing dodder to synchronize its flowering with the hosts (Shen et al., 2020). Under nutrient deficiency conditions, systemic signaling was also found to occur between host and dodder. When a dodder bridge connected a soybean plant with a cucumber (Cucumis sativus) plant and the soybean host suffered from N starvation, systemic signals from the soybean plant traveled to the cucumber plant through dodder and enhanced its N uptake (Zhang et al., 2021).
The growth and development of dodder rely entirely on the host plants, and it is very likely that these two interacting organisms constantly communicate using systemic signals, which could be the mobile mRNAs and proteins. Recently, the term mobileome/mobilome has been used to describe the collective contents of mobile macromolecules (Heeney and Frank, 2023). Here, in addition to studying the transcriptomic and proteomic responses of soybean and the parasitizing dodder to N and P starvation, we specifically focused on the mRNA and protein mobileomes between dodder and soybean hosts under normal and nutrient deficiency conditions.
2. Materials and methods 2.1. Plant growth and treatmentDodder (Cuscuta australis) and soybean (Glycine max cv William 82) were grown in a growth chamber under a 16 h/8 h light/dark period at 22–24 ℃. Soybean seeds were germinated on moist filter paper for 7 days, and then transferred to 1-L plastic pots filled with modified Hoagland solution (MHS). The MHS consisted of 1 mM CaCl2·2H2O, 2 mM MgSO4·7H2O, 10 mM KNO3, 1 mM KH2PO4, 0.02 mM C10H12FeN2NaO8, 2.86 mg/L H3BO3, 1.81 mg/L MnCl2·4H2O, 0.22 mg/L ZnSO4·7H2O, 0.08 mg/L CuSO4·5H2O, 0.02 mg/L H2MoO4·H2O, and 0.01 mg/L CoCl2·6H2O. The MHS was initially replaced every two weeks, but after one month, it was replaced every week until N or P deficiency treatment. Dodder seeds were treated with sulfuric acid for 30 min. After removing most of the sulfuric acid, the seeds were washed with tap water to remove the acid. The seeds were then transferred to a 50-ml centrifuge tube and shaken vigorously until most of the seed coats were completely removed. The seeds were sterilized with a sodium dichloroisocyanurate solution (0.02 g/ml, containing 0.2 µl/ml silwet-77) for 10 min. After being washed five times with sterile water, the seeds were placed on sterile moist filter paper in a sterile Petri dish. After four days, each seedling was used to parasitize a 10-day-old soybean seedling. After dodder infestation, far-red light was provided throughout the experiment for dodder parasitism.
One month after dodder successfully parasitized the soybeans, the soybeans were subjected to N or P stress treatments. Plants were randomly divided into three groups, and after soybean roots were washed three times with tap water, the plants were returned to three hydroponic solutions for full nutrient (FN), N deficiency (-N), or P deficiency (-P) treatment. The FN group received MHS, while the -N group was given MHS whose 10 mM KNO3 was replaced by 10 mM KCl and the -P group was given MHS whose 1 mM KH2PO4 was replaced by 1 mM KCl. For the short-term -N/-P experiment, the dodder (~5-cm segments, whose proximal ends were 1 cm away from the host stems), leaves (3rd leaves from bottom to the top) and roots were harvested 24 h after treatment, and the samples were used for transcriptome and proteome sequencing, as well as analyses of protein contents and C, N and P contents. For the long-term (5 and 10 days) -N/-P stress treatment, the nutrient solution was refreshed every week until the completion of the experiment. The soybean leaves, roots, and dodder stems were harvested for protein content and plant hormone measurements. Additionally, the dry weights of the above-ground and below-ground parts of the soybeans, as well as the dodder, were measured.
2.2. Measurement of C, N, and P contentsThe stems of soybean and dodder were sampled and dried for 72 h at 65 ℃, and then their masses were recorded. Next, dried tissues were ground into fine powder. C and N were determined on a Vario MAX CN instrument (Elementar Analyse system GmbH, Hanau, Germany). Total P was determined on an inductively coupled plasma atomic-emission spectrometer (IRIS Advantage-ER; Thermo Jarrell Ash Corporation, Franklin, MA, USA).
2.3. Measurement of photosynthetic parametersAll measurements of leaf gas exchange were done on fully mature and healthy soybean leaves by using a portable gas exchange fluorescence system (GFS-3000, Heinz-Walz Instruments, Effeltrich, Germany). Before photosynthetic measurements, the leaves of soybean were illuminated with an actinic light of 1200 μmol m−2 s−1 for approximately 20 min. The CO2 concentration in the chamber was set to 400 μmol mol−1, the temperature was 25 ℃, and the relative humidity was ~60%. Preliminary measurements showed that these settings for light intensity were above the photosynthetic light saturation point, but below the light intensity at which photoinhibition occurred. Then the light-saturated photosynthetic rate (Pn) and the stomatal conductance (gs) were detected under saturated light. At least four mature leaves from individual soybean plants were measured.
2.4. Measurement of phytohormonesMeasurement of plant hormones was performed according to a previously published method (Song et al., 2022).
2.5. RNA isolation and RT-qPCRTotal RNA was extracted using TRIzol reagent (ThermoFisher Scientific). cDNA synthesis was performed following the manufacturer's protocol (TaKaRa, PrimeScript ™ RT reagent Kit). Quantitative real-time PCR (RT-qPCR) was performed as previously described (Hettenhausen et al., 2017). Briefly, quantitative real-time PCR was performed on a CFX Connect Real-Time PCR Detection System (Bio-Rad) using iTaq Universal SYBR Green Supermix kits (Bio-Rad). Glycine max ACTIN11 was used as the internal control for normalizing cDNA concentration variations. Primer sequences are listed in Table S1.
2.6. Analysis of transcriptomesTwo samples were pooled as one replicate, forming three replicates that were used for RNA library construction and sequencing. Sequencing libraries were generated using the VAHTS Universal Plus DNA Library Prep Kit for MGI (Vazyme, Nanjing, China) following the manufacturer's recommendations, and index codes were added to attribute sequences to each sample. RNA sequencing was conducted at a depth of 5 G raw data on a DNBSEQ-T7 platform (MGI), and 12–18 million high-quality reads were obtained for each sample. Based on the genome sequences of soybean (SoyBase, https://soybase.org) and dodder (Sun et al., 2018), we used TopHat2 (Trapnell et al., 2012), FeatureCounts (Liao et al., 2014), and the DESeq2 package (Anders and Huber, 2010) to assemble the transcripts and identify DEGs (at least 2-fold increase or 50% decrease (false discovery rate, FDR ≤ 0.05)). Soybean GO terms enriched in these DEGs were identified using online tools on SoyBase (https://soybase.org) and an FDR cutoff of ≤0.05. To identify mobile mRNAs, the raw RNA-seq data from soybean and C. australis samples were mapped to their respective genomes, and the unmapped reads were then mapped against the other genome. For each group of samples, only mRNAs that appeared in at least two out of three replicates and with an average count ≥ 1 were chosen for further analyses. The relative abundance of mobile mRNAs in a given transcriptome was calculated according to the formula “total counts of mobile mRNAs/total counts of all mRNAs of the transcriptome × 100%”.
2.7. Sample preparation for proteome analysisFor protein extraction, samples (each had five biological replicates) were ground in liquid nitrogen. Six milliliters of lysis buffer (4% sodium dodecyl sulfate, 290 mM sucrose, 1 mM NaF, 1 mM NaVO4, 250 mM Tris–HCl pH 8.0 with 5 mM dithiothreitol and protein inhibitor cocktail (Roche)) were added to 1 g of each sample, and samples were vortexed and boiled in water for 5–10 min. After centrifugation at 16,000 g for 20 min at 4 ℃, the supernatants were collected, and each was mixed with 4 volumes of methanol, an equal volume of chloroform, and 3 volumes of water successively. The protein precipitate was obtained in the interphase after centrifugation. After washing twice with methanol, the precipitated proteins were air-dried.
Digestion of total protein for proteome analysis followed a previous protocol (Rappsilber et al., 2003). Briefly, an aliquot of the air-dried protein sample was resuspended in 400 μL of resuspension buffer (6 M urea, 2 M thiourea, 10 mM pH 8.0 Tris/HCl). The supernatants were collected, and protein concentrations were determined using the Bradford method (Bio-Rad Quick Start Bradford Protein Assay). Proteins (10 μg) were predigested for 3 h with endoproteinase Lys-C (Wako Chemicals, Neuss) at room temperature in a 1:100 (w/w) enzyme to protein ratio. After 4-fold dilution with 10 mM Tris–HCl (pH 8.0), trypsin (Promega, Germany) was added to a final 1:100 (w/w) concentration and kept overnight at 37 ℃. Thereafter, 1/10 volume of 10% trifluoroacetic acid (TFA) was added until the pH was below 3 to stop the digestion. The digested peptides were desalted using C18 tips (Empore™, America). The dried samples were then resuspended in 5 μL of a solution containing 0.3% formic acid and 2% acetonitrile for mass spectrometry analysis.
2.8. Protein identification and label free quantificationFor proteomic analysis, protein identification and quantification were performed using MaxQuant (v.1.6.7.0) (Cox and Mann, 2008) and the Andromeda search engine (Cox et al., 2011). UniProt (soybean database, UP000008827) (UniProt, 2023) and the dodder database (NCBI, taxonomy ID: 267555) were used for analysis of protein sequences. MaxQuant parameters were set as the follows: trypsin was specified as the digesting protease, and up to two missed cleavages were allowed. The precursor ion mass tolerance was set to 20 ppm for full scan, fragment ion mass tolerance was set to 0.5 Da, and multiplicity was set to 1. “Label-free quantitation” (Cox et al., 2014) and “match between runs” were selected and a time window of 0.7 min was set. The FDR cutoff was set to 0.01. The peptide sequences from soybean and dodder samples were first mapped respectively to the soybean and dodder proteome databases to identify the native proteins; next, the peptide sequences from soybean and dodder samples were mapped respectively to the foreign proteome database, namely dodder and soybean proteome database, to identify the candidate mobile proteins. Only proteins that appeared in at least three replicates out of the five replicates were considered to be genuine. For identification of mobile soybean or dodder proteins, the peptides from candidate mobile proteins were screened by mapping back to the native proteome database, and only candidate mobile proteins with at least one peptide that could only be found in the foreign proteome database were considered to be mobile proteins. Relative abundance of total mobile proteins in a proteome was calculated according to the formula “total abundance of mobile proteins/total abundance of all proteins × 100%”.
3. Results 3.1. Physiological response of soybean and dodder to N and P deficiencyTo investigate the physiological effects of N and P deficiency on both host and dodder plants, we established hydroponically cultivated dodder-soybean parasitization systems, allowing us to precisely control the nutritional conditions of the soybean host plants (Fig. 1A).
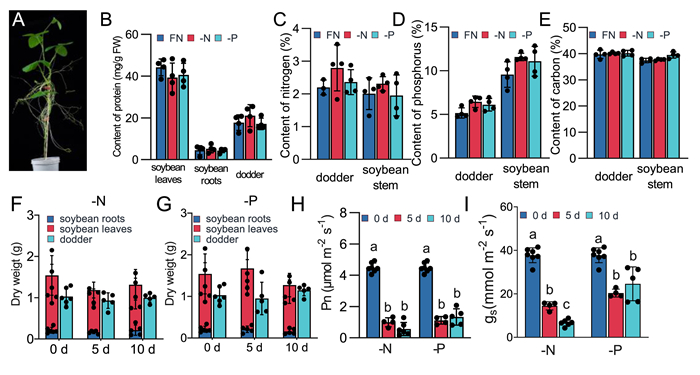
|
| Fig. 1 Physiological effects of -N and -P treatment on soybean-dodder parasitization system. A. Hydroponic culture system of soybean-dodder parasitization system. B. Protein contents in soybean leaves and roots and dodder after 24 h of -N and -P treatment. C-E. Nitrogen (C), phosphorus (D) and carbon (E) contents in soybean stems and in dodder after 24 h of -N or -P treatment. F-G. Dry weights of soybean shoots, soybean roots, and dodder after 0, 5, and 10 days of -N (F) and -P (G) treatment. H–I. Net photosynthetic rates (Pn, H) and stomatal conductance (gs, I) of soybean leaves after 0, 5, and 10 days of treatments. Different letters indicate significant differences analyzed by one-way ANOVA (P < 0.05). Data are means ± SE (n ≥ 4). |
The soybean plants were treated with normal culture medium or with N- or P- deficient culture medium (designated full nutrients (FN), -N, and -P, respectively) for 1, 5, or 10 days to determine the physiological responses of the host soybean and dodder. Protein contents in soybean leaves, roots, and dodder were unaltered in either the host or dodder on all days of treatment (Fig. 1B). Additionally, N, P, and carbon (C) contents in the soybean stem and dodder were also measured to assess nutritional changes (Fig. 1C, D, and E): no significant changes in either the soybean stem or dodder were found as well. Even after 10 days of nutrient deficiency stress, there were no significant changes in the protein contents and dry weights of either the soybean or dodder (Figs. 1F, G and S1A). However, both -N and -P treatment significantly impacted the net photosynthetic capacity (Pn) and stomatal conductance (gs) of the soybean leaves (Fig. 1H and I). Additionally, we also noted substantial increases in the levels of auxin (IAA), abscisic acid (ABA), jasmonic acid (JA), and jasmonate-isoleucine (JA-Ile) in dodder under -N and -P conditions, whereas these changes were not prominent in the soybean leaves and roots (Fig. S1B–E). Our results demonstrate that dodder may perceive and respond to host-derived nutrient systemic signaling.
3.2. Interplant mobile mRNAs between dodder and host under -N and -P deficiency conditionsTo gain insight into the signaling responses of both soybean hosts and dodder to N and P deficiency on the transcriptional level, using the same parasite system, the soybean plants were treated with FN, -N, or -P for 24 h, and subsequently the parasitizing dodder and soybean leaves and roots were harvested and used for RNA-seq analysis.
The transcriptomes of soybean leaves and roots and the parasitizing dodder grown under FN were used as the baseline to determine the differentially expressed genes (DEGs) in the respective tissues under -N and -P conditions (Fig. 2A; Dataset S1). Under -N conditions, a total of 212 and 319 DEGs were identified from the soybean leaves and roots, respectively, including the nitrate signaling-related genes “expansin-like” and “phosphate-responsive 1” in the leaves and “nitrate reductase 1”, “nitrate transporter 1.5”, “phosphate transporter 2;1”, and “phosphate-responsive 1” in the roots. Under -P conditions, only 4 and 28 DEGs were identified from soybean leaves and roots, respectively (Fig. 2A and B; Dataset S1). These DEGs included phosphate starvation-related genes “expansin A1”, “glucose-6-phosphate/phosphate translocator 2”, and “phosphate-responsive 1 family protein”. Subsequently, a total of 12 DEGs were randomly selected from the transcriptomes and used for validation by RT-qPCR. The expression patterns of these genes were confirmed to be consistent with the transcriptomes, except for one gene (Glyma.06G102000) (Figs. S2A and B), supporting the reliability of the RNA-seq data. Furthermore, no DEGs were found in dodder under either -N or -P conditions (Fig. 2A).
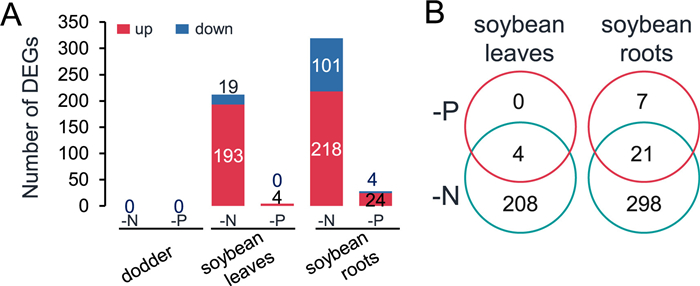
|
| Fig. 2 Changes of transcriptomes in soybean and dodder plants after -N and -P treatment. Soybean-dodder parasitization systems were treated with FN, -N, or -P nutrient regimes. After 24 h, the soybean leaves and roots and dodder stems were harvested for RNA-seq analysis. A. Numbers of DEGs in soybean leaves and roots and dodder after -N and -P treatment. B. Venn diagram analysis of DEGs in soybean leaves and roots under -N and -P conditions. FN, full nutrient; -N, no nitrate; -P, no phosphorus. |
We then analyzed the mobile mRNAs between soybean and dodder under the FN, -N, and -P conditions. Under the FN conditions, 363 mRNAs originating from dodder were identified in soybean leaves, and in response to -N stress, we detected a 1-fold increase (786) in the number of dodder mobile mRNAs, while under -P stress, we detected 57% decreased (155) number of dodder mobile mRNAs (Fig. 3A; Dataset S2). Many more mobile dodder mRNAs were identified in soybean roots: under the FN conditions, there were 2516 mobile dodder mRNAs; however, under the -N and -P conditions, the numbers of mobile dodder mRNAs decreased 66% and 82% (842 and 460), respectively (Fig. 3A). In dodder, 1966 mobile soybean mRNAs were found under the FN conditions, and after the -N and -P treatments, the numbers increased to 2645 and 2011 (Fig. 3A; Dataset S3). Venn diagram analysis of the mobile RNAs revealed large numbers of conditional mobile mRNAs: 949, 1404, and 885 mobile soybean mRNAs were specifically found in dodder under the FN, -N, and -P conditions, respectively (Fig. 3B); under the FN, -N, and -P conditions, 200, 621, and 50 dodder mRNAs were specifically found in soybean leaves and 1834, 280, and 129 dodder mRNAs were found in soybean roots (Fig. 3C). Under all three nutrient conditions, only 520, 55, and 177 mobile mRNAs were commonly identified in dodder and soybean leaves and roots, respectively (Fig. 3B and C). Gene Ontology (GO) analysis revealed that photosynthesis- and amino acid metabolism-related terms were enriched from the 520 common mobile soybean mRNAs in dodder (Table S2); “carbohydrate derivative catabolic process”, “one-carbon metabolic process”, and “cellular modified amino acid metabolic process” were enriched from the 55 common dodder mRNAs in soybean leaves, and “response to inorganic substance”, “response to salt stress”, and “response to metal ion” were enriched from the 177 common dodder mRNAs in soybean roots (Tables S2 and 3). These data strongly suggest that the interplant mobility of mRNAs between dodder and soybean hosts is specifically dependent on the nutrient conditions of the soybean host plants.
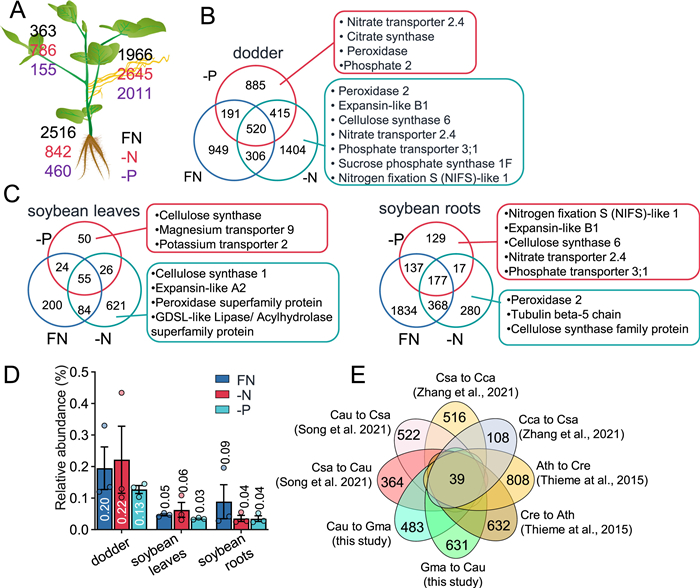
|
| Fig. 3 Changes of mRNA mobileomes in soybean and dodder plants after -N and -P treatment. Soybean-dodder parasitization systems were treated with FN, -N, or -P nutrient regimes. After 24 h, the soybean leaves and roots and dodder stems were harvested for RNA-seq analysis. A. Numbers of inter-plant mobile mRNAs identified in dodder and soybean leaves and roots under FN (numbers in black), -N (numbers in red), and -P (numbers in purple) conditions. B–C. Venn diagram analysis of the mobile mRNAs detected in dodder (B) and soybean leaves and roots (C) under different nutrient regimes. D. Relative contents of foreign/mobile mRNAs in soybean leaves and roots and in dodder. The data are means ± SE (n = 3). E. Venn diagram analysis of conserved mobile mRNAs in the Arabidopsis (Ath)-Cuscuta reflexa (Cre), cucumber (Csa)-Cuscuta campestris (Cca), and soybean (Gma)-Cuscuta australis (Cau) parasitization systems. FN, full nutrient; -N, no nitrate; -P, no phosphorus. |
Kim et al. (2014) found that 0.17%–1.1% of the total mRNAs were foreign mRNAs in the dodder-tomato and dodder-Arabidopsis parasitization system. In a dodder soybean system, approximately 1% of total mRNAs were foreign mRNAs (Liu et al., 2020). In our experimental setup, we found that 0.13%–0.22% of the total mRNAs in dodder were soybean mRNAs, and among the total mRNAs in soybean leaves and roots, 0.03%–0.06% and 0.04%–0.09% were dodder mRNAs, respectively (Fig. 3D). Using relative signal intensity ratios (RSIRs) (Liu et al., 2020), which are the ratios of the relative abundance of mobile mRNAs in the foreign plants to their relative abundance in the native plants, we quantitatively estimated the changes in the abundance of mobile mRNAs after being transferred from dodder to soybean leaves and roots (mRNAs from soybean plants to dodder were not analyzed, as the source of mobile mRNAs in dodder cannot be assigned to either leaf or root). In soybean leaves and roots, 73.2%–92.8% of the dodder mRNAs’ abundance was less than 0.1% of their native abundance in the dodder plants, and only a small proportion of the mobile dodder mRNAs (less than 4%) retained more than 10% of the native abundance in soybean (Fig. S3A). A previous study suggested that mRNAs in high concentrations tend to be mobile (Calderwood et al., 2016). We therefore compared the abundance of mRNAs with and without mobility in the dodder transcriptome, and indeed, the mRNAs of mobility had significantly higher abundance than did the mRNAs without mobility (Fig. S3B).
Next, we retrieved the previously published data of mRNAs transferred between dodder and host plants in parasitization systems (Thieme et al., 2015; Zhang et al., 2021; Song et al., 2022) and between scions and stocks in grafting systems (Thieme et al., 2015). Together with the mobile mRNAs identified in this study, these mRNA species were used for identification of their Arabidopsis homologs using OrthoFinder (Emms and Kelly, 2019), which were thereafter used for Venn diagram analysis to answer the question of which mobile mRNAs are commonly transferred between different species: 39 mRNA species were likely to be conserved in long-distance interplant transfer (Fig. 3E; Dataset S4). Among these, the GO terms “regulation of monoatomic ion transmembrane transport” and “protein binding” were enriched.
3.3. Protein transfer between soybean and dodder under different nutrient conditionsTo study how a soybean-dodder parasitization system responds to N and P starvation at the proteome level, the dodder and soybean samples harvested under the -N, -P, and FN conditions were used for mass spectrometry (MS)-based proteomics analysis and label-free quantification. In the dodder samples, ~5500 dodder proteins were identified (Dataset S5), and in soybean leaves and roots, ~3500 and ~5500 soybean proteins were found respectively (Fig. 4A; Dataset S6). To investigate how soybean and dodder native proteins are modulated under -N and -P conditions, the FN samples were again used as the baselines for screening the differentially expressed proteins (DEPs) in both soybean and dodder (Dataset S7). A total of 163 DEPs were identified in dodder under -N conditions, while almost 2-fold more DEPs (474) were detected under -P conditions (Fig. 4B; Dataset S7). Among these DEPs, “nitrite reductases” which are involved in N signaling, were downregulated under the -N conditions, while “peptide transporter 2” and “phosphate transporter traffic facilitator 1” (Landi and Esposito, 2017), which are associated with P signaling, were upregulated. Soybean leaves, under the -N and -P conditions, showed 370 and 165 DEPs, respectively. Among these the N signaling-related proteins “phosphatase 2C family protein”, “glutathione S-transferase”, and “sucrose synthase 4” were identified. In the soybean roots, the numbers of DEPs were quite similar under -N and -P conditions (Fig. 4B; Dataset S7): under the -N condition, 251 DEPs were identified, including the N signaling-related proteins “phosphatase 2C″, “glutathione S-transferase”, “peroxidase superfamily protein”, and “subtilisin-like serine endopeptidase family protein” (Landi and Esposito, 2017); under the -P condition, we found 270 DEPs, which included a few P stress-related proteins, such as “phosphate deficiency response 2” and “sucrose synthase” (Fig. 4B; Dataset S7).
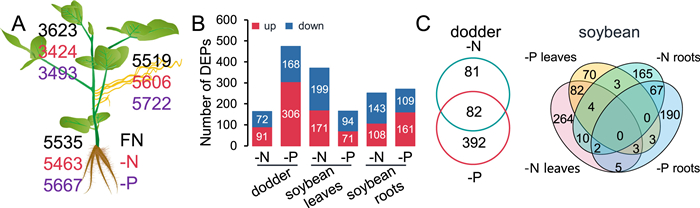
|
| Fig. 4 Effect of host nutrient status on the proteomes in soybean and dodder plants. Soybean-dodder parasitization systems were treated with FN, -N, or -P nutrient regimes. After 24 h, the soybean leaves and roots and dodder stems were harvested for proteome analysis. A. Numbers of proteins detected in dodder and soybean plants by proteomics analysis. B. Numbers of differentially expressed proteins (DEPs) in dodder and soybean leaves and roots under FN, -N, and -P conditions. C. Venn diagram analysis of DEPs under -N and -P conditions in dodder (left) and soybean leaves and roots (right). Data are means ± SE (n = 5). FN, full nutrient; -N, no nitrate; -P, no phosphorus. |
To gain further insight into the regulation of dodder and soybean proteomes in response to N and P starvation, we specifically compared the DEPs identified under -N and -P conditions in dodder and soybean (Fig. 4C; Dataset S7). In dodder, 82 DEPs were commonly regulated under both -N and -P conditions, while 81 and 392 specific DEPs were found under the -N and -P conditions, respectively (Fig. 4C). Venn diagram analysis of the DEPs in soybean leaves and roots under both -N and -P conditions revealed that most of the DEPs were tissue- or nutrient condition-specific (Fig. 4C). First, no proteins were commonly regulated in either tissues or under either nutrient stress conditions, suggesting tissue- and treatment-specific proteomic changes. Second, 264 and 70 proteins were specific for soybean leaves under -N and -P conditions, and 165 and 190 proteins were specific for soybean roots under -N and -P conditions, respectively (Fig. 4C). Finally, 89 proteins were commonly regulated in soybean leaves under -N and -P conditions, and 69 proteins were commonly regulated in soybean roots under -N and -P conditions (Fig. 4C), although no GO terms were enriched from these common proteins.
Previously it was shown that hundreds to more than a thousand proteins were exchanged in large quantities between dodder and host plants, and some of the mobile proteins still retained their biological activity (Liu et al., 2020; Shen et al., 2020). To determine whether the transfer of mobile proteins between dodder and soybean is affected by the nutritional conditions of the host, the proteome data were screened for mobile proteins. Within the five biological replicates of each group, only foreign proteins that appeared in at least three replicates were considered to be mobile. In soybean leaves and roots, under the FN conditions, 113 and 217 dodder mobile proteins were identified, respectively, while 111 and 204 (-N) and 110 and 205 (-P) dodder proteins were identified in nutrient deficient soybean leaves and roots, respectively (Fig. 5A; Dataset S5). Among these dodder mobile proteins, a bZIP transcription factor and a nitrate reductase 2 (NIA2) were identified in soybean roots under all conditions (Table S5). In dodder, 283, 270, and 264 soybean proteins were identified under FN, -N, and -P conditions, respectively (Fig. 5A; Dataset S5). Using Venn diagram analysis, we specifically inspected the effect of -N and -P on the mobile proteins. Under FN, -N, and -P conditions, 71, 51, and 47 soybean proteins were specifically transported to dodder (Figs. 5B) and 160 soybean proteins were commonly transported to dodder under all conditions (Fig. 5B), among which, 5 (2 up- and 3 down-regulated) were differentially regulated under -N conditions (Fig. 5B; Dataset S7) and 13 (8 up- and 5 down-regulated) were differentially regulated under -P conditions. In soybean, 44 dodder proteins were commonly detected in all samples (Fig. 5B). In the leaves, 14, 12, and 14 were specifically detected under FN, -N, and -P conditions; among these, 0 and 8 were up- and down-regulated under -N conditions, respectively, and 0 and 4 were up- and down-regulated under -P conditions, respectively (Fig. 5B; Dataset S7). In the soybean roots, we detected 58, 47, and 66 specific dodder proteins under FN, -N, and -P conditions, 3 and 5 were respectively up- and down-regulated under -N conditions, and 2 and 3 were respectively up- and down-regulated under -P conditions (Fig. 5B; Dataset S7). Among these mobile soybean proteins, under the -P conditions, a MYB-like transcription factor was identified in dodder (Table S5). Those results indicate that the mobile proteins are also affected by the host's nutrient conditions.
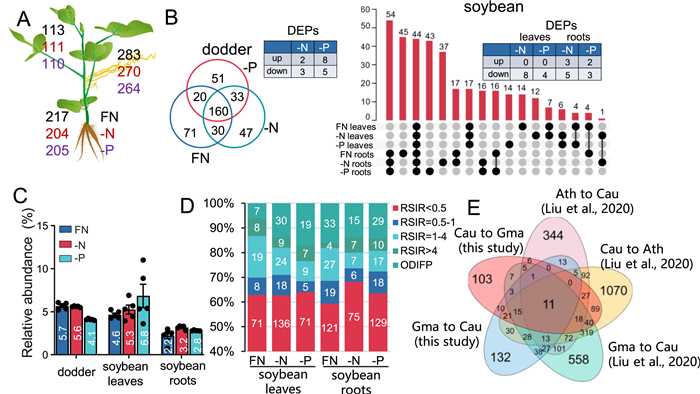
|
| Fig. 5 Effect of host nutrient status on the proteomes and protein transfer in soybean and dodder plants. Soybean-dodder parasitization systems were treated with FN, -N, or -P nutrient regimes. After 24 h, the soybean leaves and roots and dodder stems were harvested for proteome analysis. A. Numbers of mobile proteins in dodder and soybean leaves and roots. Black, red, and purple numbers indicate the numbers of mobile proteins under FN, -N, and -P conditions. B. Venn diagram analysis indicating the specific and common mobile proteins in dodder (left) and soybean leaves and roots (right) under different nutrient regimes. C. Relative contents of total mobile proteins in soybean host and dodder. Data are means ± SE (n = 5). D. Relative signal intensity ratios (RSIRs) of dodder mobile proteins. RISRs = the ratios of the dodder mobile protein abundance in soybean plants to their original abundance in dodder; ODIFP = only detected in foreign plants. E. Venn diagram analysis of the mobile proteins identified in this study and previously published Arabidopsis-Cuscuta australis (Cau) and soybean (Gma)-Cuscuta australis (Cau) systems. FN, full nutrient; -N, no nitrate; -P, no phosphorus. |
The amounts of transferred proteins were estimated based on the signal intensities from the mass spectrum information of each protein. Similar to previous findings (Liu et al., 2020), ~5% of the total proteins in soybean leaves were transferred from dodder under all three conditions (Fig. 5C). In soybean roots, dodder proteins accounted for approximately 3% of the total protein (2.2%, 3.2%, and 2.8% under FN, -N, and -P conditions, respectively) (Fig. 5C). Consistent with the highest number of transferred soybean proteins in dodder, we found that 5.7%, 5.6%, and 4.1% of the proteins in dodder were from soybean respectively under FN, -N, and -P conditions (Fig. 5C). Thus, large quantities of proteins are transferred between host plants and dodder and the transferred proteins are qualitatively and quantitatively regulated by unknown mechanisms under nutrient stress.
Furthermore, the abundance of dodder proteins with or without mobility was compared. Similar to mRNAs, our analysis indicated that proteins in relatively high concentrations tend to be mobile (Fig. S4), and this result is consistent with our previous findings supporting the possibility that protein mobility is positively correlated with concentrations (Liu et al., 2020). Moreover, RSIRs based on the mass spectrometry data were used to assess the changes of mobile dodder proteins’ abundance after they were transferred to soybean leaves and roots. We found that in soybean leaves and roots, over 30% of dodder proteins maintained at least 50% of their original abundance in dodder (Fig. 5D).
By analyzing the data from this study and previously published interplant mobile proteins in Arabidopsis, soybean, and dodder (Liu et al., 2020), we found that 11 proteins were bidirectionally transferred in all systems, including “glycine cleavage T-protein”, “TCP-1/CPN60 chaperonin family protein” and “xylose isomerase family protein” (Fig. 5E; Table S6; Dataset S8).
3.4. Correlation among interplant mobile mRNAs and proteinsTo examine the possibility that the foreign proteins are translated after the foreign mRNAs are translocated to the recipient plants, we first analyzed the correlation between mobile mRNAs and proteins in soybean and dodder plants. Under three different nutrient treatments, in dodder, 11% (26/230, -P conditions) to 20% (49/240, FN conditions) of the mobile soybean proteins had their mRNA counterparts (Fig. 6A). Similarly, in terms of the mobile dodder mRNAs in soybean leaves and roots, 8.2% (9/110, in -P leaves) to 49% (107/217, in FN roots) of the mobile proteins’ corresponding mRNAs were detected (Fig. 6B and C). These data suggest that large portions of the foreign proteins were directly transferred (mobile), instead of being the products of translation of mobile mRNAs. Taken together, the mobile mRNAs and proteins between dodder and hosts are likely independently transported, at least for most of them.
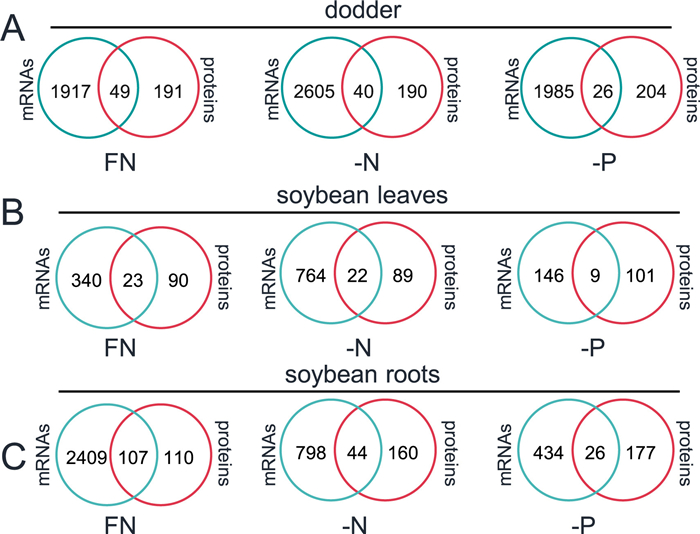
|
| Fig. 6 Correlation analysis of mobile mRNAs and proteins. Venn diagram analysis of the commonality between mobile soybean mRNAs and proteins in dodder (A), soybean leaves (B), and roots (C). |
The vasculature transports not only organic nutrients from source to sink organs but also systemic signals from local to systemic organs, activating systemic responses. Numerous studies, mostly using grafting systems, have shown that many molecules, including phytohormones, microRNAs, proteins, Ca2+, ROS, and peptides, act as systemic signals that travel between different organs during plant growth and development and in response to biotic or abiotic stresses (Gilroy et al., 2016). Given that dodder establishes phloem and xylem connections with host vasculature, when host plants suffer from nutrient stresses, dodder can perceive systemic signals from host plants and respond accordingly, although the possibility cannot be ruled out that certain changes in host nutrient status can be perceived directly by dodder, activating physiological changes. In this study, we analyzed the N and P deficiency-induced physiological response, including transcriptome and proteome changes, in a soybean-dodder parasitization system, and importantly, our study provides detailed mobileome data that reveal large-scale interplant mobile mRNAs and proteins in such a soybean-dodder system under different nutrient regimes.
Previously, it was found that cucumber plants responded to P starvation with 27 and 34 DEGs in lamina and roots after 12 h of treatment (Zhang et al., 2016). Similarly, we found that in soybean plants, -N treatment resulted in 212 and 319 DEGs in leaves and roots, while under -P conditions, only 4 and 28 DEGs were found in leaves and roots, respectively (Fig. 2A). In contrast, in soybean, we found substantially more proteins that responded to N or P starvation: up to 370 (370 in leaves and 251 in roots) and 270 (165 in leaves and 270 in roots) DEPs were found under -N and -P conditions, respectively (Fig. 4B). Thus, after short-term nutrient deficiency treatments, soybean exhibited more pronounced changes at the protein level compared to the transcript level.
In Arabidopsis and rice, transcriptome analysis indicated that a number of genes are involved in the response to both N and P starvation (Medici et al., 2015, 2019; Hu et al., 2019). Functional studies have also revealed that the NRT1.1-SPX-PHR/NLP cascade plays a central role in the N and P balance (Hu et al., 2019). Our analysis indicated that only 4 and 28 genes transcriptionally responded to -P conditions in soybean leaves and roots, respectively, although many more genes responded to -N (212 in leaves, 319 in roots) (Fig. 2A). We found that all four DEGs identified under -P stress in soybean leaves were also present in the DEGs identified under -N conditions. Similarly, in the soybean roots, 21 out of the 28 DEGs identified under -P stress were among the -N-induced DEGs (Fig. 2B). These results suggest that, at the transcriptional level, soybean exhibited minimal responses to -P than to -N and there are only a small number of common responsive genes. However, when subjected to -N and -P conditions, 89 and 69 DEPs were commonly identified in soybean leaves and roots, respectively (Fig. 4C). It would be interesting to explore whether some of these proteins function in soybean adaptation to both N and P starvation.
In this study, we specifically focused on the mRNA and protein mobileome of soybean-dodder parasitization system. We found that similar to many other parasitization systems (Thieme et al., 2015; Zhang et al., 2021; Song et al., 2022), hundreds to thousands of mobile mRNAs were translocated between dodder and soybean (Fig. 3A). However, quantitatively these mobile mRNAs only comprised very small portions of the total mRNAs of the recipient plants (at most 0.2%; Fig. 3D). In contrast, the mobile foreign proteins reached up to 6% of dodder total proteins and up to 7% of soybean total protein (Fig. 5C). Moreover, comparisons of the species of mRNAs and proteins indicated that these mobile molecules are very likely transferred independently (Fig. 6). Therefore, proteins are probably the major forms of macromolecules in the interplant mobileomes, and they likely have more important functions in local-systemic communications than do the mobile mRNAs. These data also suggest that most of the mobile proteins are not produced by translation of mobile mRNAs in the recipient plants.
We also used RSIRs to quantitatively analyze the changes of dodder mobile mRNAs and protein's abundance after they reached the soybean plants, and a dramatic difference was detected between mobile mRNAs and proteins: after reaching soybean plants, most dodder mRNAs' abundance decreased to less than 0.1% of their original abundance, whereas many proteins still retained relatively high concentrations (in soybean plants, over 30% of dodder proteins maintained at least 50% of their original abundance in dodder). The exact mechanisms underlying long-distance mRNA and protein transfer remain unclear. At least two scenarios may account for such large differences between mobile mRNAs and proteins: 1) mRNAs may also be transported in large quantities; however, soon after they leave the host–dodder interface and enter the foreign plants, mRNAs may rapidly degrade and thus leaving very low levels of mobile mRNAs; in contrast, proteins are relative much more stable; 2) mRNAs are actually relatively stable in the foreign plants, but only very small amounts of mRNAs are able to translocate across the host–dodder interface and move into the foreign plants.
What determines the mobility of mRNAs, proteins, and other macromolecular, such as peptides, is a fascinating question that remains to be answered. Previous research (Zhang et al., 2016, 2021; Song et al., 2022) and this study all indicated that inter-plant transfer of mRNAs is regulated by environmental factors. An important finding of this study is that the mobility of proteins is also strongly affected by nutrient stresses and many of these mobile proteins were induced or suppressed specifically under different nutrient conditions, suggesting that some of them may confer certain signaling functions. We propose that at least three mechanisms underlie the mobility of mRNAs and proteins. First, plant physiology: environmental factors modulate plant physiology, including the transcriptome and proteome, and thus conceivably change the mobile mRNAs and proteins. Second, regarding the abundance of mRNAs and proteins: our analysis of the abundance of mobile and non-mobile mRNAs and proteins suggested that mRNAs and proteins with high concentrations tend to be mobile (Figs. S3B and S4). Third, certain unknown mechanisms seem to play an important role in controlling the mobileome, as our data suggest that the abundance of mRNAs and proteins is not the only factor that regulates their mobility: even though statistically the abundant mRNAs and proteins tend to be mobile, many of these macromolecules in low abundance were mobile and conversely many in high abundance were non-mobile (Figs. S3B and S4). It is notable that specific mRNAs and proteins were detected in soybean leaves and roots (Figs. 3 and 5B), and two possibilities may account for such tissue/organ-specific regulation of the mobileome. Either there is a tissue/organ-specific rate of catabolism of mobile macromolecules leading to different stability of these mobile molecules in leaves and roots, or selective transport of macromolecules to different tissues/organs, which is controlled by unknown mechanisms.
Our previous biochemical and genetic analysis indicated that some of the mobile proteins are biochemically active after they are translocated to the recipient plants (Liu et al., 2020; Shen et al., 2020). Among the interplant mobile proteins identified in this study, we found a b-ZIP transcription factor, a MYB-like transcription factor, and NIA2 (related to N metabolism). Moreover, several proteins, such as “glycine cleavage T-protein”, “TCP-1/cpn60 chaperonin family protein”, and “xylose isomerase family protein”, were found to be transported bidirectionally between parasites and hosts in all parasitization systems studied so far (Fig. 5E and Table S6). Whether these transcriptional regulators and N metabolism-related proteins play a role in the recipient plants’ physiology in this soybean-dodder parasitization system and whether these seemingly conserved long-distance mobile proteins function in the communication among different organs and between hosts and parasitic plants remain to be functionally analyzed.
Taken together, the results of this study demonstrate large-scale inter-plant transfer of mRNAs and proteins between dodder and soybean, and importantly, nutrient deficiency strongly changes not only the transcriptomes and proteomes of both soybean hosts and dodder, but also dramatically reconfigures the mobileome in this parasitization system. Whether there is transfer of RNAs and proteins in other host-parasite systems, e.g., sunflower (Helianthus annuus)-broomrape (Orobanche cumana), remains unclear. Further studies are needed for understanding whether some of these mobile macromolecules confer certain physiological and/or ecological functions in the interactions between parasitic plants and hosts. This study also suggests that in an individual plant the mobileome very likely contains many RNAs and proteins, which may function in various aspects of plant physiology.
AcknowledgementsWe would like to express our gratitude to Dr. Wei Chang and Dr. Fei Li for their invaluable technical support. We thank the Service Center for Experimental Biotechnology at the Kunming Institute of Botany, CAS, for their assistance throughout the research. Furthermore, the Proteomics Facility of Life Science Center, Yunnan University, is thanked for their technical support and assistance in conducting the proteomic analysis. This work was supported by the National Natural Science Foundation of China (31970274 (J.W.), 32170272 (X.W.), 32100251 (J.Z.), 32000179 (Y.X.)), the Special Research Assistant of Chinese Academy of Sciences (J.Z. and Y.X.), China Postdoctoral Science Foundation (2022M713224 (J.Z.)), the Strategic Priority Research Program of Chinese Academy of Sciences (XDPB16 (J.W.)), the Yunnan Innovation Team Project (202105AE160013 (J.W.)), CAS “Light of West China” Program (G.S.), and Yunnan Revitalization Talent Support Program “Young Talents” Project (XDYC-QNRC-2022-0301 (J.Z.), XDYC-QNRC-2022-0001 (G.S.)), the General and Key Project of the Applied Basic Research Program of Yunnan (202001AS070021(J.W.)), Yunnan Fundamental Research Projects-General Project (202101AT070457 (S.L.)), and Yunnan Fundamental Research Projects-Youth Talent Project (202101AU070021(S.L.)).
Author Contributions
J.Z., X.W., and J.W. designed the research, J.Z., S.L., W.L., Z.F., S.Z., X.Z., Y.X., G.S., M.Z., and X.W. performed the experiments and/or analyzed the data; J.Z., X.W., and J.W. wrote the manuscript. All the authors interpreted and discussed the data.
Data availability
The RNA-seq data can be retrieved from the Beijing Institute of Genomics under the BioProject: PRJCA011995.
Declaration of competing interest
No conflict of interest declared.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.11.005.
Anders, S., Huber, W., 2010. Differential expression analysis for sequence count data. Genome Biol., 11: R106. DOI:10.1186/gb-2010-11-10-r106 |
Calderwood, A., Kopriva, S., Morris, R.J., 2016. Transcript abundance explains mRNA mobility data in Arabidopsis thaliana. Plant Cell, 28: 610-615. DOI:10.1105/tpc.15.00956 |
Chen, X., Yao, Q., Gao, X., et al., 2016. Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol., 26: 640-646. DOI:10.1016/j.cub.2015.12.066 |
Cox, J., Hein, M.Y., Luber, C.A., et al., 2014. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics, 13: 2513-2526. DOI:10.1074/mcp.M113.031591 |
Cox, J., Mann, M., 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol., 26: 1367-1372. DOI:10.1038/nbt.1511 |
Cox, J., Neuhauser, N., Michalski, A., et al., 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res., 10: 1794-1805. DOI:10.1021/pr101065j |
Emms, D.M., Kelly, S., 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol., 20: 238. DOI:10.1186/s13059-019-1832-y |
Gilroy, S., Bialasek, M., Suzuki, N., et al., 2016. ROS, calcium, and electric signals: key mediators of rapid systemic signaling in plants. Plant Physiol., 171: 1606-1615. DOI:10.1104/pp.16.00434 |
Ham, B.K., Chen, J., Yan, Y., et al., 2018. Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol., 49: 1-9. DOI:10.1016/j.copbio.2017.07.005 |
Heeney, M., Frank, M.H., 2023. The mRNA mobileome: challenges and opportunities for deciphering signals from the noise. Plant Cell, 35: 1817-1833. DOI:10.1093/plcell/koad063 |
Hettenhausen, C., Li, J., Zhuang, H., et al., 2017. Stem parasitic plant Cuscuta australis (dodder) transfers herbivory-induced signals among plants. Proc. Natl. Acad. Sci. U.S.A., 114: E6703-E6709. |
Hu, B., Jiang, Z., Wang, W., et al., 2019. Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants, 5: 401-413. DOI:10.1038/s41477-019-0384-1 |
Huang, T.K., Han, C.L., Lin, S.I., et al., 2013. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell, 25: 4044-4060. DOI:10.1105/tpc.113.115998 |
Jhu, M.Y., Ichihashi, Y., Farhi, M., et al., 2021. LATERAL ORGAN BOUNDARIES DOMAIN 25 functions as a key regulator of haustorium development in dodders. Plant Physiol., 186: 2093-2110. DOI:10.1093/plphys/kiab231 |
Kim, G., LeBlanc, M.L., Wafula, E.K., et al., 2014. Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science, 345: 808-811. DOI:10.1126/science.1253122 |
Landi, S., Esposito, S., 2017. Nitrate uptake affects cell wall synthesis and modeling. Front. Plant Sci., 8: 1376. DOI:10.3389/fpls.2017.01376 |
Lei, Y., Xu, Y., Zhang, J., et al., 2021. Herbivory-induced systemic signals are likely to be evolutionarily conserved in euphyllophytes. J. Exp. Bot., 72: 7274-7284. DOI:10.1093/jxb/erab349 |
Li, S., Zhang, J., Liu, H., et al., 2020. Dodder-transmitted mobile signals prime host plants for enhanced salt tolerance. J. Exp. Bot., 71: 1171-1184. DOI:10.1093/jxb/erz481 |
Li, Y., Yang, X., Liu, H., et al., 2022. Local and systemic responses conferring acclimation of Brassica napus roots to low phosphorus conditions. J. Exp. Bot., 73: 4753-4777. DOI:10.1093/jxb/erac177 |
Liao, Y., Smyth, G.K., Shi, W., 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30: 923-930. DOI:10.1093/bioinformatics/btt656 |
Liu, N., Shen, G., Xu, Y., et al., 2020. Extensive inter-plant protein transfer between Cuscuta parasites and their host plants. Mol. Plant, 13: 573-585. DOI:10.1016/j.molp.2019.12.002 |
Medici, A., Marshall-Colon, A., Ronzier, E., et al., 2015. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun., 6: 6274. DOI:10.1038/ncomms7274 |
Medici, A., Szponarski, W., Dangeville, P., et al., 2019. Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell, 31: 1171-1184. DOI:10.1105/tpc.18.00656 |
Nickrent, D.L., 2020. Parasitic angiosperms: how often and how many?. Taxon, 69: 5-27. DOI:10.1002/tax.12195 |
Rappsilber, J., Ishihama, Y., Mann, M., 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem., 75: 663-670. DOI:10.1021/ac026117i |
Ruffel, S., Krouk, G., Ristova, D., et al., 2011. Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Natl. Acad. Sci. U.S.A., 108: 18524-18529. DOI:10.1073/pnas.1108684108 |
Ruffel, S., Poitout, A., Krouk, G., et al., 2016. Long-distance nitrate signaling displays cytokinin dependent and independent branches. J. Integr. Plant Biol., 58: 226-229. DOI:10.1111/jipb.12453 |
Shahid, S., Kim, G., Johnson, N.R., et al., 2018. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature, 553: 82-85. DOI:10.1038/nature25027 |
Shen, G., Liu, N., Zhang, J., et al., 2020. Cuscuta australis (dodder) parasite eavesdrops on the host plants' FT signals to flower. Proc. Natl. Acad. Sci. U.S.A., 117: 23125-23130. DOI:10.1073/pnas.2009445117 |
Shen, G., Zhang, J., Lei, Y., et al., 2023. Between-plant signaling. Annu. Rev. Plant Biol., 74: 367-386. DOI:10.1146/annurev-arplant-070122-015430 |
Smith, J.D., Woldemariam, M.G., Mescher, M.C., et al., 2016. Glucosinolates from host plants influence growth of the parasitic plant Cuscuta gronovii and its susceptibility to aphid feeding. Plant Physiol., 172: 181-197. DOI:10.1104/pp.16.00613 |
Song, J., Bian, J., Xue, N., et al., 2022. Inter-species mRNA transfer among green peach aphids, dodder parasites, and cucumber host plants. Plant Divers., 44: 1-10. |
Sun, G., Xu, Y., Liu, H., et al., 2018. Large-scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat. Commun., 9: 2683. |
Thieme, C.J., Rojas-Triana, M., Stecyk, E., et al., 2015. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants, 1: 15025. |
Trapnell, C., Roberts, A., Goff, L., et al., 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc., 7: 562-578. DOI:10.1038/nprot.2012.016 |
Turck, F., Fornara, F., Coupland, G., 2008. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol., 59: 573-594. DOI:10.1146/annurev.arplant.59.032607.092755 |
Turnbull, C.G.N., Lopez-Cobollo, R.M., 2013. Heavy traffic in the fast lane: long-distance signalling by macromolecules. New Phytol., 198: 33-51. DOI:10.1111/nph.12167 |
UniProt, C., 2023. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res., 51: D523-D531. |
Vega, A., O'Brien, J.A., Gutierrez, R.A., 2019. Nitrate and hormonal signaling crosstalk for plant growth and development. Curr. Opin. Plant Biol., 52: 155-163. |
Zhang, J., Xu, Y., Xie, J., et al., 2021. Parasite dodder enables transfer of bidirectional systemic nitrogen signals between host plants. Plant Physiol., 185: 1395-1410. DOI:10.1093/plphys/kiaa004 |
Zhang, Z., Zheng, Y., Ham, B.K., et al., 2016. Vascular-mediated signalling involved in early phosphate stress response in plants. Nat. Plants, 2: 16033. |
Zhuang, H., Li, J., Song, J., et al., 2018. Aphid (Myzus persicae) feeding on the parasitic plant dodder (Cuscuta australis) activates defense responses in both the parasite and soybean host. New Phytol., 218: 1586-1596. DOI:10.1111/nph.15083 |



