b. Institute of Molecular Plant Science, School of Biological Science, University of Edinburgh, Edinburgh EH9 3BF, UK;
c. College of Science, Tibet University, Lhasa 850000, Xizang Autonomous Region, PR China
The conservation of threatened species is crucial in a time of global biodiversity loss (Lamoreux et al., 2006; Chau et al., 2013). Threatened species often possess small and highly fragmented populations, making them susceptible to extinction due to increasing anthropogenic pressures and stochastic environmental and demographic events (Lande, 1993; Wilson, 1996). Small populations typically exhibit low levels of genetic diversity, which limits their ability to adapt to environmental changes and other challenges due to a scarcity of adaptive alleles (Frankham et al., 2002; Jordan et al., 2019). In addition, fragmentation hinders or prevents gene flow between populations (Lange et al., 2010; Mantyka-pringle et al., 2012), causing small and isolated populations to accumulate deleterious alleles through inbreeding, resulting in a reduction in individual and mean population fitness (Bertorelle et al., 2022). This increase in genetic load reduces population fitness, manifesting as lower growth rates due to decreased resistance to fungal infections, maladaptation to harmful environments, and reduced seed viability (Schoen et al., 1998; Hedrick and Garcia-Dorado, 2016; Ma et al., 2022). Moreover, in species that have recently become rare due to human activity, increased genetic drift in small populations leads to faster fixation of deleterious mutations, ultimately reducing the probability that populations will survive (Lynch et al., 1993; Robinson et al., 2022).
The rapid advance of high-throughput sequencing technology has made investigating deleterious mutations, inbreeding as well as other factors increasingly feasible and robust (Allendorf et al., 2010; Manners et al., 2013; Palkopoulou et al., 2015; Bortoluzzi et al., 2020). Conservation genomic approaches have provided valuable insights into the accumulation or purging of deleterious mutations in small, endangered wild populations of non-model organisms, such as mountain gorillas (Xue et al., 2015), Alpine ibex (Grossen et al., 2020), Iberian lynx (Kleinman-Ruiz et al., 2022), Acer yangbiense (Ma et al., 2022), Rhododendron griersonianum (Ma et al., 2021) and Populus ilicifolia (Chen et al., 2020). However, few studies have focused on endangered conifers.
The Sichuan Arborvitae, Thuja sutchuenensis Franch., is an endangered conifer endemic to limestone cliffs, ridges, or steep slopes at 800–2100 m above sea level in northwestern Chongqing and the Daba Mountains in the eastern Sichuan Basin (Xiang et al., 2002; Cui et al., 2015; Tang et al., 2015). It is a tertiary relict species with a complex evolutionary history, and was once listed as extinct in the wild by IUCN but was rediscovered in 1999 (Xiang et al., 2002). Previous work surveyed its demographic history and level of genetic diversity using chloroplast simple sequence repeats (cpSSR) and restriction site-associated DNA sequencing (RAD-seq) data (Yao et al., 2021), as well as traditional molecular markers (e.g. inter-simple sequence repeat markers and single-copy nuclear loci; Liu et al., 2013; Qin et al., 2021). However, we have no knowledge regarding the level of genetic load in T. sutchuenensis. Note that genomic resequencing of conifers presents a particular challenge because of their very large genomes, which contains numerous and varied ancient repetitive sequences (Borthakur et al., 2022; Wei et al., 2022). In contrast, transcriptome sequencing (RNA-seq) technology provides ample coverage of the genome for conservation genomic studies and is ideally suited for analyzing deleterious alleles due to its focus on coding (exon) regions (Primmer, 2009; Angeloni et al., 2011; Li et al., 2020; Miao et al., 2021; Yang et al., 2022). Therefore, RNA-seq is a powerful approach to disentangle the demographic history, genetic diversity, degree of inbreeding, and intensity of purging deleterious mutations in rare conifer species such as T. sutchuenensis.
T. sutchuenensis has experienced recent human-induced decline, such as intensive logging and habitat loss (Qin et al., 2017, 2021). If reduced fitness results in decreased population size, which in turn increases inbreeding and genetic load, further exacerbating genetic diversity loss, this creates a destructive feedback loop known as the extinction vortex (Godwin et al., 2020). Consequently, conservation and management efforts should be based on robust evaluation of genetic diversity, inbreeding coefficients, and genetic load in small populations (Grossen et al., 2020; Bertorelle et al., 2022; Willi et al., 2022; Xie et al., 2022). Hence, in the present study, we have conducted a conservation genomic survey of T. sutchuenensis using high-throughput sequencing data. Our objectives are to (a) evaluate the genetic load and potential extinction risk of T. sutchuenensis; (b) estimate the genetic diversity and inbreeding, and unravel the population structure of T. sutchuenensis; (c) investigate the demographic history and gene flow among populations of T. sutchuenensis; and (d) develop more targeted conservation measures for T. sutchuenensis. These findings are critical for forecasting the fate of T. sutchuenensis populations and improving the effectiveness of current conservation programs. Moreover, these findings will contribute to our understanding of genetic load and extinction risk of endangered plant species.
2. Materials and methods 2.1. Sample collection and RNA sequencingWe sampled 67 individuals from five wild populations across the distribution region of T. sutchuenensis, plus eight Thuja plicata individuals from Lushan Botanical Garden, Chinese Academy of Sciences (Fig. 1; Tables 1 and S1). Fresh leaves were kept below −80 ℃ before extraction. The RNA library for RNA-seq was prepared using standard protocols (Grabherr et al., 2011). Specifically, mRNA was purified from total RNA using polyT and then fragmented into 300–350 bp fragments. The first strand cDNA was reverse-transcribed using fragmented RNA and dNTPs (dATP, dTTP, dCTP, and dGTP), and second strand cDNA synthesis was subsequently performed. The remaining overhangs of double-strand cDNA were converted into blunt ends via polymerase activities. After adenylation of 3' ends of DNA fragments, sequencing adaptors were ligated to the cDNA and the library fragments were purified. The template was enriched by PCR, and the PCR products were purified to obtain the final library. After library preparation and pooling of different samples, the samples underwent Illumina sequencing (Berry Genomics). The raw sequence data reported in this paper has been deposited in the Genome Sequence Archive (Chen et al., 2021) in the National Genomics Data Center (Xue et al., 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: gsa:CRA009439 and gsa:CRA007034), which are publicly accessible at https://ngdc.cncb.ac.cn/gsa. Additional transcriptome data of four Thuja species and Thujopsis dolabrata was gathered from Li et al. (2021) (Table S2).
|
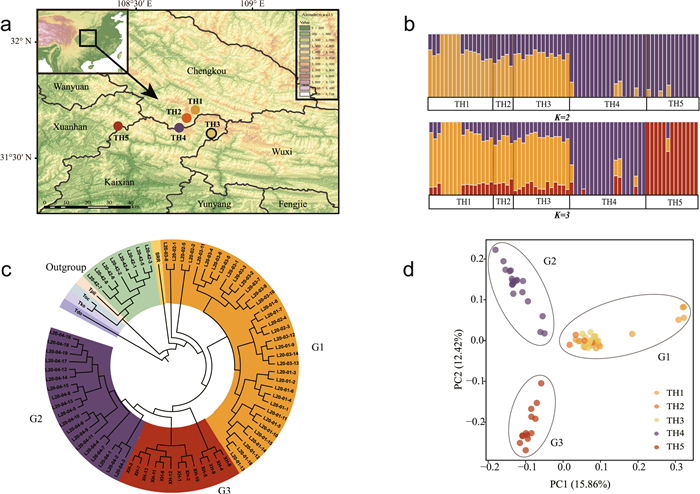
| Fig. 1 Phylogenetic and population genetic analyses of Thuja sutchuenensis based on SNP variation. (a) Map showing the geographic distribution of sampling locations for T. sutchuenensis populations. (b) Population structure plots with K = 2 and 3. The x-axis shows the different individuals of populations; the y-axis quantifies the proportion of an individual's variation from inferred ancestral populations. (c) A maximum likelihood phylogenetic tree. Abbreviations: Toc, Thuja occidentalis; Tko, Thuja koraiensis; Tpli, Thuja plicata; SRR, Thuja standishii; Tdo, Thujopsis dolabrata. (d) Principal component analysis (PCA) plot of the first two components. |
| Population | Code | N | Latitude (N) | Longitude (E) | Location |
| L20-01 | TH1 | 16 | 31.71° | 108.75° | Chengkou, Chongqing |
| L20-02 | TH2 | 5 | 31.66° | 108.72° | Chengkou, Chongqing |
| L20-03 | TH3 | 14 | 31.60° | 108.83° | Kaizhou, Chongqing |
| L20-04 | TH4 | 19 | 31.62° | 108.68° | Kaizhou, Chongqing |
| XH | TH5 | 13 | 31.63° | 108.43° | Xuanhan, Sichuan |
To obtain a high-quality reference, we built a reference transcriptome for T. sutchuenensis by assembling five sets of RNA-seq data de novo from five populations in TRINITY v.2.8.4 (Grabherr et al., 2011). The individual transcriptome was assembled in TRINITY v.2.8.4 with default parameters, and only the longest isoform of each gene was retained for downstream analyses. We used ORTHOFINDER v.2.3.12 (Emms and Kelly, 2015) to identify orthologous groups, and added the unassigned sequences to form the final reference sequence.
Assembly statistics were generated using the trinitystats. pl script within Trinity's toolkit. To obtain high-quality contigs, we used BLASTN v.2.7.1 (Altschul et al., 1997) to align sequences to the microbial genome database (http://mbgd.genome.ad.jp/). Sequences with more than 90% similarity were removed. Redundant sequences were removed using CD-HIT-EST v.4.6.8 (Fu et al., 2012). To evaluate the transcriptome completeness, we employed BUSCO v.5.4.4 (Manni et al., 2021) with default parameters, using the embryophyte conserved gene dataset (embryophyta_odb10) as the query database. Gene annotation information was predicted by TRANSDECODER v.4.6.1 (http://github.com/TransDecoder/TransDecoder/wiki/).
To obtain high-quality reads, TRIMMOMATIC v.0.36 (Bolger et al., 2014) was used to filter Illumina raw reads using the following parameters: "LEADING: 3 TRAILING: 3 SLIDINGWINDOW: 4:15 MINLEN: 5". For each individual, the quality-filtered reads were mapped to the reference transcriptome using STAR v.2.7.9 (Dobin et al., 2013) with default parameters. SAMTOOLS v.1.2 (Li et al., 2009) was used to convert Sequence Alignment/Map (SAM) files to Binary Alignment/Map (BAM) files and sort BAM files. Duplicate reads were marked and removed using Picard v.2.18.11 (http://broadinstitute.github.io/picard/). The HaplotypeCaller and GenotypeGVCFs modules from the Genome Analysis Toolkit v.4.2.0 (GATK) (DePristo et al., 2011) were used to call single nucleotide polymorphisms (SNPs) following the best practices workflow (DePristo et al., 2011). SNPs were filtered based on the GATK recommendations for hard filtering (e.g., QD < 2.0; FS > 30, Dataset 1; Fig. S1). To ensure high-quality data, we removed SNPs with genotype depths below 5, a minor allele frequency below 0.05, a significant deviation from Hardy–Weinberg equilibrium (p < 0.001) or those containing more than 20% missing data at all SNP sites, hereafter sites (Dataset 2; Fig. S1), using BCFTOOLS v.1.6 (Narasimhan et al., 2016a) and VCFTOOLS v.0.1.15 (Danecek et al., 2011).
2.3. Population genetic and phylogenetic analysesWe used VCFTOOLS v.0.1.15 to calculate the values of Tajima's D and fixation statistics (FST). As a lack of information on missing and invariant sites may cause bias when estimating nucleotide diversity (π), we used PIXY v.1.2.7. Beta1 (Korunes and Samuk, 2021) and 10-kb windows to calculate these genetic parameters on Dataset1, including invariant sites. Meanwhile, we used the R packages ggplot2 (Villanueva and Chen, 2019) and ggstatsplot (Patil, 2021) to plot and perform statistical analyses. Subsequent analyses were based on Dataset 2. Population structure was analyzed to assign individuals to variable numbers (K = 1–5) of ancestors using the program ADMIXTURE v.1.30 (Alexander and Lange, 2011). We used VCFTOOLS and PLINK v.1.90 (Purcell et al., 2007) to convert data. To complement the results of ADMIXTURE, we conducted principal component analysis (PCA) on all individuals to examine genetic structure or relatedness using the SMARTPCA module in the software EIGENSOFT v.6.1.3 (Price et al., 2006).
In our phylogenetic analysis, we used a Python script (https://github.com/edgardomortiz/vcf2phylip) to convert the SNPs database to concatenated sequences for all individuals. IQTREE v.1.6.12 (Nguyen et al., 2015) was used to construct a maximum likelihood phylogenetic tree for T. sutchuenensis with default parameters using Thujopsis dolabrata and an additional four species in Thuja as the outgroups. Bootstrap values were calculated based on 1000 replicates. The phylogenetic tree was visualized using FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
We used the KING program v.2.2.5 (Manichaikul et al., 2010) to estimate genetic relatedness (duplicate, 1st-degree relative pairs, 2nd-degree relative pairs, 3rd-degree relative pairs, and unrelated pairs) between individuals of three groups of T. sutchuenensis with the –kinship flag. We also calculated the inbreeding coefficients (FIS) in three groups and the species on a whole using the –het flag in VCFTOOLS.
2.4. Demographic history inferenceSince the accuracy of demographic inferences is affected by mutation rate (μ), we made calculations based on a fossil-calibrated approach 'μ = D*g/2T' (Kondrashov and Crow, 1993), where D is the observed frequency of pairwise differences between the two species, T is the estimated divergence time, and g is the estimated generation time. The generation time was set at 50 years following Qin et al. (2021), and the divergence time between T. sutchuenensis and T. dolabrata was set at 62.68 Mya, according to previous studies (Li et al., 2021; Qin et al., 2021). VCF2Dis (https://github.com/BGI-shenzhen/VCF2Dis) was used to calculate D and the mean difference value, and generate a pairwise differences matrix. Based on the observed frequency of pairwise differences and divergence time, we estimated a mutation rate of 1.19 × 10−8 per site per generation to infer the demographic history. Global max-likelihood estimates were derived from 100 independent runs, with 100,000 coalescent simulations and 40 likelihood maximization algorithm cycles. The Akaike's information criteria (AIC) (Akaike, 1998) was used to assess the relative fit of each model, and the best fit with the lowest AIC was chosen. The 95% confidence intervals (CIs) for the parameters from the best model were constructed using 200 parametric bootstrap replicates by simulating SFS from the maximum composite likelihood estimates.
PCA and ADMIXTURE identified three groups of T. sutchuenensis. We used fastsimcoal2 software (Excoffier et al., 2013) to estimate divergence time and gene flow between these groups, based on the site frequency spectrum (SFS). Demographic scenarios to be tested were established on the basis of the phylogenetic and population structure analyses. First, to minimize the effects induced by selection, we used SNPs (Dataset 3; Fig. S1) at those four-fold synonymous sites (4DTV, i.e., codons in which any base at the third position is translated into the same amino acid) that had no missing data across all individuals. We employed the iTools module in the ReSeqTools toolkit (https://github.com/BGI-shenzhen/Reseqtools) with the parameter "Fatools getCdsPep -4DSite" to extract 4DTV sites and used a custom script to generate 0-fold and 4-fold data. We used a Perl script to generate a folded two-dimensional site frequency spectrum for each of the three groups, inferred based on 4DTV sites (Ru et al., 2018).
In addition, we used STAIRWAY PLOT 2 (Liu and Fu, 2020) to analyze historical changes in effective population size (Ne) for the three groups of T. sutchuenensis (Dataset 3). Two hundred subsamples comprising 67% of all sites were created, the median value of the estimation was used as a final output, and the 95% confidence interval of each value was calculated.
Because both demographic methods mentioned above failed to adequately account for the most recent changes in population size, we further estimated the contemporary Ne for T. sutchuenensis. To achieve this, we used an LD (linkage disequilibrium) model estimation method with a random mating and a critical value of 0.02 that was implemented in NEESTIMATE v.2.1 (Do et al., 2014) (Data set 2). Meanwhile, we used an approximate Bayesian computation (ABC) model to estimate contemporary Ne that was implemented in ONESAMP v.1.2 (Tallmon et al., 2008) (Data set 2).
2.5. Characterization of deleterious mutationsTo compare genetic loads between T. sutchuenensis and its congeners, the target capture sequencing data from Li et al. (2021; Table S2) was used to map to the assembled reference transcriptome, using BWA v.0.7.17 (Li and Durbin, 2009) with the default parameters. This target capture sequencing data set covered three individuals of each Thuja species (15 individuals; Table S2), and captured nearly the entire exome. We used one individual from Thujopsis dolabrata as the outgroup. To obtain reliable genotypes from RNA sequencing reads, we followed the genotype calling procedure as described above. To avoid false positive results due to missing data, we applied VCFTOOLS to remove INDEL (insertion-deletion) sites, and BCFTOOLS to retain no missing sites (Dataset 4; Fig. S1) We excluded sites where the individuals contained multiple alleles. Finally, a total of 298,719 high-quality SNPs was retained. The ancestral state for each polymorphic site was inferred with outgroup sequences (Thujopsis dolabrata) using the program EST-SFS v.2.0.3 (Keightley and Jackson, 2018).
We estimated the selection pressure upon each species by calculating the nucleotide diversity (π) at nonsynonymous (zero-fold) and synonymous (four-fold, i.e., codons in which any base at the third position is translated into the same amino acid) sites (i.e., πn and πs, respectively) and the ratio of πn/πs using PIXY v.1.2.7. Beta1 (Korunes and Samuk, 2021) and 10-kb windows.
To reduce the influence of algorithm preference on the evaluation of deleterious mutations, we used three different methods to evaluate the SNPs database. SIFT (sorting intolerant from tolerant) was used to predict whether amino acid substitution (A, T, C, G) affects the protein function energy algorithm (Ng and Henikoff, 2003; Kumar et al., 2009; Sim et al., 2012). SIFT4G targeted the reference sequence by constructing a target base according to the conservation of homologous sequences of multiple species. Because of the score database of histone coding sequences, the harmfulness of nonsynonymous mutations in species can be quickly evaluated (Vaser et al., 2016). Default settings recommended by the authors were used. We first downloaded the UniRef90 protein database, following the developer's recommendations. Based on this database, we used make-SIFT-DB-all.pl script to construct the reference of T. sutchuenensis SIFT scoring database, and then used SIFT4G_Annotator.jar to build the respective nonsynonymous SNPs based on annotating the database. Any SNP mutation with a score < 0.05 in the SIFT database was defined as a deleterious mutation site (henceforth denoted as a DEL site).
Grantham score was one of the earliest algorithms developed to predict the amino acid effect of deleterious mutations (Grantham, 1974). The score varied from 0 to 215, with lower scores reflecting substitutions between amino acids that were more similar in composition, polarity, and molecular volume, whereas higher scores reflect dissimilarity (Grantham, 1974). We used the Grantham scoring matrix to score each nonsynonymous SNP, and to identify those SNPs with a Grantham score (GS) ≥ 150 which was marked as Radical in the script as moderately harmful mutation sites (Li et al., 1984) (hereafter termed 'Radical sites').
We defined any deleterious mutation identified by the above two methods as a moderate and slightly deleterious mutation. However, mutations that affect the transcription of the entire gene (stop gained, stop lost, start lost, or start gained) were highly deleterious mutations and were henceforth termed Loss of Function (LOF) mutations. To assess the accumulation of highly deleterious mutations in different species and groups, we used SNPEFF v.3.4 (Cingolani et al., 2012) to assign SNPs to different mutational types by comparing the reference and gene annotation information with parameter "-eff". The variants were grouped in three categories: (1) LOF; (2) nonsynonymous; and (3) synonymous.
To further investigate differences in genetic load between species, we sampled three individuals per species, and analyzed each species pair by selecting a single individual from each species. We then counted the number of derived mutations at each site that was variable in one individual but not in the other. From this data, we calculated the RXY ratio, which represents the difference between two populations' relative abundance of derived allele frequencies for a specific category of sites (Do et al., 2015). The RXY ratio was calculated independently for each category of mutations: Radical, DEL, and LOF. In each instance, RXY values greater than 1 or less than 1 signified that the ability to purge deleterious mutations was stronger in species X or Y, respectively (Do et al., 2015). We employed this approach to search for evidence of purifying selection. For detailed information on calculating the various types of genetic load, please refer to the Supplementary data.
2.6. Estimates of the distribution of fitness effectsThe distribution of fitness effects (DFE) from a new mutation can be independently modeled for deleterious, neutral, and beneficial mutations. For our reference transcriptome, we counted the number of polymorphic sites of each frequency class (synonymous and nonsynonymous sites, Data set 5; Fig. S1) to generate an unfolded SFS for five species and three groups. Briefly, variants in coding regions were annotated as synonymous or nonsynonymous and polarized using derived versus ancestral alleles. We inferred the distribution of fitness effects for new mutations using POLYDFE v.2 (Tataru et al., 2017; Tataru and Bataillon, 2019; Dutheil, 2020). We chose the default model, which modeled DFEs as a mixture of gamma and exponential distributions. The fitting procedure itself was run with the basin-hopping algorithm with up to 10 iterations to avoid local maxima. Using the provided postprocessing R code, the models were ranked using the Akaike-information criterion (Akaike, 1998), and parameter estimates were calculated as the AIC-weighted average. We divided the deleterious portion of the DFE in six Nes (effective population size multiply by selection coefficient) classes (-∞ to −1000, −1000 to −100, −100 to −10, −10 to −1, −1 to 0 and 0 to +∞). In addition, we used the selection coefficient (s) to create subsets of polymorphisms that were beneficial (Nes > 0), or deleterious (Nes < 0), with the latter further subdivided into different levels of severity, i.e. −1 < Nes ≤ 0, −10 < Nes ≤ −1, −100 < Nes ≤ −10, −1000 < Nes ≤ −100, and (Nes ≤ −1000), the last of which we defined as highly deleterious.
3. Results 3.1. Transcriptome sequencing and de novo assemblyThe finalized reference transcriptome of T. sutchuenensis comprised 60,102 unigenes, with an average length of 812 bp and a contig N50 of 2425 bp (Fig. S2; Table S3). The Benchmarking Universal Single-Copy Orthologs (BUSCOs) assessment indicated a quality assessment score of 90.1% and a total of 1454 complete BUSCOs. After eliminating low-quality reads, we acquired approximately 216 Gb of data from the 67 samples analyzed.
3.2. Population genetic and phylogenetic analysesAfter hard-filtering through the GATK pipeline, we obtained 3,045,724 SNPs. Of these, 636,151 SNPs were retained for population genetic and phylogenetic analyses. Phylogenetic analysis of transcriptome-wide SNPs suggested that individuals of T. sutchuenensis were clustered into three separate monophyletic clades. Principal component analysis (PCA) identified three groups along PC1 (variance explained = 15.86%) and PC2 (variance explained = 12.42%) (Fig. 1d). ADMIXTURE indicated that the population genetic structure was relatively weak (Fig. S3). When K = 3, the three identified groups clearly conformed with the results from PCA (Fig. 1b), although the shared ancestry of the groups might have caused non-monophyly in two out of three groups. The range of population level pairwise FST was between 0.0392 and 0.0729, suggesting a low to moderate level of genetic differentiation (Yang et al., 2022), especially for TH4 or TH5 between the other three populations (Table 2). Nevertheless, genetic differentiation between the three groups was moderate (FST > 0.05) (Table S4). These results indicated that we could divide all sampled population into three groups, henceforth termed groups G1, G2 and G3, and that these groups comprise populations TH1+TH2+TH3, TH4, and TH5, respectively.
| Population | TH1 | TH2 | TH3 | TH4 | TH5 |
| TH1 | – | – | – | – | – |
| TH2 | 0.0545 | – | – | – | – |
| TH3 | 0.0392 | 0.0535 | – | – | – |
| TH4 | 0.0657 | 0.0654 | 0.0545 | – | – |
| TH5 | 0.0641 | 0.0729 | 0.0583 | 0.0608 | – |
The level of nucleotide diversity was similar among the five T. sutchuenensis populations, or three groups (Tables S5 and S6). All average π (standard deviation, SD) was 0.00186 (0.00111) across 10-kb windows, which is lower than the result 0.00272 (0.0122) from Shalev et al. (2022) (transcriptome data vs. target capture sequencing data). Additionally, the values of Tajima's D were positive in each population and group, which indicates that all populations and groups might have experienced population bottleneck(s) and/or balanced selection in their evolutionary history (Table 3 and S7).
| Population | Tajima's D |
| TH1 | 0.85 |
| TH2 | 0.70 |
| TH3 | 0.88 |
| TH4 | 0.98 |
| TH5 | 0.86 |
For T. sutchuenensis populations overall, the inbreeding coefficient (FIS, 0.0209) (Table S8) that we estimated was much lower than previous study using 112 individuals showed that inbreeding coefficient in the T. plicata range-wide population was 0.331 (Shalev et al., 2022). Although sampling size (67 vs. 112) and data type (transcriptome data vs. target capture sequencing data) may have affected the estimate, mating system might have played a major role since T. plicata can be self-compatible (Shalev et al., 2022). For each of the three groups our data distinguished, FIS scores were highest in G3 and lowest in G2 (Table S8). In addition, the G1 group possessed the highest value of average kinship coefficient (0.06; Table S9). We also calculated duplicate, 1st-degree, 2nd-degree, and 3rd–degree relationships in pairs of individuals, and we found that there was a higher proportion of close relationships in G1 group (0.82; Table S9).
3.3. Demographic history and gene flowThe STAIRWAY PLOT 2 analysis showed that all groups had suffered a severe bottleneck effect during the Last Glaciation Period between around 110 and 10 thousand years ago (Kya) (Fig. 2b). Following a period of continuous population decline, the Ne of T. sutchuenensis stayed relatively stable from around 10 Kya onwards (Fig. 2b).
|
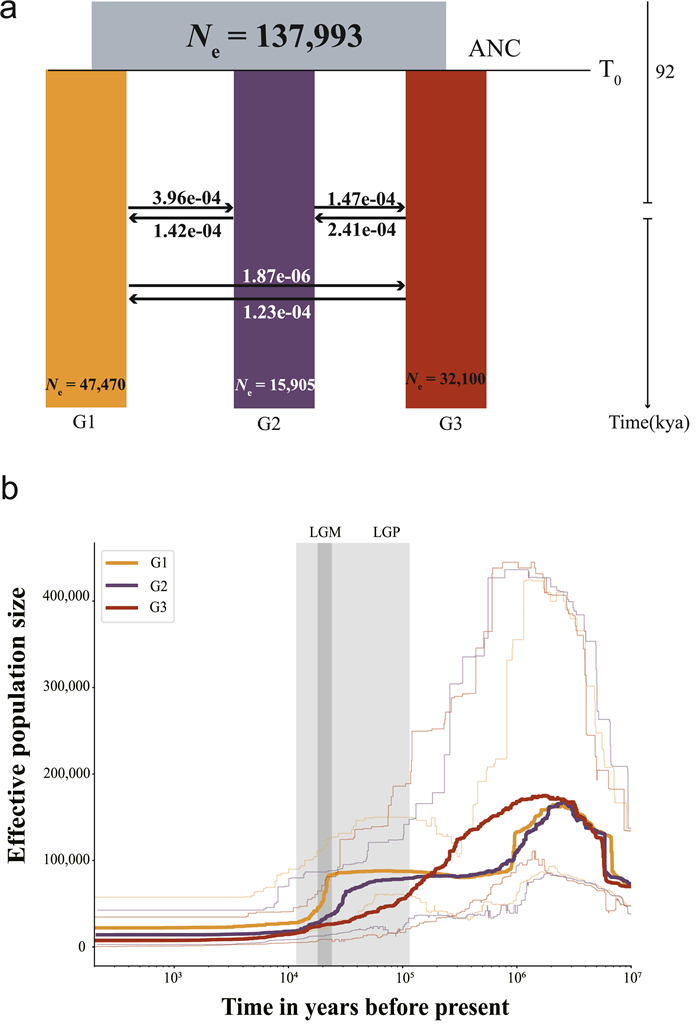
| Fig. 2 Demographic history of Thuja sutchuenensis. The light gray areas represent different glaciation events during the Pleistocene (LGP: Last Glaciation Period, LGM: Last Glaciation Maximum). (a) Schematic of demographic scenario modeled in fastsimcoal2. Estimated effective population sizes (Ne) and divergence times are indicated. The numbers next to the arrows denote the per-generation migration rate between populations. (b) Changes in effective population size (Ne) over time in T. sutchuenensis inferred by Stairway Plot method using a generation time of 50 years and an autosomal mutation rate (μ) of 1.19 × 10−8 per base pair per generation. Thick lines represent the median and thin light lines the 95% pseudo-CI defined by the 2.5% and 97.5% estimations of the SFS analysis. |
Furthermore, we set eleven models to estimate gene flow among the three T. sutchuenensis groups (G1, G2, G3) using coalescent-based simulations in fastsimcoal2. Among the eleven candidate models employed, the best-fitting model (with minimum Akaike's weight value) contained 11 parameters (e.g., ANC: common ancestor effective populations size, TDIV: divergence time, NPOP: effective population size of each group, MIG: gene flow; Table 4, the number 0, 1, 2 denotes group 1, 2, 3, respectively), including six separate periods of gene flow (Tables 4 and S10; Fig. S4). The result showed that gene flow had been continuous between the three groups since they diverged, with gene flow from G3 into other groups being greater than that into G3. Specifically, gene flow from G3 into G1 was ~65.8-fold that in the reverse direction; the gene flow from G3 into G2 was 1.6-fold that in the reverse direction (Fig. 2a).
| Parameter | Point estimation | 95% confidence intervals | |
| Lower bound | Upper bound | ||
| ANC | 137,993 | 134,790 | 142,845 |
| NPOP0 | 47,470 | 43,003 | 63,381 |
| NPOP1 | 15,905 | 14,498 | 21,790 |
| NPOP2 | 32,100 | 28,046 | 44,549 |
| TDIV | 1840 | 1619 | 5359 |
| MIG01 | 3.96E-04 | 2.11E-04 | 5.70E-04 |
| MIG10 | 1.42E-04 | 2.82E-05 | 2.25E-04 |
| MIG12 | 1.47E-04 | 2.25E-05 | 2.30E-04 |
| MIG21 | 2.41E-04 | 1.24E-04 | 3.45E-04 |
| MIG02 | 1.87E-06 | 7.40E-08 | 8.85E-05 |
| MIG20 | 1.23E-04 | 5.49E-05 | 1.98E-04 |
| Note: ANC indicates the common ancestor effective population size; NPOP0 indicates the G1 effective population size; NPOP1 indicates the G2 effective population size; NPOP2 indicates the G3 effective population size; TDIV indicates the divergence time; MIG01 indicates the gene flow from G1 to G2; MIG10 indicates the gene flow from G2 to G1; MIG12 indicates the gene flow from G2 to G3; MIG21 indicates the gene flow from G3 to G4; MIG02 indicates the gene flow from G1 to G3; MIG20 indicates the gene flow from G3 to G1. | |||
From 92 Kya onwards (95% CIs: 80.95–267.95 Kya), the three groups seemed to have a stable effective population size (Fig. 2b). Moreover, the effective population size of G2 (Ne = 15,905, 95% CIs: 14,498–21,790) was the smallest of the three groups (Table 4). Meanwhile, the estimated contemporary Ne for T. sutchuenensis was estimated to be 138.8 (95% CIs: 90.1–269.6) using NEESTIMATE v.2.1. The ratio of contemporary Ne (138.8) to the census population size (7000) was calculated to be ~0.02. At the same time, the contemporary Ne of T. sutchuenensis (138.8) was about half that of T. plicata, which was estimated to be 270.3 (transcriptome data vs. target capture sequencing data) (Shalev et al., 2022). We also used NeABC as implemented in the ONESAMP v.1.2 to estimate the effective population size. However, the results demonstrated that the mean Ne was 45.5 (95% CIs: 40.3–55.9), which was 67.2% lower than the estimate derived from NEESTIMATE (138.8).
3.4. Detection of deleterious mutationsAccording to the nearly neutral theory (Ohta, 1992), the efficacy of purifying selection depends on the effective population size (Ne). Thus, the πn/πs ratio is directly and negatively related to Ne. Moreover, a small population size could result in a low level of genetic diversity and/or selection efficacy, so that the extent of genetic load can be reflected by the ratio of nonsynonymous versus synonymous nucleotide diversity, and hence correspondingly the ratio of 0-fold and 4-fold degenerate positions in protein-coding sequences. We used 15 individuals of Thuja from a previous study (five species, three individuals per species; Li et al., 2021) to estimate πn and πs. Our analysis indicated that T. sutchuenensis had relatively higher nucleotide diversity compared with other species in 0-fold degenerate sites and the value of θπ on 4-fold degeneration sites of T. sutchuenensis was higher than that of the other 4 species (p < 0.001; the Kruskal–Wallis rank-sum test) (Figs. S5 and S6). Likewise, the πn/πs ratio of T. sutchuenensis was lower than that of other species (Table S11), suggesting a higher selection efficacy within T. sutchuenensis than in its congeners. Therefore, relative to other Thuja species, T. sutchuenensis has presumably purged more deleterious mutations and possesses a lower genetic load, which was consistent with our results of πn/πs ratio, πn and πs values (Figs. S5 and S6; Table S11). However, our result of the πn/πs ratio (0.478) of T. sutchuenensis was slightly higher than that found in recent work on T. plicata (π0/π4 ratio of 0.33) (both are target capture sequencing data) (Shalev et al., 2022).
To explore the accumulation of genetic load in each species, we used a total of 258,271 (T. occidentalis), 261,714 (T. koraiensis), 259,889 (T. plicata), 267,209 (T. standishii), and 271,524 (T. sutchuenensis) SNPs from protein-coding regions to evaluate the accumulation and purging of deleterious mutations (Table S12). The SNPEFF program annotated 5196 Loss of Function (LOF) SNPs from T. sutchuenensis, and similar numbers from the other species: 5147 from T. occidentalis, 5151 from T. koraiensis, 5158 from T. plicata, and 5201 from T. standishii (Table S12). The respective totals of Radical and DEL SNP mutations identified for each species by Grantham score and SIFT4G were 7487 and 5519 (T. occidentalis), 7557 and 5525 (T. koraiensis), 7499 and 5516 (T. plicata), 7601 and 5564 (T. standishii), and 7673 and 5586 (T. sutchuenensis) (Table S12).
The relative number of derived alleles (RXY), which was used to estimate genetic load (Xue et al., 2015; Narasimhan et al., 2016b), indicated that the proportion of all three kinds of deleterious mutations were lower in T. sutchuenensis than in any other congeneric species (Fig. 3a). This suggested a purging of genetic load, including both high and low impact deleterious alleles, in T. sutchuenensis. Among the three groups in T. sutchuenensis, RXY found the lowest proportion of LOF, Radical and DEL mutations in G1, but the highest level of LOF and Radical mutations in G3 (Fig. 3b).

|
| Fig. 3 The relative number of derived alleles at LOF (red), Radical (orange), and DEL (yellow) sites are frequent in one population and not another. The horizontal line of 1 indicates nondifference in a relative number of derived alleles. (a) Abbreviations: Toc, Thuja occidentalis; Tko, T. koraiensis; Tpli, T. plicata; Tst, T. standishii; Tsu, T. sutchuenensis. (b) Note: G1 includes TH1, TH2, and TH3; G2, TH4; G3, TH5. |
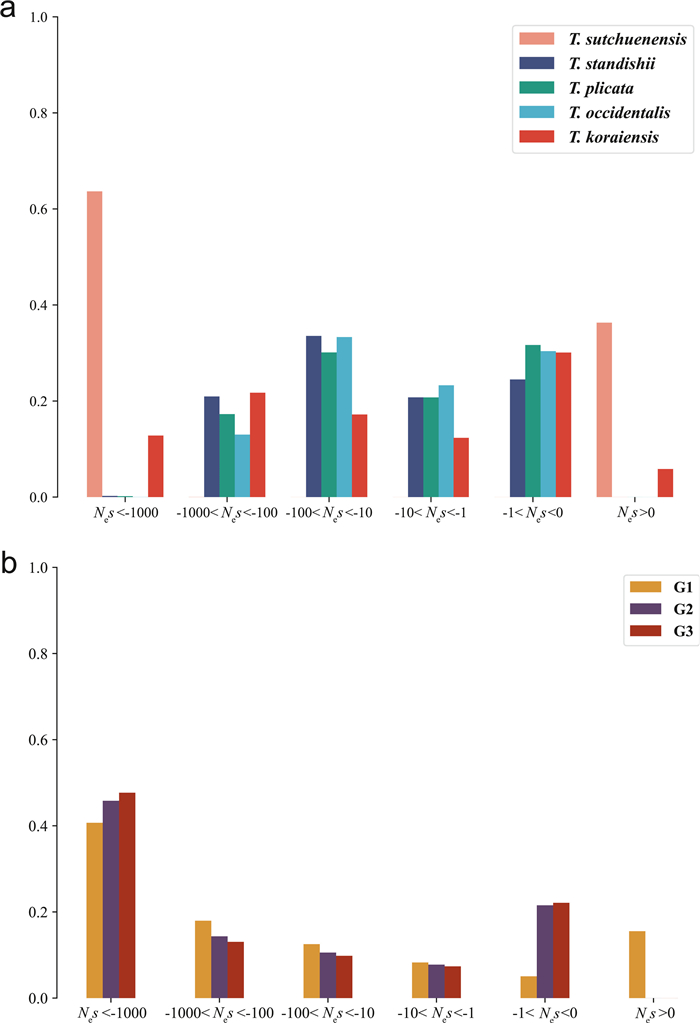
Analysis of population genetic polymorphism offered a perspective on sequence evolution over more recent timescales, notably via the distribution of fitness effects (DFEs) for segregating derived alleles (Eyre-Walker and Keightley, 2007; Tataru et al., 2017). DFE analyses revealed extraordinary differences between T. sutchuenensis and the other species (Fig. 4a). Within T. sutchuenensis, the G3 group possessed the most deleterious polymorphisms (Nes < −1000); no beneficial polymorphisms (Nes > 0) were detected in the G2 or G3 groups (Fig. 4b).
|
| Fig. 4 The DFEs in six bins of Nes (effective population size multiplied by selection coefficient) for new mutations for (a) five species in Thuja, and (b) three groups in T. sutchuenensis. |
The genetic diversity of small populations tends to be low due to genetic drift and inbreeding (Keller and Waller, 2002; Kohn et al., 2006; Li et al., 2012; Kim et al., 2022). Therefore, the genetic diversity of rare and endangered species with a narrow distribution is often lower than that of its more widespread relatives (Clegg et al., 1989; Yang et al., 2018; De Kort et al., 2021). Here, genetic diversity within T. sutchuenensis was estimated using population-level transcriptome sequencing data. The nucleotide genetic diversity (0.00186) of T. sutchuenensis was found to be relatively low compared with that of other threatened conifers such as Cupressus chengiana (0.0077) (Li et al., 2020), Cupressus gigantea (0.0027) (Yang et al., 2022), Juniperus erectopatens (0.00222) but higher than that of J. microsperma (0.00129) (Miao et al., 2021). This is consistent with two previous surveys based on six nuclear loci (Qin et al., 2021) and RAD-seq of T. sutchuenensis (Yao et al., 2021).
Our phylogenetic, PCA, and ADMIXTURE analyses suggest there are three distinct groups in T. sutchuenensis, two comprising single populations and the other (G1) comprising the remaining three. Pairwise FST values indicate moderate levels of genetic differentiation among these three groups (between 0.0570 and 0.0608; Table 2 and S4), which is higher than the reported pairwise FST value between the two management units (MUs) of YTR and NR (0.0488) of C. gigantea based on the same data type (RNA-seq; Yang et al., 2022). Hence, there are three groups in T. sutchuenensis that form natural MUs.
4.2. Demographic history and gene flow explain the genetic diversity pattern of Thuja sutchuenensisOur reconstructed demographic history of the three T. sutchuenensis groups suggested that two groups might have experienced two bottleneck events. The first bottleneck for G1 and G2 occurred around 1 Mya (Fig. 2b), with a steep drop in G1 but gradual in G2, coinciding with the Xixiabangma Glaciation (1.2–0.8 Mya) on the Qinghai-Tibet Plateau (Zheng et al., 2002). The second bottleneck occurred ~26.5 Kya in G1 and slightly earlier in G2, around the start of the Last Glacial Maximum (LGM, 26–19 Kya) (Fig. 2) (Clark et al., 2009; Cui et al., 2011; Qin et al., 2017). Conversely, group G3 contracted gradually from ~1 Mya until ~5 Kya, becoming the smallest group from ~200 Kya onwards. Hence the Quaternary climate oscillations affected this group differently. From the early Holocene (~10 Kya), G1 and G2 were stable in size, whereas G3 was stable in size from ~5 Kya (Fig. 2a). Hence, the three groups (G1, G2, G3) exhibited distinct evolutionary histories (Fig. 2). Previous findings based on extended Bayesian skyline plots suggested that T. sutchuenensis population size has not changed over time (Qin et al., 2021), although this approach predicts effective population size with less certainty (von et al., 2022). Because MUs are based on the demographic independence of populations (Palsbøll et al., 2007), we argue that our demographic reconstruction provides further support for the division of the T. sutchuenensis population into three separate MUs.
Fossil evidence from the late Pliocene in Shanxi Province, northern China (Cui et al., 2015), ca. 700 km to the northeast of the current distribution of T. sutchuenensis in the Daba Mountains, southwestern China, indicates that this species once had a wider distribution in China. According to the fastsimcoal2 analysis, the ancestral effective population size of the species at approximately 92 Kya was around 137,993 (95% CIs: 134,790–142,845). This is 8.7 to 2.6 times larger than that of the present-day groups and 997.8 times greater than the contemporary effective population size (as inferred by NEESTIMATE, which is 138.8) of this species. This suggests that the species as a whole has experienced strong bottlenecks and habitat fragmentation in the past.
In general, the effective population size (Ne) is much smaller than the census population size (Nc) (Nei et al., 1975; Charlesworth, 2009). The Nc of T. sutchuenensis was approximately 7000 (https://www.iucnredlist.org/species/32378/2816862, last accessed on 31 March 2023), much smaller than the historical Ne of any of the three groups as estimated by fastsimcoal2 (> 7000) (Fig. 2a). This finding suggests that the effective population size may have a hysteresis effect and will continue to decline. Meanwhile, given the estimated contemporary Ne of 138.8 (NEESTIMATE) or 45.5 (NeABC), and the extremely low ratio of the estimated contemporary Ne (0.02 or 0.006) to the census population size (7000), it is possible that there was a recent reduction in the effective population size of T. sutchuenensis (Turner et al., 2006).
One possible explanation for this could be that T. sutchuenensis has experienced very recent population contractions due to human impacts such as habitat destruction and logging, but these are too recent in terms of generations to have created a bottleneck effect so far. However, if no action is taken, the bottleneck effect will lead to a further decline of Ne, increasing future extinction risk of the species.
Our fastsimcoal2 analyses suggested high levels of historical gene flow between groups. Gene flow was higher from G1 or G3 to G2 than the other way, yet these were similar in magnitude. However, gene flow from G1 to G3 was 66 times smaller than the other way (Fig. 2a, Table 4). Previous studies based on nuclear loci detected similar extensive gene flow among populations of T. sutchuenensis (Qin et al., 202). It has been demonstrated that anthropogenic impacts are directly related to the loss of biodiversity and the mass extinction of species (Ceballos et al., 2015). In the case of T. sutchuenensis, trees have long been harvested by local residents for such diverse products as Buddhist beads, carvings, and furniture (Qin et al., 2017). This issue, plus habitat destruction and fragmentation, are likely to be the main causes of a reduction in its Ne and genetic diversity. These, plus the additional threat of climate change, are likely to lead to continued population shrinkage, which will ultimately lead to extinction unless suitable conservation and management measures are taken.
4.3. Low genetic load yet high extinction risk of Thuja sutchuenensisEndangered species generally suffer from a high genetic load (Díez-del-Molino et al., 2018; Bertorelle et al., 2022; Xie et al., 2022). The accumulation of deleterious mutations leads to a high genetic load (Agrawal and Whitlock, 2012; Chen et al., 2020) and can put small populations at risk of extinction (Lande, 1994; Lynch et al., 1995; Dussex et al., 2021; Khan et al., 2021). Given enough time, even a very small population may adapt to and survive through high levels of homozygosity (Qiao et al., 2010; Dussex et al., 2021; Khan et al., 2021); nevertheless, this does not apply to species that were driven to the verge of extinction by recent human activity.
Based on the relatively lower θπ(0-fold)/θπ(4-fold) ratio of T. sutchuenensis compared to other species in Thuja (Table S11), it is therefore likely to have a lower genetic load, which indicates a more robust efficiency in purifying selection, and a lower probability of extinction (Chen et al., 2017, 2019; Stewart et al., 2017). Likewise, a relatively lower ratio of RXY compared to the other congeneric species, indicates that T. sutchuenensis has been more successful in purging deleterious mutations. The efficiency of purifying selection depends on the population size and life-history traits (Agrawal and Whitlock, 2012; Chen et al., 2017, 2022), but also how long a period it has been acting. Long-lived plants with small population size can persist despite a high genetic load (Klekowski, 1988). Our analyses suggest that the long-lived conifer species, T. sutchuenensis, has suffered severe population declines during its evolutionary history, and its population size became relatively small since at least 5 Kya. Therefore, like Tupistra pingbianensis (Qiao et al., 2010), it has been naturally rare for a long time, allowing ample time for both genetic drift and purging selection to act. In support of this, we found that the majority of detected deleterious alleles within T. sutchuenensis population are heterozygous (Table S13), consistent with purifying selection acting more strongly when the deleterious recessives are expressed (Zhu et al., 2022). These results suggest that some endangered trees with small Ne tend to purge deleterious mutations whether their impacts are high, mild, or low.
Populations with small Ne are expected to deviate from their optimal fitness, due to a stronger effect of stochastic events, as these populations are less able to generate new polymorphisms that are neutral or nearly neutral. However, the likelihood of beneficial or deleterious polymorphisms increases in these populations (Galtier, 2016). DFE scores indicate that T. sutchuenensis had the highest proportion of both beneficial and deleterious polymorphisms among the five Thuja species (Fig. 4a), but the lowest proportion of neutral to nearly neutral polymorphisms. This indicates that T. sutchuenensis has suboptimal fitness, elevating its extinction risk (Robinson et al., 2019).
4.4. Conservation implicationsIt is critical to identify conservation units (CUs) for biodiversity protection, which makes managers understand the population units of endangered species, such as (a) evolutionarily significant units (ESUs), which represent different lineages that have experienced independent evolutionary history without gene flow; (b) management units (MUs), that means demographically distinct populations; and (c) adaptive units (AUs), which indicate differentiation in adaptability to landscape and climate (Funk et al., 2012; Yang et al., 2022). Our analyses have indicated that T. sutchuenensis comprises three distinct groups. We recommend that these groups be treated as independent MUs. The effective population size is lowest in G2, and highest in G1 (Fig. 2a), whereas the proportion of highly deleterious mutations is highest in G3, followed by G2 and G1 (Fig. 3b; Table S14). Nucleotide diversity was higher in G1 and G2 but lower in G3 (Table S5). To maximize the efficacy of conservation measures, we recommend prioritizing efforts in the following order: G3 > G2 > G1.
Although T. sutchuenensis appears to have persisted successfully in small populations for more than 10,000 years, two factors are critical in determining whether it will continue to do so. The first is whether the degree of recent anthropogenic shrinkage, on top of past natural population decline, has been enough to trigger a new round of genetic drift, genetic diversity loss, further reducing fitness, and potentially sending it towards an extinction vortex (Lynch et al., 1993; Agrawal and Whitlock, 2012; Teixeira and Huber, 2021). The second is how it will respond to climate change, given that its reduced genetic diversity would likely reduce its ability to adapt to new environments. Therefore, conservation strategies and measures, including habitat conservation, collecting its seeds as germplasm (Guo et al., 2019; Zhang et al., 2021), enhancing connectivity of fragmented habitats, artificial assistance of migration between populations, increasing of local population size by artificial plantation etc., should be seriously considered to minimize the extinction risk of T. sutchuenensis. Currently T. sutchuenensis is listed as 'Endangered' in the IUCN Red List (Yang et al., 2013) and "Class I" on the National Protection List of Wild Plants in China (Fu and Chin, 1992). Conservation efforts, including evaluating its conservation status, implementing in situ preservation through the establishment of nature reserves (e.g., Xuebaoshan National Nature Reserve), and propagating seedlings from artificial cuttings or seeds, have already been undertaken. Future research employing whole genome sequencing and resequencing to investigate adaptive variation and promote selective breeding should be encouraged to support the sustainable conservation of this endangered conifer (Borthakur et al., 2022; Zhao et al., 2023). In short, our study provides new insight as well as recommendations to the conservation of rare tree species, which, in combination with existing conservation approaches, will facilitate the survival of T. sutchuenensis.
AcknowledgmentsThis study was financially supported by National Natural Science Foundation of China (grant No. U20A2080, 31622015), the Institutional Research Fund from Sichuan University (2021SCUNL102) and Fundamental Research Fund for the Central Universities of China (SCU 2021D006, SCU 2022D003).
Author contributions
K.S.M. designed and supervised this study. T.Z.T. and J.L.L. managed the fieldwork and collected the materials. T.Z.T., J.L.L., H.Y., S.H.C. and S.Y.W. performed the data analysis. T.Z.T, R.I.M, J.L.L and K.S.M. prepared the manuscript. All authors read and approved the manuscript.
Data availability
The data used for the analyses in this manuscript are available from the National Genomics Data Center: https://ngdc.cncb.ac.cn/gsa/s/h3Pn1TTa and https://ngdc.cncb.ac.cn/gsa/s/HZ98AJ87.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.06.005.
Agrawal, A.F., Whitlock, M.C., 2012. Mutation load: the fitness of individuals in populations where deleterious alleles are abundant. Annu. Rev. Ecol. Evol. Syst., 43: 115-135. DOI:10.1146/annurev-ecolsys-110411-160257 |
Akaike, H., 1998. Information theory and an extension of the maximum likelihood principle. In: Parzen, E., Tanabe, K., Kitagawa, G. (Eds.), Selected Papers of Hirotugu Akaike. Springer, New York, pp. 199-213.
|
Alexander, D.H., Lange, K., 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinf., 12: 1-6. DOI:10.1186/1471-2105-12-1 |
Allendorf, F.W., Hohenlohe, P.A., Luikart, G., 2010. Genomics and the future of conservation genetics. Nat. Rev. Genet., 11: 697-709. DOI:10.1038/nrg2844 |
Altschul, S.F., Madden, T.L., Schäffer, A.A., et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25: 3389-3402. DOI:10.1093/nar/25.17.3389 |
Angeloni, F., Wagemaker, C., Jetten, M., et al., 2011. De novo transcriptome characterization and development of genomic tools for Scabiosa columbaria L. using next-generation sequencing techniques. Mol. Ecol. Resour., 11: 662-674. DOI:10.1111/j.1755-0998.2011.02990.x |
Bertorelle, G., Raffini, F., Bosse, M., et al., 2022. Genetic load: genomic estimates and applications in non-model animals. Nat. Rev. Genet., 23: 492-503. DOI:10.1038/s41576-022-00448-x |
Bolger, A.M., Lohse, M., Usadel, B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30: 2114-2120. DOI:10.1093/bioinformatics/btu170 |
Borthakur, D., Busov, V., Cao, X.H., et al., 2022. Current status and trends in forest genomics. For. Res., 2: 11. |
Bortoluzzi, C., Bosse, M., Derks, M.F., et al., 2020. The type of bottleneck matters: insights into the deleterious variation landscape of small managed populations. Evol. Appl., 13: 330-341. DOI:10.1111/eva.12872 |
Ceballos, G., Ehrlich, P.R., Barnosky, A.D., et al., 2015. Accelerated modern human–induced species losses: entering the sixth mass extinction. Sci. Adv., 1: e1400253. DOI:10.1126/sciadv.1400253 |
Charlesworth, B., 2009. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet., 10: 195-205. DOI:10.1038/nrg2526 |
Chau, M.M., Reyes, W.R., Ranker, T.A., 2013. Ecological factors influencing growth of the endangered Hawaiian fern Marsilea villosa (Marsileaceae) and implications for conservation management. Am. J. Bot., 100: 1532-1543. DOI:10.3732/ajb.1200625 |
Chen, J., Bataillon, T., Glémin, S., et al., 2022. What does the distribution of fitness effects of new mutations reflect? Insights from plants. New Phytol., 233: 1613-1619. DOI:10.1111/nph.17826 |
Chen, J., Glémin, S., Lascoux, M., 2017. Genetic diversity and the efficacy of purifying selection across plant and animal species. Mol. Biol. Evol., 34: 1417-1428. DOI:10.1093/molbev/msx088 |
Chen, J., Li, L., Milesi, P., et al., 2019. Genomic data provide new insights on the demographic history and the extent of recent material transfers in Norway spruce. Evol. Appl., 12: 1539-1551. DOI:10.1111/eva.12801 |
Chen, T., Chen, X., Zhang, S., et al., 2021. The genome sequence archive family: toward explosive data growth and diverse data types. Dev. Reprod. Biol., 19: 578-583. DOI:10.1016/j.gpb.2021.08.001 |
Chen, Z., Ai, F., Zhang, J., et al., 2020. Survival in the Tropics despite isolation, inbreeding and asexual reproduction: insights from the genome of the world's southernmost poplar (Populus ilicifolia). Plant J., 103: 430-442. DOI:10.1111/tpj.14744 |
Cingolani, P., Platts, A., Wang, L.L., et al., 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6: 80-92. DOI:10.4161/fly.19695 |
Clark, P.U., Dyke, A.S., Shakun, J.D., et al., 2009. The last glacial maximum. Science, 325: 710-714. DOI:10.1126/science.1172873 |
Clegg, M.T., Kahler, A.L., Weir, B.S., 1989. Plant Population Genetics, Breeding, and Genetic Resources. Sinauer Associates, MA.
|
Cui, Y.-M., Sun, B., Wang, H.-F., et al., 2015. Exploring the formation of a disjunctive pattern between Eastern Asia and North America based on fossil evidence from Thuja (Cupressaceae). PLoS One, 10: e0138544. DOI:10.1371/journal.pone.0138544 |
Cui, Z., Chen, Y., Zhang, W., et al., 2011. Research history, glacial chronology and origins of Quaternary glaciations in China. Quat. Sci., 31: 749-764. |
Danecek, P., Auton, A., Abecasis, G., et al., 2011. The variant call format and VCFtools. Bioinformatics, 27: 2156-2158. DOI:10.1093/bioinformatics/btr330 |
De Kort, H., Prunier, J.G., Ducatez, S., et al., 2021. Life history, climate and biogeography interactively affect worldwide genetic diversity of plant and animal populations. Nat. Commun., 12: 1-11. DOI:10.1038/s41467-020-20314-w |
DePristo, M.A., Banks, E., Poplin, R., et al., 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43: 491-498. DOI:10.1038/ng.806 |
Díez-del-Molino, D., Sánchez-Barreiro, F., Barnes, I., et al., 2018. Quantifying temporal genomic erosion in endangered species. Trends Ecol. Evol., 33: 176-185. DOI:10.1016/j.tree.2017.12.002 |
Do, C., Waples, R.S., Peel, D., et al., 2014. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour., 14: 209-214. DOI:10.1111/1755-0998.12157 |
Do, R., Balick, D., Li, H., et al., 2015. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat. Genet., 47: 126-131. DOI:10.1038/ng.3186 |
Dobin, A., Davis, C.A., Schlesinger, F., et al., 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29: 15-21. DOI:10.1093/bioinformatics/bts635 |
Dussex, N., Van Der Valk, T., Morales, H.E., et al., 2021. Population genomics of the critically endangered kākāpō. Cell Genomics, 1: 100002. DOI:10.1016/j.xgen.2021.100002 |
Dutheil, J.Y., 2020. Statistical Population Genomics. Springer Nature, New York.
|
Emms, D.M., Kelly, S., 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol., 16: 1-14. DOI:10.1186/s13059-014-0572-2 |
Excoffier, L., Dupanloup, I., Huerta-Sánchez, E., et al., 2013. Robust demographic inference from genomic and SNP data. PLoS Genet., 9: e1003905. DOI:10.1371/journal.pgen.1003905 |
Eyre-Walker, A., Keightley, P.D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet., 8: 610-618. DOI:10.1038/nrg2146 |
Frankham, R., Ballou, S.E.J.D., Briscoe, D.A., et al., 2002. Introduction to Conservation Genetics. Cambridge: Cambridge University Press.
|
Fu, L., Chin, C.M., 1992. China Plant Red Data Book. Beijing: Science Press.
|
Fu, L., Niu, B., Zhu, Z., et al., 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics, 28: 3150-3152. DOI:10.1093/bioinformatics/bts565 |
Funk, W.C., McKay, J.K., Hohenlohe, P.A., et al., 2012. Harnessing genomics for delineating conservation units. Trends Ecol. Evol., 27: 489-496. DOI:10.1016/j.tree.2012.05.012 |
Galtier, N., 2016. Adaptive protein evolution in animals and the effective population size hypothesis. PLoS Genet., 12: e1005774. DOI:10.1371/journal.pgen.1005774 |
Godwin, J.L., Lumley, A.J., Michalczyk, L., et al., 2020. Mating patterns influence vulnerability to the extinction vortex. Global Change Biol., 26: 4226-4239. DOI:10.1111/gcb.15186 |
Grabherr, M.G., Haas, B.J., Yassour, M., et al., 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol., 29: 644-652. DOI:10.1038/nbt.1883 |
Grantham, R., 1974. Amino acid difference formula to help explain protein evolution. Science, 185: 862-864. DOI:10.1126/science.185.4154.862 |
Grossen, C., Guillaume, F., Keller, L.F., et al., 2020. Purging of highly deleterious mutations through severe bottlenecks in Alpine ibex. Nat. Commun., 11: 1-12. DOI:10.1038/s41467-019-13993-7 |
Guo, J.L., Cao, W.J., Li, Z.M., et al., 2019. Conservation implications of population genetic structure in a threatened orchid Cypripedium tibeticum. Plant Divers, 41: 13-18. DOI:10.3390/ht8020013 |
Hedrick, P.W., Garcia-Dorado, A., 2016. Understanding inbreeding depression, purging, and genetic rescue. Trends Ecol. Evol., 31: 940-952. DOI:10.1016/j.tree.2016.09.005 |
Jordan, R., Breed, M.F., Prober, S.M., et al., 2019. How well do revegetation plantings capture genetic diversity?. Biol. Lett., 15: 20190460. DOI:10.1098/rsbl.2019.0460 |
Keightley, P.D., Jackson, B.C., 2018. Inferring the probability of the derived vs. the ancestral allelic state at a polymorphic site. Genetics, 209: 897-906. DOI:10.1534/genetics.118.301120 |
Keller, L.F., Waller, D.M., 2002. Inbreeding effects in wild populations. Trends Ecol. Evol., 17: 230-241. DOI:10.1016/S0169-5347(02)02489-8 |
Khan, A., Patel, K., Shukla, H., et al., 2021. Genomic evidence for inbreeding depression and purging of deleterious genetic variation in Indian tigers. Proc. Natl. Acad. Sci. U.S.A., 118: e2023018118. DOI:10.1073/pnas.2023018118 |
Kim, C., Kim, D.K., Sun, H., et al., 2022. Phylogenetic relationship, biogeography, and conservation genetics of endangered Fraxinus chiisanensis (Oleaceae), endemic to South Korea. Plant Divers, 44: 170-180. DOI:10.1016/j.pld.2021.06.004 |
Kleinman-Ruiz, D., Lucena-Perez, M., Villanueva, B., et al., 2022. Purging of deleterious burden in the endangered Iberian lynx. Proc. Natl. Acad. Sci. U.S.A., 119: e2110614119. DOI:10.1073/pnas.2110614119 |
Klekowski, E.J., 1988. Genetic load and its causes in long-lived plants. Trees (Berl.), 2: 195-203. DOI:10.1007/bf00202374 |
Kohn, M.H., Murphy, W.J., Ostrander, E.A., et al., 2006. Genomics and conservation genetics. Trends Ecol. Evol., 21: 629-637. DOI:10.1016/j.tree.2006.08.001 |
Kondrashov, A.S., Crow, J.F., 1993. A molecular approach to estimating the human deleterious mutation rate. Hum. Mutat., 2: 229-234. DOI:10.1002/humu.1380020312 |
Korunes, K.L., Samuk, K., 2021. pixy: unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol. Ecol. Resour., 21: 1359-1368. DOI:10.1111/1755-0998.13326 |
Kumar, P., Henikoff, S., Ng, P.C., 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4: 1073-1081. DOI:10.1038/nprot.2009.86 |
Lamoreux, J.F., Morrison, J.C., Ricketts, T.H., et al., 2006. Global tests of biodiversity concordance and the importance of endemism. Nature, 440: 212-214. DOI:10.1038/nature04291 |
Lande, R., 1993. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am. Nat., 142: 911-927. DOI:10.1086/285580 |
Lande, R., 1994. Risk of population extinction from fixation of new deleterious mutations. Evolution, 48: 1460-1469. DOI:10.2307/2410240 |
Lange, R., Durka, W., Holzhauer, S.I., et al., 2010. Differential threshold effects of habitat fragmentation on gene flow in two widespread species of bush crickets. Mol. Ecol., 19: 4936-4948. DOI:10.1111/j.1365-294X.2010.04877.x |
Li, H., Durbin, R., 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25: 1754-1760. DOI:10.1093/bioinformatics/btp324 |
Li, H., Handsaker, B., Wysoker, A., et al., 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25: 2078-2079. DOI:10.1093/bioinformatics/btp352 |
Li, J., Milne, R.I., Ru, D., et al., 2020. Allopatric divergence and hybridization within Cupressus chengiana (Cupressaceae), a threatened conifer in the northern Hengduan Mountains of western China. Mol. Ecol., 29: 1250-1266. DOI:10.1111/mec.15407 |
Li, J., Zhang, Y., Rusham, M., et al., 2021. Seeing through the hedge: phylogenomics of Thuja (Cupressaceae) reveals prominent incomplete lineage sorting and ancient introgression for Tertiary relict flora. Cladistics, 38: 187-203. |
Li, W.H., Wu, C.I., Luo, C.C., 1984. Nonrandomness of point mutation as reflected in nucleotide substitutions in pseudogenes and its evolutionary implications. J. Mol. Evol., 21: 58-71. DOI:10.1007/BF02100628 |
Li, Y.Y., Guan, S.M., Yang, S.Z., et al., 2012. Genetic decline and inbreeding depression in an extremely rare tree. Conserv. Genet., 13: 343-347. DOI:10.1007/s10592-011-0286-x |
Liu, J., Shi, S., Chang, E., et al., 2013. Genetic diversity of the critically endangered Thuja sutchuenensis revealed by ISSR markers and the implications for conservation. Int. J. Mol. Sci., 14: 14860-14871. DOI:10.3390/ijms140714860 |
Liu, X., Fu, Y.X., 2020. Stairway Plot 2: demographic history inference with folded SNP frequency spectra. Genome Biol., 21: 1-9. DOI:10.1186/s13059-019-1906-x |
Lynch, M., Bürger, R., Butcher, D., et al., 1993. The mutational meltdown in asexual populations. J. Hered., 84: 339-344. DOI:10.1093/oxfordjournals.jhered.a111354 |
Lynch, M., Conery, J., Burger, R., 1995. Mutation accumulation and the extinction of small populations. Am. Nat., 146: 489-518. DOI:10.1086/285812 |
Ma, H., Liu, Y., Liu, D., et al., 2021. Chromosome-level genome assembly and population genetic analysis of a critically endangered rhododendron provide insights into its conservation. Plant J., 107: 1533-1545. DOI:10.1111/tpj.15399 |
Ma, Y., Liu, D., Wariss, H.M., et al., 2022. Demographic history and identification of threats revealed by population genomic analysis provide insights into conservation for an endangered maple. Mol. Ecol., 31: 767-779. DOI:10.1111/mec.16289 |
Manichaikul, A., Mychaleckyj, J.C., Rich, S.S., et al., 2010. Robust relationship inference in genome-wide association studies. Bioinformatics, 26: 2867-2873. DOI:10.1093/bioinformatics/btq559 |
Manners, V., Kumaria, S., Tandon, P., 2013. SPAR methods revealed high genetic diversity within populations and high gene flow of Vanda coerulea Griff ex Lindl (Blue Vanda), an endangered orchid species. Gene, 519: 91-97. DOI:10.1016/j.gene.2013.01.037 |
Manni, M., Berkeley, M.R., Seppey, M., et al., 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol., 38: 4647-4654. DOI:10.1093/molbev/msab199 |
Mantyka-pringle, C.S., Martin, T.G., Rhodes, J.R., 2012. Interactions between climate and habitat loss effects on biodiversity: a systematic review and meta-analysis. Global Change Biol., 18: 1239-1252. DOI:10.1111/j.1365-2486.2011.02593.x |
Miao, J., Farhat, P., Wang, W., et al., 2021. Evolutionary history of two rare endemic conifer species from the eastern Qinghai-Tibet Plateau. Ann. Bot., 128: 903-918. DOI:10.1093/aob/mcab114 |
Narasimhan, V., Danecek, P., Scally, A., et al., 2016a. BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics, 32: 1749-1751. DOI:10.1093/bioinformatics/btw044 |
Narasimhan, V.M., Hunt, K.A., Mason, D., et al., 2016b. Health and population effects of rare gene knockouts in adult humans with related parents. Science, 352: 474-477. DOI:10.1126/science.aac8624 |
Nei, M., Maruyama, T., Chakraborty, R., 1975. The bottleneck effect and genetic variability in populations. Evolution, 29: 1-10. DOI:10.2307/2407137 |
Ng, P.C., Henikoff, S., 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res., 31: 3812-3814. DOI:10.1093/nar/gkg509 |
Nguyen, L.T., Schmidt, H.A., Von Haeseler, A., et al., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol., 32: 268-274. DOI:10.1093/molbev/msu300 |
Ohta, T., 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Evol. Syst., 23: 263-286. DOI:10.1146/annurev.es.23.110192.001403 |
Palkopoulou, E., Mallick, S., Skoglund, P., et al., 2015. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr. Biol., 25: 1395-1400. DOI:10.1016/j.cub.2015.04.007 |
Palsbøll, P.J., Berube, M., Allendorf, F.W., 2007. Identification of management units using population genetic data. Trends Ecol. Evol., 22: 11-16. DOI:10.1016/j.tree.2006.09.003 |
Patil, I., 2021. Visualizations with statistical details: the 'ggstatsplot' approach. J. Open Source Softw., 6: 3167. DOI:10.21105/joss.03167 |
Price, A.L., Patterson, N.J., Plenge, R.M., et al., 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38: 904-909. DOI:10.1038/ng1847 |
Primmer, C.R., 2009. From conservation genetics to conservation genomics. Ann. N. Y. Acad. Sci., 1162: 357-368. DOI:10.1111/j.1749-6632.2009.04444.x |
Purcell, S., Neale, B., Todd-Brown, K., et al., 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81: 559-575. DOI:10.1086/519795 |
Qiao, Q., Zhang, C.Q., Milne, R.I., 2010. Population genetics and breeding system of Tupistra pingbianensis (Liliaceae), a naturally rare plant endemic to SW China. J. Syst. Evol., 48: 47-57. DOI:10.1111/j.1759-6831.2009.00064.x |
Qin, A., Ding, Y., Jian, Z., et al., 2021. Low genetic diversity and population differentiation in Thuja sutchuenensis Franch., an extremely endangered rediscovered conifer species in southwestern China. Global Ecol. Conserv., 25: e01430. DOI:10.1016/j.gecco.2020.e01430 |
Qin, A., Liu, B., Guo, Q., et al., 2017. Maxent modeling for predicting impacts of climate change on the potential distribution of Thuja sutchuenensis Franch., an extremely endangered conifer from southwestern China. Global Ecol. Conserv., 10: 139-146. DOI:10.1016/j.gecco.2017.02.004 |
Robinson, J., Kyriazis, C.C., Yuan, S.C., et al., 2022. Deleterious variation in natural populations and implications for conservation genetics. Annu. Rev. Anim. Biosci., 11: 93-114. |
Robinson, J.A., Räikkönen, J., Vucetich, L.M., et al., 2019. Genomic signatures of extensive inbreeding in Isle Royale wolves, a population on the threshold of extinction. Sci. Adv., 5: eaau0757. DOI:10.1126/sciadv.aau0757 |
Ru, D., Sun, Y., Wang, D., et al., 2018. Population genomic analysis reveals that homoploid hybrid speciation can be a lengthy process. Mol. Ecol., 27: 4875-4887. DOI:10.1111/mec.14909 |
Schoen, D.J., David, J.L., Bataillon, T.M., 1998. Deleterious mutation accumulation and the regeneration of genetic resources. Proc. Natl. Acad. Sci. U.S.A., 95: 394-399. DOI:10.1073/pnas.95.1.394 |
Shalev, T.J., El-Dien, O.G., Yuen, M.M., et al., 2022. The western redcedar genome reveals low genetic diversity in a self-compatible conifer. Genome Res., 32: 1952-1964. |
Sim, N.L., Kumar, P., Hu, J., et al., 2012. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res., 40: W452-W457. DOI:10.1093/nar/gks539 |
Stewart, G.S., Morris, M.R., Genis, A.B., et al., 2017. The power of evolutionary rescue is constrained by genetic load. Evol. Appl., 10: 731-741. DOI:10.1111/eva.12489 |
Tallmon, D.A., Koyuk, A., Luikart, G., et al., 2008. Computer programs: onesamp: a program to estimate effective population size using approximate Bayesian computation. Mol. Ecol. Resour., 8: 299-301. DOI:10.1111/j.1471-8286.2007.01997.x |
Tang, C.Q., Yang, Y., Ohsawa, M., et al., 2015. Community structure and survival of tertiary relict Thuja sutchuenensis (Cupressaceae) in the subtropical Daba mountains, Southwestern China. PLoS One, 10: e0125307. DOI:10.1371/journal.pone.0125307 |
Tataru, P., Bataillon, T., 2019. polyDFEv2.0: testing for invariance of the distribution of fitness effects within and across species. Bioinformatics, 35: 2868-2869. DOI:10.1093/bioinformatics/bty1060 |
Tataru, P., Mollion, M., Glémin, S., et al., 2017. Inference of distribution of fitness effects and proportion of adaptive substitutions from polymorphism data. Genetics, 207: 1103-1119. DOI:10.1534/genetics.117.300323 |
Teixeira, J.C., Huber, C.D., 2021. The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. U.S.A., 118: e2015096118. DOI:10.1073/pnas.2015096118 |
Turner, T.F., Osborne, M.J., Moyer, G.R., et al., 2006. Life history and environmental variation interact to determine effective population to census size ratio. Proc. R. Soc. A B, 273: 3065-3073. DOI:10.1098/rspb.2006.3677 |
Vaser, R., Adusumalli, S., Leng, S.N., et al., 2016. SIFT missense predictions for genomes. Nat. Protoc., 11: 1-9. DOI:10.1038/nprot.2015.123 |
Villanueva, R.A.M., Chen, Z.J., 2019. ggplot2: Elegant Graphics for Data Analysis. Taylor & Francis, Beijing.
|
Wei, J., Pei, X., Hu, X., et al., 2022. Applications of transcriptome in conifer species. Plant Cell Tissue Organ Cult., 150: 511-525. DOI:10.1007/s11240-022-02322-4 |
Willi, Y., Kristensen, T.N., Sgrò, C.M., et al., 2022. Conservation genetics as a management tool: the five best-supported paradigms to assist the management of threatened species. Proc. Natl. Acad. Sci. U.S.A., 119: e2105076119. DOI:10.1073/pnas.2105076119 |
Wilson, E.O., 1996. In: Matthew, A.C., Rory, O. 'B. (Eds.), The Diversity of Life, Thinking about the Environment. Routledge, England, pp. 193-195.
|
Xiang, Q., Fajon, A., Zhenyu, L., et al., 2002. Thuja sutchuenensis: a rediscovered species of the Cupressaceae. Bot. J. Linn. Soc., 139: 305-310. DOI:10.1046/j.1095-8339.2002.00055.x |
Xie, H.X., Liang, X.X., Chen, Z.Q., et al., 2022. Ancient demographics determine the effectiveness of genetic purging in endangered lizards. Mol. Biol. Evol., 39: msab359. DOI:10.1093/molbev/msab359 |
Xue, Y., Bao, Y., Zhang, Z., et al., 2022. Database resources of the national genomics data center, China national center for bioinformation in 2023. Nucleic Acids Res., 51: 18-28. DOI:10.1117/12.2610559 |
Xue, Y., Prado-Martinez, J., Sudmant, P.H., et al., 2015. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science, 348: 242-245. DOI:10.1126/science.aaa3952 |
Yang, H., Li, J., Milne, R.I., et al., 2022. Genomic insights into the genotype–environment mismatch and conservation units of a Qinghai-Tibet Plateau endemic cypress under climate change. Evol. Appl., 15: 919. DOI:10.1111/eva.13377 |
Yang, Y., Li, N., Christian, T., et al., 2013. Thuja sutchuenensis. The IUCN Red List of Threatened Species 2013: e. T32378A2816862. https://doi.org/10.2305/IUCN.UK.2013-1.RLTS.T32378A2816862.en. (Accessed 15 May 2023).
|
Yang, Y., Ma, T., Wang, Z., et al., 2018. Genomic effects of population collapse in a critically endangered ironwood tree Ostrya rehderiana. Nat. Commun., 9: 5449. DOI:10.1038/s41467-018-07913-4 |
Yao, Z., Wang, X., Wang, K., et al., 2021. Chloroplast and nuclear genetic diversity explain the limited distribution of endangered and endemic Thuja sutchuenensis in China. Front. Genet., 12: 801229. DOI:10.3389/fgene.2021.801229 |
Zhang, X.J., Liu, X.F., Liu, D.T., et al., 2021. Genetic diversity and structure of Rhododendron meddianum, a plant species with extremely small populations. Plant Divers., 43: 472-479. DOI:10.1016/j.pld.2021.05.005 |
Zhao, Y.J., Yin, G.S., Gong, X., 2023. RAD-sequencing improves the genetic characterization of a threatened tree peony (Paeonia ludlowii) endemic to China: implications for conservation. Plant Divers., 45: 513-522. DOI:10.1016/j.pld.2022.07.002 |
Zheng, B., Xu, Q., Shen, Y., 2002. The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quat. Int., 97: 93-101. |
Zhu, M., Cheng, Y., Wu, S., et al., 2022. Deleterious mutations are characterized by higher genomic heterozygosity than other genic variants in plant genomes. Genomics, 114: 110290. DOI:10.1016/j.ygeno.2022.110290 |



