b. School of Ecology, Sun Yat-Sen University, Shenzhen 510275, China;
c. School of Geography and Remote Sensing, Guangzhou University, Guangzhou 510006, China;
d. College of Life and Environmental Science, Wenzhou University, Wenzhou 325035, China;
e. Centre for Future Landscapes and Department of Environment and Genetics, La Trobe University, Bundoora, VIC 3083, Australia
Patterns of species diversity are uneven around the world due to highly diverse and heterogeneous environmental conditions (Jenkins et al., 2013). Understanding these geographic variations in species diversity at large scales is a core goal of macroecology and biogeography (Fine, 2015). At a global or regional scale, species diversity has clear latitudinal and elevational gradients (Kreft and Jetz, 2007; Quintero and Jetz, 2018). Along these gradients, dispersal limitation and environmental filtering cause species composition among sites (i.e., taxonomic beta diversity) to change greatly with an increase of geographical distance and climatic difference (Qian and Ricklefs, 2007). In recent years, species turnover at the tips, or basal nodes, of a dated mega phylogenetic tree of plant species (i.e., phylogenetic beta diversity) (Zanne et al., 2014) has allowed researchers to ask whether variations in species composition are influenced by historical events (e.g., mountain building periods or past climatic fluctuations) in deep time (Graham and Fine, 2008; Xu et al., 2023). Thus, the calculation of beta diversity not only provides insight into relationships between species diversity and contemporary environmental conditions, it can also reveal the historical and ecological processes underlying these diversity patterns (Graham and Fine, 2008).
Multiple indices have been developed to measure both taxonomic and phylogenetic beta diversity (Si et al., 2017). Studies on beta diversity have commonly adopted the Jaccard and Sørensen dissimilarity index (e.g., Chao et al., 2005). These two indices measure total beta diversity between localities (pairwise differences), which can be further partitioned into two components: turnover and nestedness, allowing researchers to distinguish the relative importance of processes of species replacement and species loss in creating the differences between the two localities (Baselga, 2010). Dispersal limitations and niche conservatism of species generally led to the replacement of some or even all species between sites. Such species turnover independent of gradients of species richness, is commonly associated with significant environmental gradients (Jankowski et al., 2009). In contrast, changes in species nestedness between sites explicitly consider species loss or gain; where the lower number of species in a site is a subset of the species of the richer site, this reveals the contribution of ordered extinction or colonization to patterns of compositional changes (Baselga, 2010). The relative proportion of total beta diversity then reveals the relative contribution of gradients inducing turnover in composition from extinction and colonization processes. For taxonomic beta diversity, the turnover component tends to dominate compositional change, particularly at biogeographical scales (Soininen et al., 2018).
Species dissimilarity usually occurs along contemporary environmental gradients (Jankowski et al., 2009). Two well-known, large-scale climatic gradients are the decreases in mean annual temperature and precipitation from the coast to inland regions that are influenced by annual monsoons and local topography. Accordingly, species composition among sites varies along this climatic gradient, which has been most widely tested in East Asia and North America (Pinto-Ledezma et al., 2018; Qian et al., 2020). Comparable patterns are common along elevational gradients, particularly those created by high mountains (i.e., habitat island systems), such as the Andes, Himalaya and Hengduan Mountains. Due to the large elevational range and topographic heterogeneity involved, high mountain ranges can create dozens of vegetation zones, producing large species compositional changes over a very short distance (Zeng et al., 2022). Moreover, past geological and climatic events, can also affect species composition among localities due to species diversification, extinction or colonization (González-Trujillo et al., 2021). In particular, recent Quaternary climatic fluctuations have resulted in the loss of many species at higher latitudinal or elevational areas and colonization of lower areas, which has altered species nestedness in these regions (Xu et al., 2023; Du et al., 2022).
Because beta diversity changes greatly in areas of vegetation transition, an important objective of calculating beta diversity patterns in a region is to delineate zones of regional biota (Daru et al., 2020). Previous studies that have delineated biogeographical regions have mostly relied on expert experience and knowledge (qualitative) or occasionally indirect quantification based on environmental information (Zheng, 2008; Wu et al., 2010). Recent studies have begun to use species distribution and phylogenetic data to identify floristic regions (Ye et al., 2019). Compared to biogeographical regionalization using taxonomic beta diversity indices, the classification of biota using phylogenetic beta diversity indices can reflect the relative importance of evolutionary processes in shaping current diversity patterns of a flora (Graham and Fine, 2008), especially for regions with a complex evolutionary history, such as the flora of the Qinghai-Tibet Plateau (QTP). The QTP has experienced extensive and multiple uninterrupted mountain uplifts during the Neogene (Deng and Ding, 2015; Spicer et al., 2020), as well as the subsequent effects of Quaternary climatic fluctuations (Zheng et al., 2002). Thus, revealing the floristic subregions of the QTP may aid our understanding of the evolutionary history of the flora associated with these geological and climatic events in this region. In addition, the QTP has clear environmental gradients due to its complex topography and the East Asian and Indian monsoons. These influential geological and environmental conditions make the QTP an ideal model for investigating the beta diversity of plant species.
In this study, we used a phylogenetic approach to reveal the beta diversity and floristic subregions of the QTP. Specifically, we determined whether taxonomic and phylogenetic beta diversity, as well as their turnover and nestedness components, differed for all plants, endemic plants, and alpine plants in the QTP. We also identified how geographic distance and climatic difference influence beta diversity and its components. In addition, we identified dominant beta diversity patterns of the flora of the QTP and the relative importance of various factors, including current climate and topography, as well as historical conditions, in creating these patterns. Finally, we used phylogenetic data to quantitatively delimit the flora of the QTP.
2. Materials and methods 2.1. Study areaThe QTP is the largest and highest plateau in the world, covering an area of about 2.5 million square kilometers (Zhang et al., 2002; Fig. 1). It has a large elevational range (100–8848 m, average elevation > ca. 4000 m) with an elevational gradient extending from the southeast to the northwest. Mean annual temperature and precipitation decrease along this same elevation gradient (Fig. 1). The QTP region harbors two recognized biodiversity hotspots, the mountains of southwest China (mainly corresponding to the Hengduan Mountains) and the eastern Himalayas (Mittermeier et al., 2011), both of which are considered potential evolutionary cradles of many temperate taxa in Eurasia (Lu et al., 2018; Ding et al., 2020).
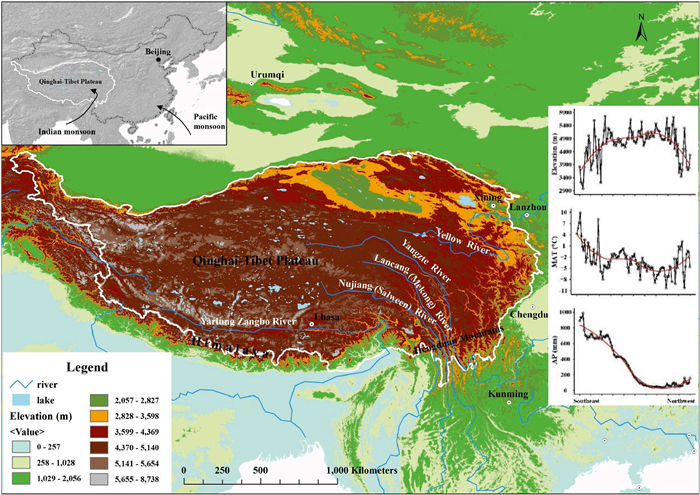
|
| Fig. 1 Map of the Qinghai-Tibet Plateau. The QTP is affected by topography, as well as the Indian and Pacific monsoons. Elevation, mean annual temperature (MAT) and annual precipitation (AP) increase or decrease from the south-east to north-west. |
We used a dataset compiled from our previous studies (Yu et al., 2020, 2021), which comprises a checklist of native seed plant species of the QTP and their geographical distribution at a county-level. The data set, which was derived from a series of monographs (e.g., Flora of China; The Vascular Plants and Their Eco-geographical Distribution of the Qinghai-Tibet Plateau), online databases (e.g., CVH, Chinese Virtual Herbarium; GBIF, Global Biodiversity Information Facility), primary literature (Zhang et al., 2016) and field vegetation surveys gathered between 2010 and 2020, also records the elevational range of each species and whether the species is endemic to the QTP or alpine plant (above the tree line). For consistency in species nomenclature, the combined plant species list was based on Flora of China with unresolved or synonymous species names being excluded. For angiosperms, we followed the APG IV classification system (The Angiosperm Phylogeny Group et al., 2016), and for gymnosperms, we followed an updated classification proposed by Yang et al. (2022).
We divided the QTP into 1126 grid cells at a resolution of 50 km × 50 km and obtained the elevational range of each grid using digital elevation maps (DEMs) in ArcGIS 10.3 (ESRI). County-level distributions for each species were converted to grid-level distributions. Species distributions were filtered by elevation; if the elevational range of the species fell entirely or partially within the elevational range of the grid, we recorded the species as present in this grid cell. Ultimately, our database was created using R package 'spaa' (https://github.com/helixcn/spaa) and comprised three species presence/absence matrices for all plant species, endemic plant species and alpine plant species from the grid data.
2.3. Phylogenetic treeThe QTP includes 12,091 seed plant species, 5457 endemic species and 3320 alpine species. The names of these species were standardized according to The Plant List (TPL; http://www.theplantlist.org). To generate phylogenetic trees for the three groups of species, we used R packages V.PhyloMaker2 and U.PhyloMaker (scenarios S3; Jin and Qian, 2019, 2022, 2023). The two packages use the dated mega-phylogeny GBOTB (Smith and Brown, 2018) as a backbone, and further updates that extend this (designated GBOTB.extended.TPL.tre, see Jin and Qian, 2022 for details). At the species level, most seed plant species in our dataset were included in this megatree. For any unmatched species (277 species), we used the most closely related species according to the SoTree database (http://www.darwintree.cn/; Chen et al., 2016) included in the megatree. Finally, we obtained phylogenetic trees corresponding to all seed plants, endemic plants and alpine plants of the QTP (Appendix A).
2.4. Beta diversityBecause taxonomic and phylogenetic beta diversity usually display some differences in pattern, we calculated beta diversity indices for both, using the Sørensen dissimilarity index (βsor). Following Baselga (2010), we partitioned βsor into two components: the Simpson dissimilarity index (βsim), which reflects species turnover between sites, and the species nestedness index (βnes), which reflects species replacement and species loss or gain. Pairwise total beta diversity and its turnover and nestedness components were calculated as follows (Baselga, 2010).
1) βsor = (b + c)/(2a + b + c)
2) βsim = min (b, c)/[a + min (b, c)]
3) βnes = βsor - βsim
Where, for taxonomic beta diversity, a is the number of species shared by the two sites and b, c are the number of species unique to the respective sites. For phylogenetic beta diversity, a and b, c are instead the shared and unique branch lengths among sites. To differentiate taxonomic from phylogenetic beta diversity, we added 'phy' to the subscript for each, using βsor.phy, βsim.phy and βnes.phy, to indicate total, turnover and nestedness components of phylogenetic beta diversity respectively.
To calculate the pairwise taxonomic and phylogenetic beta diversity based on the species presence/absence matrix and phylogenies for our three different plant groupings, we used the functions 'beta.pair' and 'phylo.beta.pair' in R package 'betapart' (Baselga and Orme, 2012). Then, to build the patterns of beta diversity, we used a neighborhood approach (Qian et al., 2020) to calculate beta diversity between each focal cell and its 24 first- and second-order neighboring cells (i.e., the 8 adjacent cells and the 16 adjacent to those; Fig. S1).
2.5. Environmental dataBeta diversity patterns are closely associated with contemporary climatic and topographic conditions, as well as past climate changes (Graham and Fine, 2008). Here, we chose six predictors to represent current climatic and topographic, and historical variables. For contemporary climatic conditions, we used mean annual temperature (MAT), annual precipitation (AP) (Qian et al., 2007) and a composite climatic gradient (CG) (Brown et al., 2014). The MAT, AP and other 17 bioclimatic variables were downloaded from the WorldClim v2.1 database (http://worldclim.org/; corresponding to bio1 and bio12, respectively; Fick and Hijmans, 2017). These variables were clipped to the extent of the QTP region in ArcGIS 10.3 (ESRI) (Fig. S2a and b) and then standardized to a range 0–1 based on the min–max normalization. To create a composite predictor of the CG, we calculated a principal component analysis (PCA) for all 19 bioclimatic variables and used the first principal component (PCs) to represent the CG (Fig. S2c).
To quantify topographic variation, we used DEMs obtained from the Consortium for Spatial Information (http://www.cgiar-csi.org/) with a resolution of 90 m × 90 m. From this, elevation range (ER; Fig. S2d) was extracted from the maximum and minimum elevation for each grid cell using ArcGIS 10.3. Topographic heterogeneity (TH; Fig. S2e) was quantified as the standard deviation of elevation within a target cell and the eight neighboring cells using the SDMToolbox v.2.4 (Brown et al., 2017).
More recent, climatic modifications, such as Quaternary climatic oscillations, have affected plant diversity patterns in the QTP (Yu et al., 2019). Climatic instability (CI) usually results in species extinction or dispersal, with a clear influence on the beta diversity in a region. We quantified CI using four time periods of bioclimatic data (Last Interglacial, Last Glacial Maximum, Mid-Holocene, and current conditions; 2.5 min resolution). We first clipped these bioclimatic variables based on the QTP's extent using ArcGIS 10.3 and excluded any with high collinearity (Pearson's r > 0.95), retaining 10 variables (bio1, bio2, bio3, bio4, bio6, bio10, bio12, bio14, bio15, bio17) using the SDMToolbox v.2.4 (Brown et al., 2017). We then calculated the standard deviation of each bioclimatic variable throughout the four time periods, summing these to create a spatial layer representing climatic instability (Fig. S2f).
We also extracted the centroid coordinates for the 1126 grid cells and calculated a matrix of geographical distance between all grid cells in ArcGIS 10.3. Based on the standard 19 bioclimatic variables, we used the Euclidean distance to measure a climatic distance matrix between all sites.
2.6. Biogeographical regionalizationBased on the matrices of phylogenetic beta diversity (Sørensen and Simpson dissimilarity indices) for all plant species, endemic plant species and alpine plant species, we determined an optimal number of clusters using hierarchical dendrograms of dissimilarity and non-metric multidimensional scaling (NMDS) ordination in R package 'phyloregion' (Daru et al., 2020). We then used the function 'phyloregion' to regionalize the flora of the QTP, visualizing the floristic subregion boundaries in ArcGIS 10.3.
2.7. Statistical analysesGeographical distance and climatic difference between sites can affect large-scale beta diversity (Qian and Ricklefs, 2007). Here, we calculated Pearson's correlation coefficients between (phylo-) beta diversity matrices and geographical distance and climatic distance and partitioned the effects of geographical distance and climatic distance to (phylo-) beta diversity using the 'varpart' function in the R package 'vegan' (Oksanen et al., 2013).
To explore the associations between taxonomic and phylogenetic beta diversity and each explanatory variable, we first used ordinary least squares (OLS) regression. To control any influence of spatial autocorrelation in these associations, we further applied spatial auto regression (SAR). We performed a hierarchical partitioning analysis to account for the relative contribution of each variable to the beta diversity patterns using R package 'rdacca.hp' (Lai et al., 2022). This method can effectively resolve the collinearity problems between variables and quickly obtain results even with large data. Prior to analysis, all explanatory variables were standardized to 0–1 using the min–max normalization. All statistical analyses were conducted in R 4.1.3 (R Core Team, 2019) and SAM v.4.0 (Rangel et al., 2010).
3. Results 3.1. Effects of geographical distance and climatic difference on multifaceted beta diversityTaxonomic and phylogenetic beta diversity for each of the plant groups in the QTP were significantly correlated with both geographical distance and climatic difference (Tables 1 and S1; P < 0.001 for all cases). The strongest correlations were between total beta diversity for all groups (0.3 < Pearson's r < 0.42, Table 1). However, associations with turnover and nestedness differed among groups. In alpine plants, turnover was strongly associated with geography and climate, whereas in endemics nestedness was strongly associated with these factors (Table 1). Partial regression analyses further suggested that the greater correlation of total phylogenetic beta diversity with geography and climate was due to their shared, rather than independent effects (Fig. 2). However, partitioning explained less than 6% of total taxonomic and phylogenetic beta diversity and the components of turnover and nestedness for any plant group (Figs. 2 and S3).
| Beta diversity | Geographic distance | Climatic distance |
| sor.phy_all | 0.30 | 0.42 |
| sor.phy_end | 0.36 | 0.40 |
| sor.phy_alp | 0.30 | 0.32 |
| sim.phy_all | 0.25 | 0.16 |
| sim.phy_end | 0.12 | 0.14 |
| sim.phy_alp | 0.27 | 0.18 |
| sne.phy_all | 0.05 | 0.21 |
| sne.phy_end | 0.18 | 0.18 |
| sne.phy_alp | 0.03 | 0.07 |
| sor.phy: phylogenetic Sørensen dissimilarity index; sim.phy: phylogenetic Simpson dissimilarity index; sne.phy: phylogenetic nestedness index; end: endemic; alp: alpine. | ||
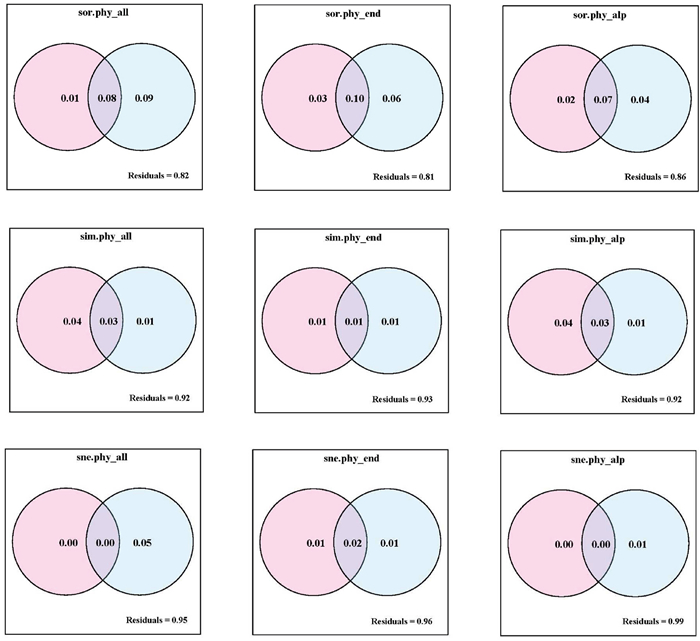
|
| Fig. 2 The individual and shared contributions of geographical distance (pink) and climatic difference (blue) to aspects of phylogenetic beta diversity. For abbreviations, see Table 1. |
Geographic patterns of total beta diversity and the components of turnover and nestedness for distinct plant groups showed similar patterns in the QTP, although the values were higher for turnover than for nestedness (Figs. 3 and S4). In general, beta diversity was high in the southern, southeastern and northeastern parts of the QTP and low within the inner plateau.
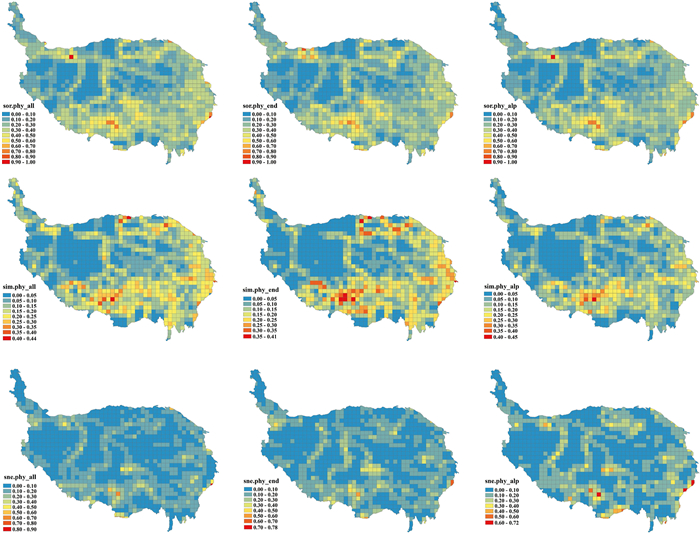
|
| Fig. 3 Patterns of phylogenetic beta diversity and its turnover and nestedness components for all species, endemic and alpine plants in the Qinghai-Tibet Plateau. Hotter colours indicate higher beta diversity. |
The single-variable ordinary least squares and corresponding spatial autoregressive models showed similar results, although with some interesting exceptions (Tables 2 and S2). Based on relative magnitude of coefficients, both models showed that the most influential predictor of total and nestedness beta diversity across all plant groups was AP (Table 2). However, beta diversity turnover for all plant groups was most associated with CG, with AP having limited influence, especially in the autoregressive model. Differences among plant groups were also more apparent in the SAR model coefficients, where MAT was more influential on alpine that other groups, particularly for turnover (Table 2). Turnover and total beta diversity for all plant groups was strongly and negatively affected by CI, which showed no effect on nestedness. Taken together, these models suggest that beta diversity patterns of plant species in the QTP have been affected by contemporary climate and topography, as well as historical climate changes.
| MAT | AP | CG | ER | TH | CI | ||
| sor.phy_all | Coef.OLS | 0.14*** | 0.26*** | 0.17*** | 0.13*** | 0.14*** | −0.12*** |
| ROLS2 | 0.02 | 0.06 | 0.05 | 0.02 | 0.04 | 0.03 | |
| Coef.SAR | −0.01 | 0.14 | 0.10 | 0.04 | 0.08 | −0.09 | |
| AICSAR | −1209.71 | −1261.70 | −1255.83 | −1223.23 | −1242.36 | −1241.70 | |
| sor.phy_end | Coef.OLS | 0.20*** | 0.28*** | 0.13*** | 0.10*** | 0.11*** | −0.13*** |
| ROLS2 | 0.04 | 0.07 | 0.03 | 0.01 | 0.02 | 0.04 | |
| Coef.SAR | 0.09 | 0.18 | 0.07 | 0.02 | 0.06 | −0.11 | |
| AICSAR | −1264.90 | −1303.26 | −1259.63 | −1240.52 | −1254.20 | −1275.34 | |
| sor.phy_alp | Coef.OLS | 0.21*** | 0.30*** | 0.11*** | 0.04 | 0.08*** | −0.10*** |
| ROLS2 | 0.04 | 0.08 | 0.02 | 0.00 | 0.01 | 0.02 | |
| Coef.SAR | 0.12 | 0.22 | 0.05 | −0.04 | 0.03 | −0.07 | |
| AICSAR | −1264.84 | −1313.21 | −1241.43 | −1225.71 | −1232.69 | −1248.42 | |
| sim.phy_all | Coef.OLS | 0.10*** | 0.17*** | 0.23*** | 0.03 | 0.09*** | −0.12*** |
| ROLS2 | 0.01 | 0.03 | 0.05 | – | 0.04 | 0.08 | |
| Coef.SAR | −0.03 | −0.01 | 0.09 | 0.09 | 0.09 | −0.09 | |
| AICSAR | −2361.63 | −2361.63 | −2419.59 | −2409.55 | −2407.74 | −2455.53 | |
| sim.phy_end | Coef.OLS | 0.16*** | 0.19*** | 0.22*** | 0.01 | 0.09*** | −0.12*** |
| ROLS2 | 0.03 | 0.04 | 0.05 | – | 0.04 | 0.08 | |
| Coef.SAR | 0.01 | −0.01 | 0.09 | 0.09 | 0.09 | −0.09 | |
| AICSAR | −2362.11 | −2360.28 | −2414.10 | −2405.99 | −2402.95 | −2449.50 | |
| sim.phy_alp | Coef.OLS | −0.01 | 0.10*** | 0.16*** | 0.06*** | 0.04*** | −0.07*** |
| ROLS2 | – | 0.01 | 0.02 | 0.02 | 0.01 | 0.04 | |
| Coef.SAR | −0.08 | −0.04 | 0.07 | 0.07 | 0.06 | −0.06 | |
| AICSAR | −2665.65 | −2665.62 | −2693.04 | −2684.51 | −2677.53 | −2704.80 | |
| sne.phy_all | Coef.OLS | 0.10*** | 0.24*** | 0.13*** | 0.10*** | 0.05*** | – |
| ROLS2 | 0.01 | 0.06 | 0.02 | 0.04 | 0.01 | – | |
| Coef.SAR | 0.01 | 0.16 | 0.02 | −0.03 | −0.01 | 0.01 | |
| AICSAR | −2095.25 | −2155.48 | −2098.60 | −2091.13 | −2091.13 | −2091.13 | |
| sne.phy_end | Coef.OLS | 0.08** | 0.19*** | 0.09** | 0.00 | 0.03 | −0.02 |
| ROLS2 | 0.01 | 0.04 | 0.01 | – | – | – | |
| Coef.SAR | 0.01 | 0.11 | 0.01 | −0.04 | −0.01 | −0.01 | |
| AICSAR | −2308.19 | −2345.85 | -2309.53 | −2305.09 | −2305.09 | −2305.96 | |
| sne.phy_alp | Coef.OLS | 0.21*** | 0.26*** | 0.10*** | 0.00 | 0.04* | −0.04** |
| ROLS2 | 0.05 | 0.07 | 0.01 | – | 0.01 | 0.01 | |
| Coef.SAR | 0.11 | 0.17 | 0.01 | −0.06 | −0.01 | −0.01 | |
| AICSAR | −2110.72 | −2138.68 | −2066.47 | −2063.68 | −2063.68 | −2067.60 | |
| *P < 0.05, **P < 0.01, ***P < 0.001. "-": value < 0.001. The two (or one) highest R2 values and two lowest AIC values in each category are deepened. MAT, mean annual temperature; AP, annual precipitation; CG, climatic gradients; ER, elevation range; TH, topographic heterogeneity; CI, climate instability. For other abbreviations, see Table 1. | |||||||
Although individual variable only explains a small portion of the variation in taxonomic and phylogenetic beta diversity patterns, hierarchical partitioning (Figs. 4 and S5) indicates that some variables have a larger influence on these patterns. Specifically, patterns of total beta diversity and nestedness were best explained by AP (3–5%), whereas patterns of turnover were best explained by CI (4–7%). As expected, the results of hierarchical partitioning were broadly supportive of those of regression analyses.
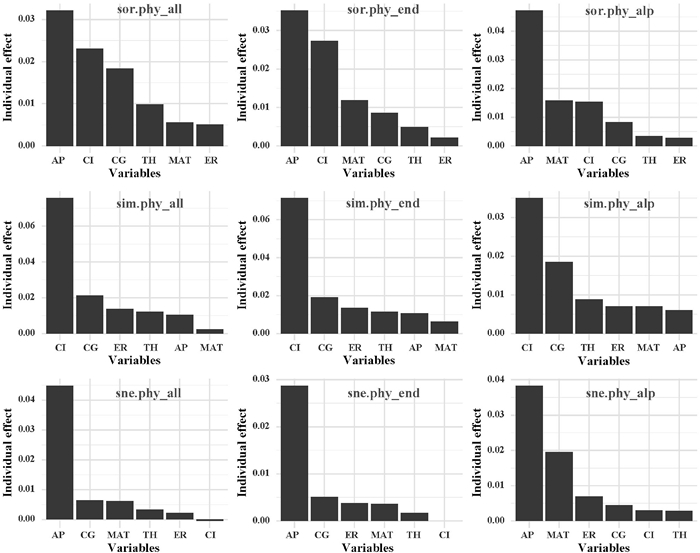
|
| Fig. 4 The individual contribution of each variable to phylogenetic beta diversity patterns. MAT, mean annual temperature; AP, annual precipitation; CG, climatic gradients; ER, elevation range; TH, topographic heterogeneity; CI, climate instability. |
A total of ten floristic subregions of the QTP were identified based on total phylogenetic beta diversity (Fig. 5). The boundaries of each subregion were broadly consistent, irrespective of the plant group used to delineate the boundaries, although there was one obvious exception, i.e., the southeastern part of the QTP. Analysis of alpine plant data divided this region into two subregions; however, analyses of all plants and endemic plants each divided the region into four subregions. These latter delimitations are more similar to the qualitative divisions by Wu et al. (2010) and the quantitative divisions by Zhang et al. (2016). The origins of these inconsistent patterns and fewer subregions based on alpine plants were identified due to differences in species turnover (Fig. S6). Here, we favor the ten subregions delimited using total phylogenetic beta diversity for all and endemic plants, given the more precise delineation and consistency with past works. These subregions are as follows: 1) South Hengduan Mountains; 2) North Hengduan Mountains; 3) southern region of East Himalaya; 4) South Qinghai Plateau; 5) East Chang Tang Plateau; 6) South Tibet; 7) West Himalaya-West Chang Tang Plateau; 8) Qilian Mountains; 9) Qaidam Basin; 10) West Kunlun Mountains-Kashgar.

|
| Fig. 5 Floristic subregions of the Qinghai-Tibet Plateau based on total phylogenetic beta diversity. Eco-geographical regions produced by Zheng (2008) and floristic subregions produced by Wu et al. (2010) and Zhang et al. (2016) of the QTP are cited here for comparison. The subregions are 1) South Hengduan Mountains; 2) North Hengduan Mountains; 3) southern region of East Himalaya; 4) South Qinghai Plateau; 5) East Chang Tang Plateau; 6) South Tibet; 7) West Himalaya-West Chang Tang Plateau; 8) Qilian Mountains; 9) Qaidam Basin; 10) West Kunlun Mountains-Kashgar. |
In this study, we used a phylogenetic approach to characterize patterns of multifaceted beta diversity and delimit flora subregions of the QTP. We also explored the effects of environmental variables (climate, topography and historical climate changes) on phylogenetic beta diversity and its major components (i.e., turnover and nestedness). Finally, we show how the environmental gradient from the southeastern to northwestern parts of the QTP have affected the phylogenetic and taxonomic beta diversity of plants, and ultimately shaped the current patterns of the flora of the QTP.
4.1. Patterns of multifaceted beta diversityAt large scales, taxonomic, functional and phylogenetic beta diversity for different taxa usually present congruent patterns (Zeng et al., 2022), because they are all in part a function of the common environmental background. We observed this for the QTP, where geographic patterns of taxonomic and phylogenetic beta diversity for all plants, as well as for endemic and alpine plants, were similar (Figs. 3 and S4). High levels of beta diversity were identified within the peripheral mountainous areas of the QTP, particularly in the Himalaya and Hengduan Mountains. These vast mountains have abundant elevational and climatic gradients, and obvious climatic variation between different slopes, which can result in fast species turnover within a short distance. Thus, these mountains not only harbor high levels of alpha species diversity, but also high levels of beta diversity (Li et al., 2019).
We also found that patterns of species turnover and nestedness, two components of total beta diversity, differed in the QTP. Higher species turnover occurred among sites in the eastern and southern areas of the QTP, whereas species turnover was lower on the inner plateau. In comparison, species nestedness among sites was generally not high across the QTP. This indicates that the flora of the QTP harbors high species turnover across environmental gradients that had not apparently resulted in species nestedness structures. Accordingly, regression coefficients for ER between turnover and nestedness had opposite signs (positive and negative, respectively) that were consistent across plant groups. Mountain areas, which are highly heterogenous, usually harbor many small-ranged endemic species, generating fast species turnover along elevational gradients (Buckley and Jetz, 2008). However, mountain areas have relatively stable climatic conditions and many dispersal barriers, resulting in low levels of species extinction and colonization (Rumpf et al., 2019), which may explain the observed lower species nestedness in these areas and negative coefficient values.
The inner plateau, which has an average elevation of > 4000 m, is relatively flat, with little fluctuation in climate, two conditions that have generated low species turnover among sites. In addition, this area was not covered by large ice sheets during the Quaternary glaciations, when it may have provided many microrefugia (Liu et al., 2012). Accordingly, sites in the inner plateau rarely show nestedness. The small coefficient values for climatic instability and nestedness suggest nestedness is not widespread in the QTP, implying that the impacts of Quaternary glaciations on species diversity of the QTP were weak.
Patterns of phylogenetic beta diversity for plant species in the QTP differed in some small ways from that of taxonomic beta diversity. Usually, phylogenetic beta diversity patterns are derived from the exchange of phylogenetically close species across space (Qian et al., 2020). In this study, the mega-phylogeny for calculating phylogenetic beta diversity was at a genus level, hindering the detection of changes among closely related species, and contributing to relatively low phylogenetic, compared with taxonomic beta diversity. Beta diversity could be more accurately calculated with the construction of species-level mega phylogenetic trees, which would reveal the evolutionary history of biota in a region at higher resolution (Graham and Fine, 2008).
4.2. Distance and environmental effects on beta diversityAs with past studies (e.g., Qian et al., 2020), we found that plant beta diversity in the QTP was significantly related to both geographical distance and climatic difference (Tables 1 and S1). As the greatest and highest plateau in the world, the QTP spans vast west-east (ca. 2900 km) and north-south (ca. 1500 km) distances and has a remarkable climatic gradient from the southeast (warm and humid) to northwest (cold and dry) (Zhang et al., 2002). This largely accounts for significant patterns of beta diversity along geographical distance and climatic difference. However, these two factors only explain limited variation in beta diversity (Figs. 2 and S3), while greater explanatory power has been found in large-scales studies (e.g., global scale; Mazel et al., 2017). Compared to a continental or global scale, the smaller scope of the QTP may have limited the effects of geography and climate on beta diversity. Species composition across the QTP mainly reflects plateau features, resulting in species that are the same or closely related found in places located far apart. These effects decrease variation of beta diversity along geographic and climatic gradients. This was particularly evident in alpine plants (Fig. 2), because many of these plants have a cosmopolitan distribution across the QTP.
We found that the patterns of beta diversity for different plant groups in the QTP were predominantly driven by AP, MAT, CG and CI (Tables 2 and S2; Fig. 4). Among these, AP was a consistently strong predictor of beta diversity. Few studies have found that AP greatly influences regional biodiversity patterns and vegetation distribution (Greve et al., 2011; Liu et al., 2019; Hu et al., 2021; Li et al., 2022). Each summer, the Indian and East Asian monsoon decreases AP along a southeastern to northwestern gradient in the QTP. This AP gradient generates four major vegetation types: mountain forest, alpine meadow, alpine steppe, and alpine desert (The China Vegetation Atlas Editorial Committee, 2001). In turn, this has affected patterns of plant beta diversity in the QTP. MAT also gradually decreases along this elevation gradient. Thus, patterns of beta diversity are likely determined by the combined effects of AP (i.e., climatic gradient) and MAT. Climate-related variables also affected the patterns of two components of beta diversity (turnover and nestedness). This suggests that species sorting is related to differential climatic tolerances conserved across lineages (i.e., phylogenetic niche conservatism). This may play an important role in structuring the assemblage of plants in the QTP, consistent with evidence from previous studies (Losos, 2008; Crisp and Cook, 2012). Although the Quaternary climate changes impacted the demographic history of species and diversity patterns in the QTP (Qiu et al., 2011; Yu et al., 2019), microrefugia across the QTP, where many cold-tolerant species survived at higher elevational zones (Liu et al., 2012), allowed past climate changes to consolidate the distributions of different tolerant plants (the effect of niche conservatism) and ultimately influenced the patterns of species turnover among sites (Fig. 4).
4.3. Floristic subregions of the QTPApart from understanding the origins of the pattern itself, another application of beta diversity is quantitatively delimiting floristic regions (Daru et al., 2020). The flora of the QTP has previously been qualitatively divided into four floristic regions and seventeen subregions by Wu and Wu (1996), and quantitatively divided into four floristic regions and twelve subregions by Zhang et al. (2016) based on taxonomic beta diversity. In this study, we used a phylogenetic perspective to divide the flora of the QTP into ten subregions (Fig. 5), which is generally consistent with Zhang's division, with the only difference found in the scope of several subregions. For example, in this study, we merged four subregions from Zhang et al. (2016) into two subregions, i.e., the eastern Qinghai and western Qinghai subregions were merged into the South Qinghai Plateau subregion, and the eastern Tibet and middle Tibet subregions were merged into the East Chang Tang Plateau. These mergers of floristic subregions are possibly related to the resolution of the mega-phylogenetic tree (genus level) used for calculating beta diversity, which results in more closely related species being gathered in a subregion. For example, the genera of Pedicularis L., Potentilla L., Meconopsis Vig. etc. are mostly distributed on the South Qinghai Plateau, while the genera of Artemisia L., Kobresia Willd., Stipa L. etc. usually grow in the East Chang Tang Plateau. When using alpine plants to divide subregions, the disparity that arises is the combination of Hengduan Mountains and South Qinghai Plateau within a subregion. For plants above the tree line in this subregion, the species composition is quite similar, and the deviation of beta diversity patterns is very low.
5. ConclusionsThe QTP harbors plentiful seed plants, that demonstrate clear beta diversity patterns. These patterns are more closely related to climatic gradients (coupled with the effects of annual precipitation and temperature) from south-east to north-west, but are also associated with past climatic changes. These contemporary environmental and historical factors together determine the patterns of species turnover among sites from both taxonomic and phylogenetic perspectives. Ten floristic subregions were identified in the QTP based on phylogenetic beta diversity, revealing that species in these subregions could have similar evolutionary histories. Few constraints on the resolution of species distribution (county level) and mega phylogeny (genus level) may produce clearer beta diversity patterns, and finer delineations of floristic subregions are likely to be identified. Thus, future studies should emphasize data resolution (e.g., constructing a species-level phylogeny and applying the Grade of Membership approach) to conduct beta diversity research, which is important to further understand the evolutionary history of the QTP's biota, as well as to inform biodiversity conservation of the QTP.
AcknowledgementsThis study was funded by the National Natural Science Foundation of China (grant no. 31901212) and Talent Start-up Foundation of Guangzhou University (grant no. RP2020079).
Author contributions
H.Y. conceived the research, collected data, analyzed data and wrote the paper; Z.L., M.Y. and H.Y. collected data and analyzed data; W.W. and X.Y. analyzed data and generated maps; F.Y., Y.Z., J.H. and D.D. reviewed and revised the paper.
Data availability statement
The species list, distributions and dated phylogeny of Tibetan seed plants for this study can be found in our previous studies (
Declaration of competing interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2023.07.006.
Baselga, A., 2010. Partitioning the turnover and nestedness components of beta diversity. Global Ecol. Biogeogr., 19: 134-143. DOI:10.1111/j.1466-8238.2009.00490.x |
Baselga, A., Orme, C.D.L., 2012. betapart: an R package for the study of beta diversity. Methods Ecol. Evol., 3: 808-812. DOI:10.1111/j.2041-210X.2012.00224.x |
Brown, J.L., Cameron, A., Yoder, A.D., et al., 2014. A necessarily complex model to explain the biogeography of the amphibians and reptiles of Madagascar. Nat. Commun., 5: 5046. DOI:10.1038/ncomms6046 |
Brown, J.L., Bennett, J.R., French, C.M., 2017. SDMtoolbox 2.0: the next generation Python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. PeerJ, 5: e4095. DOI:10.7717/peerj.4095 |
Buckley, L.B., Jetz, W., 2008. Linking global turnover of species and environments. Proc. Natl. Acad. Sci. U.S.A., 105: 17836-17841. DOI:10.1073/pnas.0803524105 |
Chao, A., Chazdon, R.L., Colwell, R.K., et al., 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett., 8: 148-159. DOI:10.1111/j.1461-0248.2004.00707.x |
Chen, Z., Yang, T., Lin, L., et al., 2016. Tree of life for the genera of Chinese vascular plants. J. Syst. Evol., 54: 277-306. DOI:10.1111/jse.12219 |
Crisp, M.D., Cook, L.G., 2012. Phylogenetic niche conservatism: what are the underlying evolutionary and ecological causes?. New Phytol., 196: 681-694. DOI:10.1111/j.1469-8137.2012.04298.x |
Daru, B.H., Karunarathne, P., Schliep, K., 2020. phyloregion: R package for biogeographical regionalization and macroecology. Methods Ecol. Evol., 11: 1483-1491. DOI:10.1111/2041-210x.13478 |
Deng, T., Ding, L., 2015. Paleoaltimetry reconstructions of the Tibetan plateau: progress and contradictions. Natl. Sci. Rev., 2: 417-437. DOI:10.1093/nsr/nwv062 |
Ding, W.N., Ree, R.H., Spicer, R.A., et al., 2020. Ancient orogenic and monsoon-driven assembly of the world's richest temperate alpine flora. Science, 369: 578-581. DOI:10.1126/science.abb4484 |
Du, W., Jia, P., Du, G., 2022. Current patterns of plant diversity and phylogenetic structure on the Kunlun Mountains. Plant Divers., 44: 30-38. DOI:10.1016/j.pld.2021.04.007 |
Fick, S.E., Hijmans, R.J., 2017. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol., 37: 4302-4315. DOI:10.1002/joc.5086 |
Fine, P.V., 2015. Ecological and evolutionary drivers of geographic variation in species diversity. Annu. Rev. Ecol. Evol. Syst., 46: 369-392. DOI:10.1146/annurev-ecolsys-112414-054102 |
González-Trujillo, J.D., Saito, V.S., Petsch, D.K., et al., 2021. Historical legacies and contemporary processes shape beta diversity in Neotropical montane streams. J. Biogeogr., 48: 101-117. DOI:10.1111/jbi.13986 |
Graham, C.H., Fine, P.V., 2008. Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecol. Lett., 11: 1265-1277. DOI:10.1111/j.1461-0248.2008.01256.x |
Greve, M., Lykke, A.M., Blach-Overgaard, A., et al., 2011. Environmental and anthropogenic determinants of vegetation distribution across Africa. Global Ecol. Biogeogr., 20: 661-674. DOI:10.1111/j.1466-8238.2011.00666.x |
Hu, Y., Fan, H., Chen, Y., et al., 2021. Spatial patterns and conservation of genetic and phylogenetic diversity of wildlife in China. Sci. Adv., 7: eabd5725. DOI:10.1126/sciadv.abd5725 |
Jankowski, J.E., Ciecka, A.L., Meyer, N.Y., et al., 2009. Beta diversity along environmental gradients: implications of habitat specialization in tropical montane landscapes. J. Anim. Ecol., 78: 315-327. DOI:10.1111/j.1365-2656.2008.01487.x |
Jenkins, C.N., Pimm, S.L., Joppa, L.N., 2013. Global patterns of terrestrial vertebrate diversity and conservation. Proc. Natl. Acad. Sci. U.S.A., 110: 2602-2610. |
Jin, Y., Qian, H., 2019. V.PhyloMaker: an R package that can generate very large phylogenies for vascular plants. Ecography, 42: 1353-1359. DOI:10.1111/ecog.04434 |
Jin, Y., Qian, H., 2022. V.PhyloMaker2: an updated and enlarged R package that can generate very large phylogenies for vascular plants. Plant Divers., 44: 335-339. DOI:10.1016/j.pld.2022.05.005 |
Jin, Y., Qian, H., 2023. U.PhyloMaker: an R package that can generate large phylogenetic trees for plants and animals. Plant Divers., 45: 347-352. DOI:10.1016/j.pld.2022.12.007 |
Kreft, H., Jetz, W., 2007. Global patterns and determinants of vascular plant diversity. Proc. Natl. Acad. Sci. U.S.A., 104: 5925-5930. DOI:10.1073/pnas.0608361104 |
Lai, J., Zou, Y., Zhang, J., et al., 2022. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca.hp R package. Methods Ecol. Evol., 13: 782-788. DOI:10.1111/2041-210x.13800 |
Li, F., Lu, H., Wang, G., et al., 2022. Zoning of precipitation regimes on the Qinghai-Tibet Plateau and its surrounding areas responded by the vegetation distribution. Sci. Total Environ., 838: 155844. DOI:10.1016/j.scitotenv.2022.155844 |
Li, N., Chu, H., Qi, Y., et al., 2019. Alpha and beta diversity of birds along elevational vegetation zones on the southern slope of Altai Mountains: implication for conservation. Glob. Ecol. Conserv., 19: e00643. |
Liu, J., Lindenmayer, D.B., Yang, W., et al., 2019. Diversity and density patterns of large old trees in China. Sci. Total Environ., 655: 255-262. DOI:10.1016/j.scitotenv.2018.11.147 |
Liu, J.Q., Sun, Y.S., Ge, X.J., et al., 2012. Phylogeographic studies of plants in China: advances in the past and directions in the future. J. Syst. Evol., 50: 267-275. DOI:10.1111/j.1759-6831.2012.00214.x |
Losos, J.B., 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett., 11: 995-1003. DOI:10.1111/j.1461-0248.2008.01229.x |
Lu, L., Mao, L., Yang, T., et al., 2018. Evolutionary history of the angiosperm flora of China. Nature, 554: 234-238. DOI:10.1038/nature25485 |
Mazel, F., Wüest, R.O., Lessard, J.P., et al., 2017. Global patterns of β-diversity along the phylogenetic time-scale: the role of climate and plate tectonics. Global Ecol. Biogeogr., 26: 1211-1221. DOI:10.1111/geb.12632 |
Mittermeier, R.A., Turner, W.R., Larsen, F.W., et al., 2011. Global biodiversity conservation: the critical role of hotspots. In: Zachos, F.E., Habel, J.C. (Eds.), Biodiversity Hotspots. Springer Publishers, London, pp. 3e22.
|
Oksanen, J., Blanchet, F.G., Kindt, R., et al., 2013. Package 'vegan'. Community Ecology package, version, 2: 1-295. |
Pinto-Ledezma, J.N., Larkin, D.J., Cavender-Bares, J., 2018. Patterns of beta diversity of vascular plants and their correspondence with biome boundaries across North America. Front. Ecol. Evol., 6: 194. DOI:10.3389/fevo.2018.00194 |
Qian, H., Jin, Y., Leprieur, F., et al., 2020. Geographic patterns and environmental correlates of taxonomic and phylogenetic beta diversity for large-scale angiosperm assemblages in China. Ecography, 43: 1706-1716. DOI:10.1111/ecog.05190 |
Qian, H., Ricklefs, R.E., 2007. A latitudinal gradient in large-scale beta diversity for vascular plants in North America. Ecol. Lett., 10: 737-744. DOI:10.1111/j.1461-0248.2007.01066.x |
Qiu, Y.X., Fu, C.X., Comes, H.P., 2011. Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world's most diverse temperate flora. Mol. Phylogenet. Evol., 59: 225-244. DOI:10.1016/j.ympev.2011.01.012 |
Quintero, I., Jetz, W., 2018. Global elevational diversity and diversification of birds. Nature, 555: 246-250. DOI:10.1038/nature25794 |
R Core Team, 2019. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. Available online at: http://www.R-project.org/.
|
Rangel, T.F., Diniz-Filho, J.A.F., Bini, L.M., 2010. SAM: a comprehensive application for spatial analysis in macroecology. Ecography, 33: 46-50. DOI:10.1111/j.1600-0587.2009.06299.x |
Rumpf, S.B., Hülber, K., Wessely, J., et al., 2019. Extinction debts and colonization credits of non-forest plants in the European Alps. Nat. Commun., 10: 4293. DOI:10.1038/s41467-019-12343-x |
Si, X., Zhao, Y., Chen, C., et al., 2017. Beta-diversity partitioning: methods, applications and perspectives. Biodivers. Sci., 25: 464-480. DOI:10.17520/biods.2017024 |
Smith, S.A., Brown, J.W., 2018. Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot., 105: 302e314. |
Soininen, J., Heino, J., Wang, J.J., 2018. A meta-analysis of nestedness and turnover components of beta diversity across organisms and ecosystems. Global Ecol. Biogeogr., 27: 96-109. DOI:10.1111/geb.12660 |
Spicer, R.A., Farnsworth, A., Su, T., 2020. Cenozoic topography, monsoons and biodiversity conservation within the Tibetan Region: an evolving story. Plant Divers., 42: 229-254. DOI:10.1016/j.pld.2020.06.011 |
The Angiosperm Phylogeny Group, Chase, M.W., Christenhusz, M., et al., 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc., 181: 1-20. DOI:10.1111/boj.12385 |
The China Vegetation Atlas Editorial Committee, Chinese Academy of Sciences, 2001. Atlas of China Vegetation (1:1,000,000). Science Press, Beijing.
|
Wu, Z., Sun, H., Zhou, Z., et al., 2010. Floristics of Seed Plants from China. Beijing: Science Press.
|
Wu, Z.Y., Wu, S.G., 1996. A proposal for a new floristic kingdom (realm): the East Asiatic Kingdom, its delineation and characteristics. In: Zhang, A.L., Wu, S.G. (Eds.), Floristic Characteristics and Diversity of East Asian Plants, Proceedings of the First International Symposium on Floristic Characteristics and Diversity of East Asian Plants. China Higher Education Press, Beijing, pp. 3–42.
|
Xu, W.B., Guo, W.Y., Serra-Diaz, J.M., et al., 2023. Global beta-diversity of angiosperm trees is shaped by Quaternary climate change. Sci. Adv., 9: eadd8553. DOI:10.1126/sciadv.add8553 |
Yang, Y., Ferguson, D.K., Liu, B., et al., 2022. Recent advances on phylogenomics of gymnosperms and an updated classification. Plant Divers., 44: 340-350. DOI:10.1016/j.pld.2022.05.003 |
Ye, J., Lu, L., Liu, B., et al., 2019. Phylogenetic delineation of regional biota: a case study of the Chinese flora. Mol. Phylogenet. Evol., 135: 222-229. DOI:10.1016/j.ympev.2019.03.011 |
Yu, H., Deane, D.C., Sui, X., et al., 2019. Testing multiple hypotheses for the high endemic plant diversity of the Tibetan Plateau. Global Ecol. Biogeogr., 28: 131-144. DOI:10.1111/geb.12827 |
Yu, H., Deane, D.C., Zhang, Y., et al., 2021. Integrating multiple indices of geobiodiversity reveals a series of regional species-rich areas worthy of conservation in the region of the Qinghai-Tibet Plateau. Biol. Conserv., 261: 109238. DOI:10.1016/j.biocon.2021.109238 |
Yu, H., Miao, S., Xie, G., et al., 2020. Contrasting floristic diversity of the Hengduan mountains, the Himalayas and the Qinghai-Tibet Plateau sensu stricto in China. Front. Ecol. Evol., 8: 136. DOI:10.3389/fevo.2020.00136 |
Zanne, A.E., Tank, D.C., Cornwell, W.K., et al., 2014. Three keys to the radiation of angiosperms into freezing environments. Nature, 506: 89-92. DOI:10.1038/nature12872 |
Zeng, C., Li, W., Ding, P., et al., 2022. A landscape-level analysis of bird taxonomic, functional and phylogenetic β-diversity in habitat island systems. J. Biogeogr., 49: 1162-1175. DOI:10.1111/jbi.14384 |
Zhang, D.C., Ye, J.X., Sun, H., 2016. Quantitative approaches to identify floristic units and centres of species endemism in the Qinghai-Tibetan Plateau, south-western China. J. Biogeogr., 43: 2465-2476. DOI:10.1111/jbi.12819 |
Zhang, Y., Li, B., Zheng, D., 2002. A discussion on the boundary and area of the Tibetan Plateau in China. Geogr. Res., 21: 1-8. |
Zheng, B., Xu, Q., Shen, Y., 2002. The relationship between climate change and Quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quat. Int., 97: 93-101. |
Zheng, D., 2008. Ecogeographical Regional System in China. Beijing: Commercial Press.
|



